Abstract
Plant pathogenic bacteria, such as Pseudomonas syringae pv. tomato strain DC3000, the causative agent of tomato bacterial speck disease, grow to high levels in the apoplastic space between plant cells. Colonization of plant tissue requires expression of virulence factors that modify the apoplast to make it more suitable for pathogen growth or facilitate adaptation of the bacteria to the apoplastic environment. To identify new virulence factors involved in these processes, DC3000 Tn5 transposon insertion mutants with reduced virulence on Arabidopsis thaliana were identified. In one of these mutants, the Tn5 insertion disrupted the malate:quinone oxidoreductase gene (mqo), which encodes an enzyme of the tricarboxylic acid cycle. mqo mutants do not grow to wild-type levels in plant tissue at early time points during infection. Further, plants infected with mqo mutants develop significantly reduced disease symptoms, even when the growth of the mqo mutant reaches wild-type levels at late stages of infection. Mutants lacking mqo function grow more slowly in culture than wild-type bacteria when dicarboxylates are the only available carbon source. To explore whether dicarboxylates are important for growth of DC3000 in the apoplast, we disrupted the dctA1 dicarboxylate transporter gene. DC3000 mutants lacking dctA1 do not grow to wild-type levels in planta, indicating that transport and utilization of dicarboxylates are important for virulence of DC3000. Thus, mqo may be required by DC3000 to meet nutritional requirements in the apoplast and may provide insight into the mechanisms underlying the important, but poorly understood process of adaptation to the host environment.
One important aspect of interactions between plant pathogens and their hosts is the ability of the pathogen to obtain nutrients within the plant tissue. Nutrient acquisition is essential for growth within the host, since both cell division and DNA replication can be influenced by nutrient availability. Bacterial plant pathogens differ in the strategies they use to get necessary nutrients during infection. Some pathogens, such as Agrobacterium tumefaciens, elicit production of specific carbon and nitrogen sources by the plant (1). Other pathogens may rely on metabolites that are readily available in the plant apoplast or may stimulate the release of water or nutrients from surrounding plant cells (27).
Little is known about how pathogenic Pseudomonas syringae strains acquire nutrients when growing in their hosts. P. syringae strains are gram-negative gammaproteobacteria, which as a group cause disease on many agriculturally important plants. For example, P. syringae pv. tomato strain DC3000 causes disease on tomato, A. thaliana, and several agriculturally important Brassicas, such as turnip, mustard, collard, and cauliflower (8, 51). Initially, DC3000 colonizes plant surfaces and then enters the plant tissue through natural openings (such as stomata) or wounds (34, 38). DC3000 then establishes itself in the plant apoplast, the intercellular space between plant cells (38). Once in the apoplast of susceptible hosts, DC3000 multiplies to high levels, and the infected plants develop disease symptoms, including chlorosis (yellowing) of the leaf tissue and necrotic spots or patches called lesions (38, 49). Pseudomonads, such as P. aeruginosa and P. fluorescens, preferentially utilize tricarboxylic acid (TCA) cycle intermediates (20, 29, 33, 44), and DC3000 utilizes these carbon sources in culture (19). Some studies to investigate nutrient acquisition of DC3000 have been carried out (3, 7, 40); however, it is not clear what carbon sources DC3000 utilizes when growing in plant tissue.
Several virulence factors are necessary for DC3000 to enter, grow inside the plant, and cause disease. Like many other bacterial pathogens, DC3000 uses a type III secretion system (TTSS) (15), which is encoded by the hrp/hrc genes, to inject effector proteins into plant cells (16, 27). Many of these effectors suppress host defenses, and it is likely that some may be involved in modulating the apoplastic environment or nutrient acquisition (16). DC3000 also produces the phytotoxin coronatine, which promotes entry of the bacteria into the plant apoplast by stimulating the opening of stomata (34) and is required for bacterial growth in the apoplast by suppressing salicylic acid (SA)-dependent host defenses (4, 45). Coronatine also promotes disease symptom development via an SA-independent mechanism (4). While much emphasis has been placed on exploring how type III-secreted effectors and coronatine promote DC3000 virulence, other factors are also likely to be important during pathogenesis.
To identify additional factors involved in pathogenesis, we undertook a genetic screen to identify novel virulence factors (5, 24). DC3000 mutants with reduced virulence were identified by assaying for their ability to elicit disease symptoms on A. thaliana and tomato plants (24, 37). One of these mutants, AK4C9, had reduced virulence on both hosts. The gene disrupted in this mutant is the malate:quinone oxidoreductase gene (mqo), which encodes an enzyme of the TCA cycle. mqo mutants grow more slowly than wild-type DC3000 in planta and in culture when dicarboxylates are the only carbon source, suggesting that dicarboxylates are important for the growth of DC3000 in the apoplast. In the present study, we explore the role of Mqo and a dicarboxylate transporter, DctA1, in DC3000 pathogenesis.
MATERIALS AND METHODS
Plant growth and infections.
A. thaliana ecotype Columbia plants were grown from seed in a growth chamber with 8 h of light at 22°C and 75% relative humidity. For dip-inoculation infections, 3.5-4-week-old plants were dipped in a solution of bacteria with an optical density at 600 nm (OD600) between 0.25 and 0.4, containing 10 mM MgCl2 and 0.02% Silwet (26). The plants were covered with a clear plastic dome for 24 h after the dip treatment. For day 0, the leaves were removed 2 h after inoculation. Leaves collected on day 0 were surfaced sterilized with a solution of 15% hydrogen peroxide for 5 min and then washed three times in sterile distilled water, prior to manual grinding of tissue in 10 mM MgCl2. For dctA1::KO infections, surface sterilization was also included on day 1 where indicated. A similar protocol was used to dip inoculate tomato plants.
For mqo::KO infections on sid2 mutant plants, sid2-2 (50) plants were used.
Hypersensitive response (HR) assays were performed as described previously (49), where a range of bacterial concentrations (5 × 106, 1 × 107, 5 × 107, and 1 × 108 CFU/ml) resuspended in 10 mM MgCl2 were used to syringe infiltrate tobacco leaves (Nicotiana tabacum cv. Xanthi N/N). Leaves were scored for macroscopic tissue collapse, a finding indicative of an HR, at 18 h after inoculation.
Bacterial strains and vectors.
The bacterial strains and plasmids used in the present study are shown in Table 1. P. syringae strains were routinely grown at 28 to 30°C in King's broth (KB) medium (23) unless otherwise described. Escherichia coli strains were grown on Luria-Bertani (LB) medium (42) at 37°C. Antibiotics were used at the following concentrations (in μg/ml) for P. syringae strains: rifampin, 100; kanamycin, 25; spectinomycin, 100; and tetracycline, 16. Antibiotics were used at the following concentrations for E. coli strains: kanamycin, 25 μg/ml; spectinomycin, 80 μg/ml; chloramphenicol, 20 μg/ml; and tetracycline, 10 μg/ml. Plasmids were transferred from E. coli DH5α (containing λpir) (35) to P. syringae strains by triparental mating (11) with the helper strain MM294A(pRK600) (12).
TABLE 1.
Bacterial strains and vectors used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. syringae | ||
| DC3000 | Derivative of NCPPB1106; Rifr | 8 |
| AK4C9 | mqo::Tn5 uidA; Rifr Kmr | This study |
| mqo::KO | mqo gene replaced with Sm/Specr gene; Rifr Specr | This study |
| DC3000 pMQO | DC3000 with pMQO clone; Rifr Tetr | This study |
| DC3000 pME6031 | DC3000 with pME6031; Rifr Tetr | This study |
| mqo::KO pMQO | mqo::KO with pMQO clone; Rifr Specr Tetr | This study |
| mqo::KO pME6031 | mqo::KO with pME6031; Rifr Specr Tetr | This study |
| dctA1::KO | dctA1 gene replaced with Sm/Specr gene; Rifr Specr | This study |
| dctA2::pJP5603 | dctA2::disrupted with pJP5603; Rifr, Kmr | This study |
| dctA1::KO dctA2::pJP5603 | dctA1::KO; dctA2::pJP5603; Rifr Specr Kmr | This study |
| E. coli | ||
| DH5α λpir | recA, lacZΔM15, λpir | 35 |
| MM294A (pRK600) | Triparental mating helper strain; Cmr | 12 |
| Plasmids | ||
| pHP45Ω | Source of Ω cassette; Sm/Specr gene | 37a |
| pRK415 | Broad-host-range vector; Tetr | 21a |
| pJP5603 | Suicide vector; Kmr | 36a |
| pmqo-pRK415 | Construct used to make mqo::KO; Tetr | This study |
| pmqo::Spec-pRK415 | Construct used to make mqo::KO; Specr Tetr | This study |
| pmqo::KO | Construct used to make mqo::KO strain; Specr Kmr | This study |
| pdctA1up-pJP5603 | Construct used to make dctA1::KO plasmid; Tetr | This study |
| pdctA1down-TOPO | Construct used to make dctA1::KO plasmid; Kmr | This study |
| pdctA1::KO | Construct used to make dctA1::KO strain; Specr Tetr | This study |
| pdctA2::pJP5603 | Construct used to make dctA2::pJP5603 strain; Kmr | This study |
| pCR-Blunt II TOPO | Cloning vector; Kmr | Invitrogen |
| pME6031 | Replicating vector, stable in P. syringae; Tetr | 18 |
| pMQO | Wild-type mqo gene in pME6031; Tetr | This study |
| pDctA1 | Wild-type dctA1 gene in pME6031; Tetr | This study |
Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; Rifr, rifampin resistance; Specr, spectinomycin resistance; Kmr, kanamycin resistance.
DNA manipulation and sequencing.
Plasmid DNA was isolated by using a Promega Wizard miniprep kit, and bacterial genomic DNA was isolated by using the genomic DNA purification kit (Promega, Madison, WI). Sequencing reactions were performed by using ABI BigDye v3.1, with 4 μl of BigDye mix used in a 12-μl volume reaction (Advanced Biosystems, Inc., Foster City, CA). A total of 150 to 300 ng of template was used for each sequencing reaction. Automated sequencing was performed on an ABI sequencer at the Washington University Biology Department sequencing facility (Washington University, St. Louis, MO). For semiquantitative PCR assays of gene expression, DC3000 RNA was isolated from cultures grown in KB or minimal medium by using the Qiagen RNeasy minikit (Qiagen, Hilden, Germany). cDNA was synthesized by using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA).
To identify the location of the Tn5 insertion in the AK4C9 mutant, the genomic fragment containing the Tn5 was identified by digesting total AK4C9 genomic DNA with PstI, followed by ligation of these fragments into the cloning vector pBluescript KS(+) cut with PstI. The resulting plasmids were introduced into E. coli, and the transformants were selected for kanamycin resistance (Kanr) provided by the aph gene on the Tn5 (24). An ∼4-kb genomic DNA fragment was identified that contained the Tn5 transposon and was sequenced to identify the location of the Tn5 transposon.
To make the mqo::KO strain, 2 kb of genomic sequence flanking each side of the mqo coding sequence was amplified with the following primers (restriction enzyme sites are underlined): mqo3EcoRIf, 5′-ATA GGA ATT CGG CGA TAC CAA ACG CTA CGG-3′; mqo3KpnIr, 5′-TCA GGA TCG TGG TAC CGG TCT TCG CGC AGT TCA TCC-3′; mqo5KpnIf, 5′-CGC GAA GAC CGG TAC CAC GAT CCT GAG GCT GGA TGC-3′; and mqo5XbaIr, 5′-TAT GTC TAG AGG AGT TGA TCG ACG GCT TGG-3′. The resulting PCR products (which contained 26 nucleotides of overlapping sequence allowing them to anneal to each other) were mixed together and amplified 10 times, using the mqo3EcoRIf and mqo5XbaIr primers, to produce a 4-kb fragment containing both flanking regions without the mqo coding sequence in between. The PCR product was digested with EcoRI and XbaI and ligated into the pRK415 vector, also digested with EcoRI and XbaI, to generate pmqo-pRK415 (Table 1). The spectinomycin resistance (Specr) cassette from the omega fragment was amplified from pHP45Ω with the primers Spec1f (5′-GTC CGG TAC CCC AAG CTC TCG GGT AAC ATC-3′) and Spec1r (5′-GTC CGG TAC CTT CGC CAA CTA TTG CGA TAA C-3′) containing the KpnI recognition site (underlined) on the 5′ end of each primer and inserted in between the mqo flanking DNA sequences to generate pmqo::Spec-pRK415 (Table 1). The fragment containing the mqo flanking sequence and Specr gene was removed from pRK415 by digestion with EcoRI and XbaI and inserted into the suicide vector pJP5603 cut with EcoRI and XbaI, to generate pmqo::KO (Table 1). This plasmid was introduced into DC3000 by triparental mating using the helper strain MM294A (Table 1). DC3000 transconjugates were screened for Specr and kanamycin sensitive (Kans), and gene-specific replacement of the mqo gene with the Specr gene was verified by PCR.
To construct the mqo complementing clone, pMQO, the following primers were used to amplify the mqo coding sequence and promoter from genomic DNA: Mqosmallreverse, 5′-ATA CCT CGA GTC ATC GCT ATA ATC TTC CCC CGA TT-3′ (XhoI site underlined); and mqo+promf1, 5′-ATA TGA ATT CAG CTC TGA CGG CAG CAA CCA AAA AG-3′ (EcoRI site underlined). The resulting 2.3-kb PCR product was digested with EcoRI and XhoI and cloned into the shuttle vector pME6031 (18) cut with the same enzymes. The clone was sequenced and found to have two nucleotide changes, one in the coding sequence but not predicted to change the amino acid sequence, and one T-to-C change 138 bp upstream of the mqo start codon. Because these changes would most likely not affect Mqo function, this clone was used in complementation tests and was demonstrated to confer wild-type Mqo activity (see Results).
The dctA1::KO strain was made using a strategy similar to that used to generate the mqo::KO mutant. A 1-kb region directly upstream of the dctA1 gene and a 1-kb region directly downstream of dctA1 were cloned into the pJP5603 suicide vector. These regions were amplified by using the following primers: dcta1upF, 5′-TCA TAA AGC TTG CGA CAG CTT GAG TGA AGA GCA T-3′; dcta1upR, 5′-ATT GGA TCC GTC GTC GGC ATC TTC ACC TGA T-3′ (with the HindIII and BamHI restriction sites, respectively, underlined); dcta1downF, 5′-TAT GGA TCC TCT GAT AGC TTC AGG CCA TGA AAA A-3′; and dcta1downR, 5′-TAA GAA TTC GTA CTC CAC ACC CAG CTC TTT CAG A-3′ (with the BamHI and EcoRI restriction sites, respectively, underlined). The Specr cassette from the omega fragment was cloned in between these two fragments. The resulting plasmid, pdctA1::KO, was introduced into DC3000 by triparental mating as described described above. DC3000 transconjugates were screened for Specr and Kans, and gene-specific replacement of the dctA1 gene with the Specr gene was verified by PCR.
The dctA1 complementing clone (pDctA1, Table 1) was constructed similar to the way the mqo complementing clone was made. The following primers were used to amplify the dctA1 coding sequence and promoter from genomic DNA: dctA1F, 5′-GAA AGC TAT GGC GCA CAA CTG AC-3′; and dctA1R, 5′-CTG ATG TTC AGC GGC GAG AC-3′. The resulting 1.9-kb PCR product was cloned into the pCR-Blunt II TOPO vector and then subcloned into the shuttle vector pME6031 by using HindIII and XhoI restriction enzymes (18). The clone was sequenced and found to have no nucleotide changes.
To generate pdctA2::pJP5603, a 667-bp fragment internal to the dctA2 coding sequence was amplified with the primers Dcta2KOF (5′-AAA ACT TCC AGG TCC CGC TGG TAC G-3′) and Dcta2KOR (5′-CGA GTG ACA GCA ACG TAC CAA TTC C-3′) and cloned into the pCR-Blunt II TOPO vector. The PCR product was digested out of the TOPO vector with BamHI and XbaI and ligated into pJP5603 cut with the same enzymes. The resulting plasmid, pdctA2::pJP5603, was introduced into DC3000 by triparental mating using the helper strain MM294A (Table 1) to make strain dctA2::pJP5603. DC3000 transconjugants were screened for Kanr, and insertional disruption of the dctaA2 gene with pJP5603 was verified by PCR. To make the dctA1::KO dctA2::pJP5603 double mutant strain, pdctA2::pJP5603 was introduced into dctA1::KO by triparental mating.
Bioinformatics, nucleotide sequences, and accession numbers.
BLASTP searches were performed with the National Center for Biotechnology Information (NCBI) servers (version 2.2.18) to search nonredundant databases and DC3000 specific sequences (2). The NCBI BLAST 2 sequences program (version 2.2.18) was used for BLASTP comparisons of amino acid sequences. DC3000 sequence information was obtained from Pseudomonas-Plant Interaction (PPI; http://pseudomonas-syringae.org [31]) and The J. Craig Venter Institute comprehensive microbial resource (http://cmr.jcvi.org/cgi-bin/CMR/CmrHomePage.cgi). The accession numbers for the following proteins were as follows: Pseudomonas syringae pv. tomato strain DC3000 Mqo, AAO54665; P. aeruginosa PAO1 MqoA, AAG06840; P. aeruginosa PAO1 MqoB, AAG08028; P. citronellolis Mqo, AAW88350; Sinorhizobium meliloti Mdh, AAG41996; P. syringae pv. tomato strain DC3000, ME, AAO57390; E. coli strain K-12 substrain MG1655 ME, NP_415996; S. meliloti DctA, P20672; P. syringae pv. tomato strain DC3000 DctA1, AAO55202; and P. syringae pv. tomato strain DC3000 DctA2, AAO57520.
Growth of DC3000 in minimal medium cultures.
To assess the ability of the mqo::KO mutant to grow on different carbon sources, mutant and wild-type DC3000 strains were grown first in KB medium with rifampin in overnight cultures. Cells were precipitated from 3 ml of each overnight culture by centrifugation and washed once with 10 mM MgCl2, and then approximately 8 × 107 bacteria were resuspended in 5 ml of minimal medium with an appropriate carbon source. For each time point, 100 μl of each culture was removed and added to a microtiter well. The OD600 was measured by using a SPECTRAmax Plus spectrophotometer (Molecular Devices, Union City, CA).
HS minimal medium was prepared as described previously (43), with appropriate carbon sources added in place of sucrose. d,l-Malate (Sigma); citric acid, trisodium, and dihydrate (Amresco, Solon, OH); succinic acid (Sigma); d-sucrose (Fisher Scientific, Pittsburgh, PA); and sodium acetate (Fisher Scientific, Pittsburgh, PA) were added to the prepared Hoitink-Sinden (HS) media to a final concentration of 10 mM, and the pH was adjusted to 6.5. The medium was then filter sterilized and used for growth assays.
RESULTS
DC3000 Mqo is required for wild-type growth and disease symptom production in A. thaliana.
To identify novel virulence factors that promote DC3000 pathogenesis, a Tn5 transposon mutagenesis screen was undertaken (5, 24). One of the DC3000 mutants identified, AK4C9, had reduced virulence on A. thaliana plants. Growth of AK4C9 in planta was reduced compared to the wild type (Fig. 1a). Plants inoculated with AK4C9 developed significantly fewer disease lesions and very little chlorosis compared to plants infected with wild-type DC3000 (data not shown). AK4C9 also had reduced virulence on tomato plants (data not shown). The AK4C9 mutant elicited a normal HR on tobacco, indicating that type III secretion is not impaired in this mutant (data not shown).
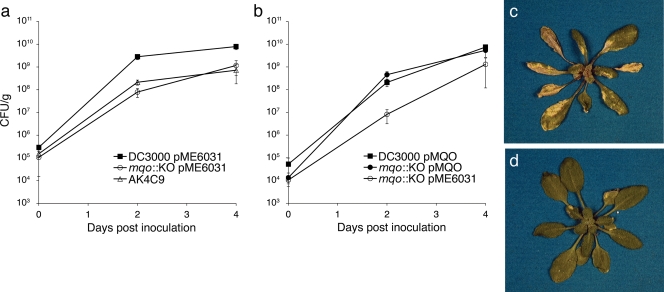
FIG. 1.
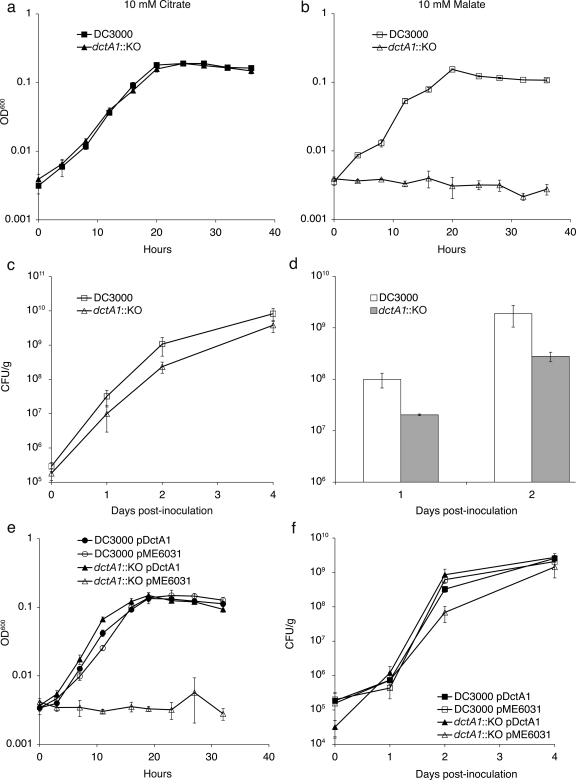
In planta growth and disease symptom phenotypes of DC3000 mqo mutants. mqo is required for normal growth and disease symptom production by P. syringae strain DC3000 on A. thaliana. (a) Growth of wild-type, mqo::KO, and AK4C9 bacteria (in CFU/gram of leaf tissue) on A. thaliana plants. DC3000 and mqo::KO strains contained the empty vector pME6031. Four samples were used for each time point. (b) Growth of wild-type and the mqo::KO mutant carrying the pMQO complementing clone, and growth of mqo::KO with the empty vector control, pME6031. Three samples were used for each time point. (c and d) Disease symptoms of a plant dip inoculated with wild-type bacteria (c) and a plant infected with the mqo::KO mutant (d) at 5 days after inoculation. Note that in panel a the mqo::KO and AK4C9 strains did not reach wild-type levels of growth on day 4, but in panel b the mqo::KO strain approached wild-type levels of growth on day 4. Error bars represent the standard error of the mean. Similar results were obtained for at least three additional independent experiments.
The Tn5 transposon insertion in AK4C9 was mapped to a region of the DC3000 genome ∼200 bp upstream of the PSPTO_1136 coding region, which is predicted to encode malate:quinone oxidoreductase (Mqo; Fig. 2). Mqo is an enzyme that can function as part of the TCA cycle, converting malate to oxaloacetate. DC3000 Mqo exhibits 77% amino acid identity and 86% similarity to P. aeruginosa MqoB. A presumptive FAD binding site and a βαβ fold ADP-binding motif, found in other Mqos (see Fig. S1 in the supplemental material) (25, 36), are present in the amino-terminal region of the DC3000 Mqo protein. Another enzyme capable of catalyzing this reaction is malate dehydrogenase (Mdh). Bacterial species differ on whether they have both a Mqo and Mdh, or just one of the two enzymes. For instance, E. coli has both an Mdh and an Mqo enzyme (46), while P. aeruginosa has two Mqos, MqoA, and MqoB, but no Mdh (13, 25). P. syringae pv. syringae B728a and P. syringae pv. phaseolicola 1448A also appear to have two Mqos, in contrast to the single Mqo in DC3000. Bioinformatic searches failed to identify a mdh gene in DC3000, suggesting that the single Mqo in DC3000 may be the primary enzyme responsible for conversion of malate to oxaloacetate.
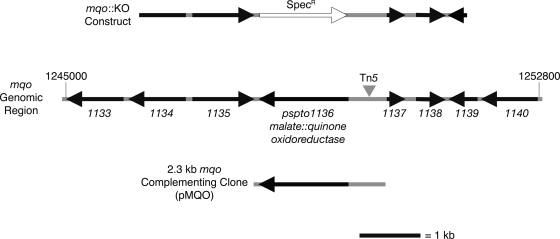
FIG. 2.
Region of DC3000 genome containing the mqo gene. A map of the P. syringae pv. strain DC3000 genomic region containing the mqo coding sequence is shown. The italicized numbers (e.g., 1133) are abbreviations for the PSPTO-number for each gene. The Tn5 insertion location upstream of the mqo coding sequence is indicated. The mqo::KO construct shows the regions of the DC3000 genome flanking the mqo coding sequence that were cloned on either side of the Specr gene into pJP5603. The genomic DNA included in the 2.3-kb mqo complementing clone (pMQO) is also shown. A size bar indicates the scale of the figure, and coordinates indicate the genomic position of the region in nucleotide bases (1245000 to 1252800).
To confirm that the Tn5 transposon insertion in the presumptive mqo promoter region was responsible for the reduced virulence phenotype of the AK4C9 mutant, we created a gene replacement knockout strain (mqo::KO) of DC3000, in which the coding sequence of mqo was replaced by a Specr gene (Table 1 and Fig. 2). Replacement of the mqo gene was unlikely to cause any polar effects on adjacent genes, since mqo does not appear to be part of an operon. The mqo::KO strain had the same reduced virulence phenotype observed for the AK4C9 mutant (Fig. 1). We also observed that in some experiments mqo::KO reached wild-type levels of growth by 4 days postinoculation (Fig. 1b). Interestingly, disease symptoms never approached the severity of those caused by wild-type DC3000, even in experiments in which the mqo::KO bacteria grew to the same level as wild-type (Fig. 1c and d). Virulence of the mqo::KO mutant could be restored to wild-type levels, both at the level of in planta growth (Fig. 1b) and disease symptom production (data not shown) by introduction of a complementing clone containing the mqo coding region and upstream regulatory sequences (pMQO; Fig. 2), indicating that the mqo gene is required for DC3000 virulence.
To examine the possibility that reduced virulence of the mqo::KO mutant could be due to a defect in suppression or evasion of SA-dependent defenses in the host, we inoculated SA-deficient sid2-2 (49) plants with the mqo::KO mutant. Growth of the mqo::KO mutant was not restored to wild-type levels 48 h after inoculation onto sid2-2 plants, indicating that mqo is not involved in suppression of SA-dependent defenses. Thus, it is likely that the reduced virulence of the mqo mutant is due to a defect in some other aspect of pathogenesis.
Mqo is required for wild-type growth in media containing dicarboxylates.
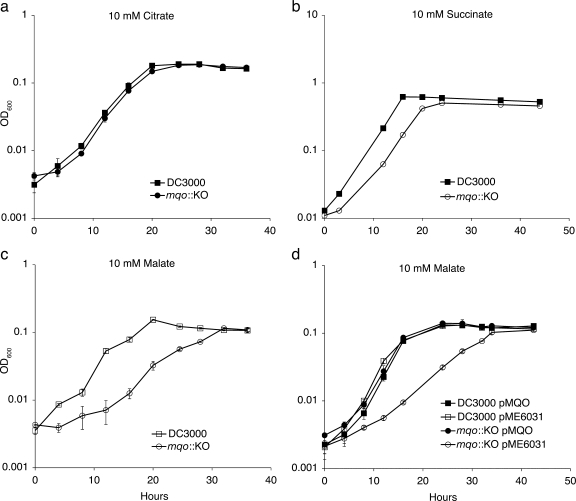
Because the Mqo enzyme is a component of the TCA cycle, it is possible that the mqo mutants are impaired for growth on specific carbon sources. To investigate this, we assayed the growth of the mqo::KO mutant in culture using various carbon sources. Growth of wild-type DC3000 and the mqo::KO mutant was indistinguishable in rich media (KB; data not shown). Several types of minimal media have been developed for studies on plant-pathogenic bacteria in an effort to develop culture conditions that are similar to those encountered in the plant apoplast. We chose to work with the defined minimal HS medium, since both coronatine biosynthesis genes and hrp/hrc pathogenesis genes are induced in this minimal medium (43). The mqo::KO strain was indistinguishable from wild-type bacteria when grown in HS minimal medium supplemented with either sucrose, citrate, or oxaloacetate as the only carbon source (Fig. 3a, data not shown). Thus, mqo::KO does not have a general growth defect in rich medium or in minimal medium with sucrose, citrate, or oxaloacetate. However, when grown in minimal medium containing malate or succinate as the only carbon source, mqo::KO grew at a slower rate than did wild-type bacteria (Fig. 3b, c, and d). The slower growth rate was more pronounced on malate than on succinate. When cultured in minimal medium with malate as the sole carbon source, wild-type bacteria grew with an approximate doubling time of 3.5 h, while the mqo::KO strain grew with a doubling time of ∼5.5 h. The slow-growth phenotype of the mqo::KO mutant in malate was restored to wild-type levels by introduction of the pMQO complementing clone (Fig. 3d). The growth defect of the mutant in culture was similar to what we observed in planta, in that there was slower growth than with the wild type initially but that the number of bacteria eventually reached wild-type levels in many experiments.
FIG. 3.
Growth of wild-type DC3000 and mqo::KO in culture. Wild-type DC3000 and the mqo::KO mutant were grown in HS media with a 10 mM concentration of the indicated carbon source: citrate (a), succinate (b), and malate (c and d). Similar results were obtained for at least two additional independent experiments. In panels a, c, and d, three replicate cultures were used for each growth curve, and the error bars represent the standard deviation of the mean. In panel b, each time point represents a single sample.
For the pseudomonads P. aeruginosa and P. citronellolis the Mqo enzyme is required for the glyoxylate pathway, for utilization of ethanol, acetate, or linear terpenes as a carbon source (13, 25). In contrast, we found that DC3000 did not grow in culture when acetate was the only available carbon source (data not shown), indicating that the glyoxylate pathway is not likely to be an important carbon utilization pathway for DC3000.
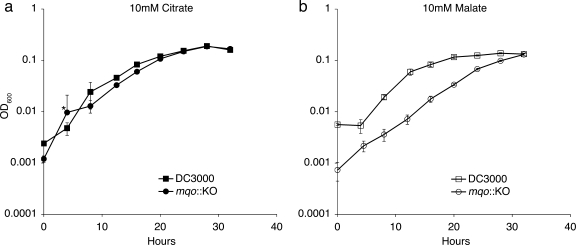
The mqo::KO mutant is still able to use dicarboxylates such as malate as carbon sources; however, its growth rate is slower than that of wild-type bacteria when relying solely on these compounds. One explanation for this slower growth rate is that the mqo::KO mutant needs time to induce alternative pathways for dicarboxylate utilization, but once these pathways are active, the mutant can begin to grow. For instance, DC3000 is predicted to have a NAD+-dependent malic enzyme (ME; PSPTO_3924), which has 64% amino acid identity and 79% similarity to the NAD+-dependent malic enzyme from E. coli. Malic enzyme can convert malate to pyruvate, possibly bypassing the need for Mqo. Thus, in the absence of functional Mqo, DC3000 may need time to induce this alternative malate/dicarboxylate utilization gene. Alternatively, the mqo::KO mutant may simply be less efficient than wild-type bacteria at producing the energy it needs for growth on malate or succinate. For example, the growth rate of the mqo mutant on dicarboxylates may be reduced due to the reduction in the number of pathways available to convert malate to oxaloacetate and complete the TCA cycle. If the first scenario were true, and mqo::KO exhibits an initial lag in growth as it needs to induce alternative malate utilization genes, we would expect that mqo::KO bacteria from a mid-log-phase culture would grow at a wild-type rate when subcultured into fresh malate medium. If, however, mqo::KO is simply less efficient when growing on malate, the bacteria would still exhibit a slower growth rate than the wild type when introduced into a new malate culture. When we took mid-log-phase bacteria from cultures grown in HS with malate and used them to reinoculate fresh HS with malate cultures, we found that the mqo::KO bacteria still had a slower growth rate compared to wild-type bacteria. Thus, the slow-growth phenotype of the mqo::KO mutant is most likely due to less efficient utilization of malate (Fig. 4).
FIG. 4.
Growth of wild-type DC3000 and the mqo::KO mutant after subculturing. Wild-type DC3000 and the mqo::KO mutant were grown in HS media with a 10 mM concentration of citrate or malate, and at mid-log (OD600 of 0.07 to 0.09) approximately 8 × 107 cells from each culture were used to inoculate a fresh (a) citrate or (b) malate culture. The graphs show the growth of the strains after subculturing. Three replicate cultures were used for each growth curve, and the error bars represent standard deviation of the mean. An asterisk at the 4-h data point for the mqo::KO growing in citrate indicates that the error bar below the mean for this time point is not shown (standard deviation = 0.011).
DctA1 is required for growth in malate cultures and for normal virulence on A. thaliana.
Given that mqo is required for both virulence of DC3000, and wild-type growth in culture with dicarboxylates such as malate and succinate, we reasoned that DC3000 may use dicarboxylates as carbon sources when growing in the plant apoplast. Thus, we predicted that a DC3000 mutant that cannot take up dicarboxylates would also show impaired growth in planta. Rhizobiaceae utilize dicarboxylates provided by their plant hosts as carbon sources during their symbiotic relationship with legume plants (32), and in S. meliloti the DctA transporter is required for this (9, 48). DC3000 has two potential orthologs of the S. meliloti dctA gene, dctA1 (PSPTO_1682) and dctA2 (PSPTO_4063). The DC3000 dctA1 gene encodes a protein with 60% identity and 78% similarity to S. meliloti DctA, while DC3000 dctA2 encodes a protein with 50% identity and 75% similarity to S. meliloti DctA. We were unable to identify DC3000 orthologs of the dctB and dctD genes in the vicinity of dctA1. These genes encode the two-component regulators that control expression of the DctA transporter in Rhizobiaceae and are found adjacent to dctA (41, 47). Transcript levels of the DC3000 dctA1 gene were increased within 60 min after the bacteria were transferred to minimal medium containing malate, compared to when they were grown on sucrose or rich media (data not shown). In contrast, the dctA2 gene was expressed at a very low level (data not shown) in minimal medium containing malate. Thus, we reasoned that DctA1 may be involved in dicarboxylate uptake. We generated a dctA1 marker replacement mutant, replacing the dctA1 gene with a Specr gene. This strain, dctA1::KO, was unlikely to have any polar effects on adjacent genes, since dctA1 does not appear to be part of an operon (Fig. 5). The strain was used to determine whether dctA1 is required for growth on dicarboxylates in culture and/or for growth in A. thaliana.
FIG. 5.
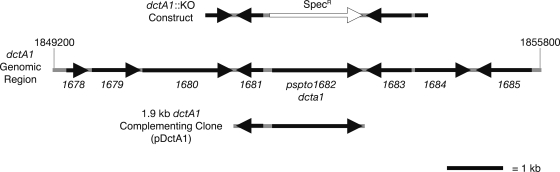
Region of DC3000 genome containing the dctA1 gene. The DC3000 genomic region containing the dctA1 (PSPTO_1682) coding sequence is shown. The italicized numbers (e.g., 1678) are abbreviations for the PSPTO number for each gene. The regions flanking dctA1 used to create the dctA1::KO construct are indicated. A representation of the genomic sequence used in the dctA1 complementing clone (pDctA1) is also shown. A size bar indicates the scale of the figure, and coordinates indicate the genomic position of the region in nucleotide bases (1849200 to 1855800).
The dctA1::KO strain grew similar to wild-type bacteria in HS supplemented with citrate but failed to grow in HSS medium containing only malate (Fig. 6a and b), demonstrating that DctA1 is likely to be the primary transporter use for uptake of malate when DC3000 is growing in minimal media. When the dctA1::KO strain was used to infect plants, the mutant strain reproducibly exhibited a small growth defect. Growth of the mutant was fivefold lower than that of the wild type on day 1 and/or day 2 after inoculation (Fig. 6c and d). Both the lack of growth of dctA1::KO in minimal medium with malate cultures and the reduced growth of dctA1::KO in planta could be restored to wild-type levels of growth by introduction of the pDctA1 complementing clone (Fig. 6e and f). This demonstrates that the DctA1 transporter is required for normal growth in planta, and suggests that dicarboxylates (e.g., malate) are an important carbon source for DC3000 at early stages of pathogenesis. Because the growth defect of the dctA1::KO is relatively small, we wondered whether one or more additional dicarboxylate transporters might also be involved in malate uptake and growth of DC3000 in planta. We constructed a mutant, dctA2::pJP5603, in which the gene encoding the second potential DctA transporter in DC3000, dctA2, was disrupted. We also constructed a double knockout of dctA1 and dctA2. The dctA2 mutant exhibited wild-type growth in A. thaliana, and the dctA1::KO dctA2::pJP5603 double mutant strain did not show any further reduction in virulence over the dctA1::KO single mutant (data not shown), indicating that dctA2 does not compensate for lack of dctA1 in planta.
FIG. 6.
Growth of the DC3000 dctA1::KO mutant in minimal media cultures and in planta. The dctA1::KO mutant and wild-type strains were grown in citrate (a) or malate (b). Three replicate cultures were used for each growth curve, and the error bars represent standard deviation of the mean. Similar results were obtained for at least two additional independent experiments. Two representative growth curves show growth of the dctA1::KO mutant in A. thaliana. (c) Seven samples were used for each time point, and error bars represent the standard errors of the mean. Student t tests with unpaired equal variation and two-tailed distribution gave P values of 0.24 for wild type and dctA1::KO at day 1 and 0.19 at day 2. Each strain contained the empty vector pME6031. (d) Three samples were used for each time point, error bars represent the standard error of the mean, and Student t tests with unpaired equal variation and two-tailed distribution gave P values of 0.07 for day 1 and 0.13 for day 2. Similar results were obtained for at least 3 additional independent experiments. (e and f) The growth of wild-type and the dctA1::KO mutant carrying the pDctA1 complementing clone is shown. Panel e shows growth of these stains in minimal media with 10 mM malate, and panel f shows growth of the strains in A. thaliana. In panel e, three replicate cultures were used for each growth curve, and error bars represent the standard deviations. Similar results were obtained for two independent experiments. In panel f, six samples were used for each time point (except for day 1 for strain dctA1::KO/pME6031, which had five samples), error bars represent the standard error of the mean, and Student t tests with unpaired equal variation and two-tailed distribution gave P values of 0.01 for day 2 between DC3000/pME6031 and dctA1::KO/pME6031.
DISCUSSION
We identified a novel DC3000 virulence gene, mqo, which unexpectedly encodes an enzyme that is part of a central metabolic pathway, the TCA cycle. To investigate the role of mqo in promoting virulence, we explored the impact of the mqo mutation on several aspects of host-pathogen interactions. We first explored whether the mqo mutant was impaired in type III secretion. The AK4C9 (mqo::Tn5) mutant elicited a normal HR on tobacco (data not shown), indicating that type III secretion is not impaired in this mutant. Thus, mqo is not required for hrp/hrc gene function. In addition, expression of mqo is not coregulated with type III secretion genes (10). Further, mqo is not directly regulated by the HrpL RNA polymerase sigma factor, which governs expression of the hrp/hrc genes, since there is no HrpL binding site (hrp box) upstream of the mqo coding sequence (10, 14).
We next looked at whether mqo was involved in suppressing host defenses. Several DC3000 virulence factors, including TTSS effector proteins and coronatine, promote virulence by suppressing SA-mediated defenses. Our finding that growth of the mqo::KO mutant was not restored to wild-type levels on sid2-2 plants indicates that mqo is not involved in suppression of SA-dependent defenses (data not shown). These data, combined with the finding that mqo mutants are not impaired for TTSS, suggests that Mqo is required for a novel aspect of virulence. We hypothesize that Mqo is required for virulence because DC3000 utilizes this enzyme to grow efficiently on dicarboxylates, a carbon source present in the plant apoplast (30, 40).
Consistent with this hypothesis, the mqo::KO bacteria exhibited growth defects in culture when provided with dicarboxylates (e.g., malate and succinate). It is important to note that the growth of the mqo::KO mutant was not impaired on rich medium or on minimal medium containing other carbon sources (e.g., sucrose or citrate), indicating that this mutant does not have a general metabolic or growth defect. This also suggests that one or more dicarboxylates may be important carbon sources during growth in the plant. Various studies have indicated that dicarboxylates such as malate are available in both the A. thaliana and tomato plant apoplast (28, 30). However, sucrose is also available in the apoplast (21, 39). Sucrose is the primary transport sugar in many plants and has been proposed to be a carbon source for apoplast-dwelling pathogens (6, 21). A study investigating which DC3000 metabolic pathways were active in tomato apoplast extracts indicated that, despite the availability of sucrose, sucrose utilization pathways were not induced (40). Thus, DC3000 may preferentially utilize dicarboxylates over other sugars available in the plant apoplast. Other studies have also suggested that TCA cycle intermediates are preferred carbon sources for DC3000, as indicated by the increased growth rate on TCA cycle intermediates (19).
One intriguing characteristic of the mqo mutants is that their growth defect in the plant is more pronounced at early time points, with the number of bacteria often reaching wild-type levels by 4 days postinoculation (Fig. 1b). However, the disease symptoms exhibited by plants infected with the mqo mutants never reach the severity of those caused by wild-type bacteria (Fig. 1c and d). The observation that wild-type levels of growth at later stages in infection does not result in normal symptom production raises the possibility that the number of bacteria must reach a certain threshold early during the infection for disease symptoms to develop normally. It is unclear why the in planta growth of mqo mutants eventually reaches wild-type levels in many experiments. One possibility is that other carbon sources become available later in infection, allowing DC3000 to grow without relying on dicarboxylates.
We have also shown that the dctA1 gene, which encodes a dicarboxylate transporter, is required for growth of DC3000 in medium containing malate. In contrast, the dctA1::KO mutant exhibited only slightly reduced growth on A. thaliana compared to wild-type bacteria. There are several possible explanations for why the dctA1::KO does not have a more severe phenotype in planta. One possibility is that, while DctA1 is the primary transporter for dicarboxylates in culture, there may be other transporters that are active when the bacteria are growing in plant tissue. Several other potential dicarboxylate transporters are encoded by the DC3000 genome, including two potential tellurite-resistance/dicarboxylate transporter transporters (PSPTO_4613 and PSPTO_3541) (17) and a tripartate ATP-independent periplasmic transporter consisting of three subunits encoded by the PSPTO_1049, PSPTO_1050, and PSPTO_1051 genes (22). One or more of these other transporters may be active in planta, but do not function during growth in minimal medium. Another possibility is that DC3000 uses dicarboxylates at early stages of colonization but then switches to one or more other carbon sources as they become available later in infection. Finally, we cannot rule out the possibility that mqo is required for other as-yet-unknown functions that contribute to pathogenesis, in addition to its role in the TCA cycle. For example, mqo could be responsible for generating or detecting a signal regulating carbon source utilization and/or expression of other DC3000 virulence factors.
The genomes of several pathogenic and nonpathogenic Pseudomonas species, including P. syringae B728a, P. syringae pv. phaseolicola, P. putida KT2440, and P. fluoresens Pf-5, are also predicted to encode one or more mqo genes, suggesting that mqo may have important functions in other pathogenic and nonpathogenic bacteria. Future studies on the regulation and function of mqo will provide insight regarding the role of this novel virulence factor during pathogenesis and will further our understanding of the strategies used by DC3000 and other pathogens for nutrient acquisition and adaptation to growth in host tissue.
Supplementary Material
Acknowledgments
We thank Gail Preston for helpful comments on the manuscript. We are also grateful to Petra Levin for critical discussion of our experiments. Agnes Demianski, Surobhi Lahiri, and Andrew Mutka also provided useful discussion and commentary.
This study was supported by a Washington University W. M. Keck Postdoctoral Program in Molecular Medicine Fellowship to E.M.M., NSF grant IBN-0130693 and NSF grant IOB-0620469 to B.N.K., and a Life Sciences Research Foundation DOE Energy Biosciences Research Fellowship to A.P.K.
Footnotes
Published ahead of print on 27 February 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aloni, R., and C. I. Ullrich. 2008. Biology of crown gall tumors, p. 565-591. In T. Tzfira and V. Citovsky (ed.), Agrobacterium: from biology to biotechnology. Springer, New York, NY.
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 4473-88. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, D. M., C. L. Bender, and B. N. Kunkel. 2005. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6629-639. [DOI] [PubMed] [Google Scholar]
- 5.Brooks, D. M., G. Hernandez-Guzman, A. P. Kloek, F. Alarcon-Chaidez, A. Sreedharan, V. Rangaswamy, A. Penaloza-Vazquez, C. L. Bender, and B. N. Kunkel. 2004. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 17162-174. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, B., W. Gruissem, and R. Jones. 2000. Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, MD.
- 7.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 10010181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato Appl. Environ. Microbiol. 51323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelke, T., D. Jording, D. Kapp, and A. Puhler. 1989. Identification and sequence analysis of the Rhizobium meliloti dctA gene encoding the C4-dicarboxylate carrier J. Bacteriol. 1715551-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira, A. O., C. R. Myers, J. S. Gordon, G. B. Martin, M. Vencato, A. Collmer, M. D. Wehling, J. R. Alfano, G. Moreno-Hagelsieb, W. F. Lamboy, G. DeClerck, D. J. Schneider, and S. W. Cartinhour. 2006. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis clement, and identifies novel coregulated genes. Mol. Plant-Microbe Interact. 191167-1179. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 is dependent of a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finan, T. M., B. N. Kunkel, G. F. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 16766-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forster-Fromme, K., and D. Jendrossek. 2005. Malate:quinone oxidoreductase (MqoB) is required for growth on acetate and linear terpenes in Pseudomonas citronellolis. FEMS Microbiol. Lett. 24625-31. [DOI] [PubMed] [Google Scholar]
- 14.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. L. Gwinn, S. G. Lazarowitz, N. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 992275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 2841322-1328. [DOI] [PubMed] [Google Scholar]
- 16.Grant, S. R., E. J. Fisher, J. H. Chang, B. M. Mole, and J. L. Dangl. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60425-449. [DOI] [PubMed] [Google Scholar]
- 17.Grobler, J., F. Bauer, R. E. Subden, and H. J. Van Vuuren. 1995. The mae1 gene of Schizosaccharomyces pombe encodes a permease for malate and other C4 dicarboxylic acids. Yeast 111485-1491. [DOI] [PubMed] [Google Scholar]
- 18.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13232-237. [DOI] [PubMed] [Google Scholar]
- 19.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 2451374-1377. [DOI] [PubMed] [Google Scholar]
- 20.Hylemon, P. B., and P. V. Phibbs, Jr. 1972. Independent regulation of hexose catabolizing enzymes and glucose transport activity in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 481041-1048. [DOI] [PubMed] [Google Scholar]
- 21.Joosten, M. H. A. J., L. J. M. Hendrickx, and P. J. G. M. De Wit. 1990. Carbohydrate composition of apoplastic fluids isolated from tomato leaves inoculated with virulent or avirulent races of Cladosporium fulvum (syn. Fulvia fulva). Eur. J. Plant Pathol. 96103-112. [Google Scholar]
- 21a.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, D. J., and G. H. Thomas. 2001. The tripartate ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25405-424. [DOI] [PubMed] [Google Scholar]
- 23.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstratoin of pyocyanin and fluorescein. J. Lab. Clin. Med. 44301-307. [PubMed] [Google Scholar]
- 24.Kloek, A. P., D. M. Brooks, and B. N. Kunkel. 2001. A dsbA mutant of Pseudomonas syringae exhibits reduced virulence and partial impairment of type III secretion. Mol. Plant Pathol. 1139-150. [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar, U., A. Ruckert, J. Jeoung, and H. Gorisch. 2002. Malate:quinone oxidoreductase is essential for growth on ethanol or acetate in Pseudomonas aeruginosa. Microbiology 1483839-3847. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel, B. N., A. F. Bent, D. Dahlbeck, R. W. Innes, and B. J. Staskawicz. 1993. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel, B. N., and Z. Chen. 2006. Virulence strategies of plant pathogenic bacteria, p. 421-440. In M. Dworkin et al. (ed.), The prokaryotes, vol. 2. Springer, New York, NY. [Google Scholar]
- 28.Lee, M., Y. Choi, B. Burla, Y. Kim, B. Jeon, M. Maeshima, J. Yoo, E. Martinoia, and Y. Lee. 2008. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. doi: 10.1038/ncb1782. [DOI] [PubMed]
- 29.Lessie, T. G., and P. V. Phibbs. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38359-388. [DOI] [PubMed] [Google Scholar]
- 30.Li, X.-Z., A. N. Starratt, and D. A. Cuppels. 1998. Identification of tomato leaf factors that activate toxin gene expression in Pseudomonas syringae pv. tomato DC3000. Phytopathology 881094-1100. [DOI] [PubMed] [Google Scholar]
- 31.Lindeberg, M., C. Collmer, and A. Collmer. 2006. The PPI website: information hub for genome viewing and analysis, Hop nomenclature, and ongoing annotation of three Pseudomonas syringae genomes. Phytopathology 96S68. [Google Scholar]
- 32.Lodwig, E., and P. Poole. 2003. Metabolism of rhizobium bacteroids. Crit. Rev. Plant Sci. 2237-78. [Google Scholar]
- 33.Lynch, W. H., and M. Franklin. 1978. Effect of temperature on diauxic growth with glucose and organic acids in Pseudomonas fluorescens. Arch. Microbiol. 118133-140. [DOI] [PubMed] [Google Scholar]
- 34.Melotto, M., W. Underwood, J. Koczan, K. Nomura, and S. Y. He. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126969-980. [DOI] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molenaar, D., M. E. Van Der Rest, and S. Petrovic. 1998. Biochemical and genetic characterization of the membrane-associated malate dehydrogenase (acceptor) from Corynebacterium glutamicum. Eur. J. Biochem. 254395-403. [DOI] [PubMed] [Google Scholar]
- 36a.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118145-146. [DOI] [PubMed] [Google Scholar]
- 37.Preiter, K., D. M. Brooks, A. Penaloza-Vazquez, A. Sreedharan, C. L. Bender, and B. N. Kunkel. 2005. Novel virulence gene of Pseudomonas syringae pv. tomato strain DC3000. J. Bacteriol. 1877805-7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Prentki, P., and H. M. Kirsch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 38.Preston, G. M. 2000. Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol. Plant Pathol. 1263-275. [DOI] [PubMed] [Google Scholar]
- 39.Reyes-Diaz, M., N. Ulloa, A. Zuniga-Feest, A. Gutierrez, M. Gidekel, M. Alberdi, L. J. Corcuera, and L. A. Bravo. 2006. Arabidopsis thaliana avoids freezing by supercooling. J. Exp. Bot. 573687-3696. [DOI] [PubMed] [Google Scholar]
- 40.Rico, A., and G. M. Preston. 2008. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant-Microbe Interact. 21269-282. [DOI] [PubMed] [Google Scholar]
- 41.Ronson, C. W., P. M. Astwood, T. B. Nixon, and F. M. Ausubel. 1987. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 157921-7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Sreedharan, A., A. Penaloza-Vazquez, B. N. Kunkel, and C. L. Bender. 2006. CorR regulates multiple components of virulence in Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 19768-779. [DOI] [PubMed] [Google Scholar]
- 44.Tiwari, N. P., and J. J. Campbell. 1969. Enzymatic control of the metabolic activity of Pseudomonas aeruginosa grown in glucose or succinate media. Biochim. Biophys. Acta 192395-401. [DOI] [PubMed] [Google Scholar]
- 45.Uppalapati, S. R., Y. Ishiga, T. Wangdi, B. N. Kunkel, A. Anand, K. S. Mysore, and C. L. Bender. 2007. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant-Microbe Interact. 20955-965. [DOI] [PubMed] [Google Scholar]
- 46.Van Der Rest, M. E., C. Frank, and D. Molenaar. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J. Bacteriol. 1826892-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson, R. J. 1990. Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB, and dctD. Mol. Plant-Microbe Interact. 3174-181. [DOI] [PubMed] [Google Scholar]
- 48.Watson, R. J., Y. K. Chan, R. Wheatcroft, A. F. Yang, and S. H. Han. 1988. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J. Bacteriol. 170927-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whalen, M. C., R. W. Innes, A. F. Bent, and B. J. Staskawicz. 1991. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 349-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wildermuth, M. C., J. Dewdney, G. Wu, and F. M. Ausubel. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414562-565. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, Y., J. P. Damicone, D. H. Demezas, V. Rangaswamy, and C. L. Bender. 2000. Bacterial leaf spot of leafy crucifers in Oklahoma caused by Pseudomonas syringae pv. maculicola. Plant Dis. 841015-1020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.