Abstract
Streptococcus pneumoniae is a causative agent of high morbidity and mortality. Although sugar moieties have been recognized as ligands for initial contact with the host, only a few exoglycosidases have been reported to occur in S. pneumoniae. In this study, a putative β-galactosidase, encoded by the bgaC gene of S. pneumoniae, was characterized for its enzymatic activity and virulence. The recombinant BgaC protein, expressed and purified from Escherichia coli, was found to have a highly regiospecific and sugar-specific hydrolysis activity for the Galβ1-3-GlcNAc moiety of oligosaccharides. Interestingly, the BgaC hydrolysis activity was localized at the cell surface of S. pneumoniae, indicating that BgaC is expressed as a surface protein although it does not have a typical signal sequence or membrane anchorage motif. The surface localization of BgaC was further supported by immunofluorescence microscopy analysis using an antibody raised against BgaC and by a reassociation assay with fluorescein isothiocyanate-labeled BgaC. Although the bgaC deletion mutation did not significantly attenuate the virulence of S. pneumoniae in vivo, the bgaC mutant strain showed relatively low numbers of viable cells compared to the wild type after 24 h of infection in vivo, whereas the mutant showed higher colonization levels at 6 and 24 h postinfection in vivo. Our data strongly indicate for the first time that S. pneumoniae bgaC encodes a surface β-galactosidase with high substrate specificity that is significantly associated with the infection activity of pneumococci.
The human pathogen Streptococcus pneumoniae (the pneumococcus) is a gram-positive coccus and a member of the lactic acid bacterium group. S. pneumoniae is the major causative agent of lobar pneumonia, otitis media, and meningitis and is carried in the nasopharynx of healthy individuals, a major reservoir for pneumococcal infections (16). In the human host, S. pneumoniae encounters a variety of glycoconjugates, including host defense molecules, mucin, and binding sites exposed on the epithelial surface. Similar to other pathogenic microbes, S. pneumoniae produces both secreted and surface-associated glycosidases that may modify glycoconjugates in the host environment (2, 6, 41). Oligosaccharides are recognized as receptors for invasion or as dietary substrates for the maintenance of colonial microflora. Recent studies have shown that deglycosylation of human glycoconjugates by the sequential actions of exoglycosidases, including neuraminidase (NanA), β-galactosidase (BgaA), and N-acetylglucosaminidase (StrH), is essential for S. pneumoniae colonization and pathogenesis (19, 33). Furthermore, S. pneumoniae can utilize monosaccharides liberated from human glycoconjugates to sustain growth by sequential deglycosylation of host glycoconjugates through the activities of these exoglycosidases (NanA, BgaA, and StrH) and another neuraminidase (NanB) (5).
The enzyme β-galactosidase, classified as EC 3.2.1.23, hydrolyzes the terminal nonreducing galactose from oligosaccharides. It is ubiquitous and present in all living organisms, ranging from bacteria to plants and mammals. Most prokaryotic β-galactosidases are large proteins (more than 120 kDa) that are primarily homologous to Escherichia coli β-galactosidase LacZ and involved in lactose metabolism (4, 26, 27). On the other hand, mammalian lysosomal β-galactosidases are smaller proteins capable of cleaving both β1,3- and β1,4-linked galactoses from glycoproteins and glycolipids and function optimally at acidic pHs (9). In contrast to typical β-galactosidases, which are generally cytoplasmic proteins, the bgaA gene of S. pneumoniae encodes a surface-associated β1,4-galactosidase with hydrolysis activity for N-linked glycans from glycoproteins. The S. pneumoniae bgaA product is synthesized as a β-galactosidase precursor composed of 2,235 amino acid residues and has been studied extensively for its expression and regulation, physiological function, and application for glycan analysis (5, 17, 19, 41). S. pneumoniae BgaA has a putative signal sequence at its N terminus and is surface exposed by anchoring to the cell wall via sortase-mediated cleavage at the LPXTG motif (41). The expression of bgaA is modulated via regulation of an upstream phosphotransferase system (PTS)-encoding operon and is important for S. pneumoniae adherence during colonization of the nasopharynx, on which no glucose is normally available (17).
The whole-genome sequence of S. pneumoniae R6 (13) has suggested the presence of another putative β-galactosidase gene, bgaC, which was annotated to encode a putative β-galactosidase 3 protein composed of 595 amino acid residues. Currently, whole-genome sequences of seven S. pneumoniae strains, S. pneumoniae ATCC 700669, S. pneumoniae G54, S. pneumoniae CGSP14, S. pneumoniae Hungary19A-6, S. pneumoniae D39, S. pneumoniae R6, and S. pneumoniae TIGR4, are available in public databases (13, 21, 32). Even though both bgaA and bgaC genes exist in all of these strains, the biochemical characteristics and function of the bgaC product have not yet been reported. Furthermore, the bgaC genes in these strains share similar genomic contexts in which the bgaC gene is clustered with putative genes involved in sugar transport (Fig. 1A). Here, we found out for the first time that S. pneumoniae BgaC is a surface-associated β-galactosidase with a specific hydrolysis activity for the Galβ1-3GlcNAc moiety of oligosaccharides that could contribute significantly to the adherence and invasion of pneumococci in vivo and in vitro. These features may provide a foundation for evaluating the role of BgaC relative to the physiology and pathogenesis of pneumococcus.
FIG. 1.
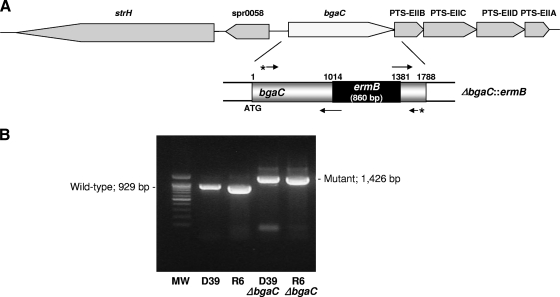
Schematic representation of the genomic context around the bgaC gene on the S. pneumoniae R6 chromosome and construction of a ΔbgaC::ermB mutant allele. (A) (Top) Shaded boxes represent genes and the directions of their transcription. The genes are as follows: strH, encoding β-N-acetylhexosaminidase; spr0058, encoding a hypothetical protein homologous to the GntR transcriptional regulator; bgaC, encoding β-galactosidase 3; PTS-EIIB, encoding PTS component IIB; PTS-EIIC, encoding PTS component IIC; PTS-EIID, encoding PTS component IID; and PTS-EIIA, encoding PTS component IIA. (Bottom) To construct ΔbgaC::ermB mutant strains, an 860-bp ermB cassette was inserted between the upstream and downstream fragments of the bgaC gene by sequential PCR and introduced into the chromosome of the S. pneumoniae R6 or D39 strain by homologous recombination as described in Materials and Methods. (B) The ΔbgaC::ermB deletion mutants were identified by PCR. After selection of erythromycin-resistant colonies, colony PCR was used to confirm insertion of the ermB cassette. A set of primers used for PCR amplification is shown by asterisked arrows in panel A. The wild type shows a 929-bp PCR product, whereas the ΔbgaC::ermB mutant product is 1,426 bp. MW, molecular weight size marker.
MATERIALS AND METHODS
Bacterial strains and cell culture conditions.
The bacterial strains used in this work are presented in Table 1. S. pneumoniae encapsulated strain D39 (type 2) and its nonencapsulated strain R6 were grown in brain heart infusion broth or Todd Hewitt-yeast extract (THY) broth as described previously (20). To create an insertion-deletion mutation of bgaC (ΔbgaC::ermB) in S. pneumoniae, an 860-bp ermB cassette (nucleotides [nt] 117 to 976) (37) was amplified with a set of primers, comprising prs3 (5′-CCGGGCCCAAAATTTGTTTGAT-3′) and prs4 (5′-AGTCGGCAGCGACTCATAGAAT-3′), from chromosomal DNA of erythromycin-resistant E. coli and used to disrupt bgaC. A 245-bp fragment (bgaC-up; nt 771 to 1015) containing part of bgaC and the 5′ end of ermB was amplified with a set of primers, comprising keh3 (5′-GGAATTGGCAGATGCAGT-3′) and keh1 (5′-ATCAAACAATTTTGGGCCCGGGTGGTTCCAACTGCGGATAC-3′), from S. pneumoniae D39 chromosomal DNA. A 321-bp fragment (bgaC-down; nt 1380 to 1700) containing part of the bgaC sequence and the 3′ terminus of ermB was amplified with keh2 (5′ATTCTATGAGTCGCTGCCGACTAGCGGATACGCAACGTAAGG-3′) and keh4 (5′-ATGATACGGTTGGCACCTTCC-3′) from S. pneumoniae D39 chromosomal DNA. The three PCR products were used as a mixed template for PCR with keh3 and keh4 to produce a 1.42-kb fragment with a 365-bp deletion of bgaC (nt 1015 to 1380) that was replaced by the ermB gene. The tripartite 1.42-kb fragment was subsequently introduced into the S. pneumoniae R6 or D39 strain by transformation, and recipient bacteria that had integrated the recombinant fragment into the chromosome by homologous recombination were selected by resistance to erythromycin. Transformants were screened for the correct deletion by PCR and immunoblot analysis. The D39 and R6 ΔbgaC mutants (KEH001 and KEH002, respectively) contained the correct deletion within bgaC and were used for further studies. Competence was controlled by appropriate addition of the competence-specific peptide and quantitated as erythromycin-resistant transformants obtained after exposure of cells to DNA in culture medium, as described previously (20). For selection of pneumococcal transformants, erythromycin was added to growth medium at a concentration of 2.5 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and cell lines used in this study
| Strain, plasmid, or cell line | Genotype or relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | recA1 endA1 gyrA96 thi1 hsdR17 supE44 relA1 lacZΔM15 | Laboratory stock |
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 Sam7 nin5]) | Novagen |
| S. pneumonia strains | ||
| D39 | Encapsulated, type 2 | 3 |
| R6 | Nonencapsulated, type 2 | 13 |
| KEH001 | D39; ΔbgaC::ermB Err | This study |
| KEH002 | R6; ΔbgaC::ermB Err | This study |
| Plasmid pET28a+ | 5.4 kb; Kanr | Novagen |
| Cell lines | ||
| A549 | Lung carcinoma epithelial cell (adherent) | ATCC CCL-185 |
| HEp-2 | Larynx carcinoma epithelial cell | ATCC CCL-23 |
Err, erythromycin resistance.
The human lung epithelial carcinoma A549 cell line and the larynx carcinoma epithelial HEp-2 cell line were obtained from the American Type Culture Collection. A549 and HEp-2 cells were cultured at 37°C in the presence of 95% air-5% CO2 in Dulbecco's modified Eagle's medium with 4.5 g/liter glucose, 10% fetal bovine serum (Gibco BRL), 100 U/ml penicillin G, and 100 μg/ml streptomycin.
Overexpression and purification of recombinant BgaC.
To construct the plasmid expressing the His6-tagged version of BgaC, chromosomal DNA of S. pneumoniae R6 was used as the template in PCRs with the primer pair comprising BgaC-f (5′-CGCTAGCATGACACGATTTGAGATACGAG-3′) and BgaC-r (5′-GGAAGCTTTCATAAGTTTTCCCCCTTTATATG-3′). The 1.8-kb PCR product was digested with NheI and HindIII and ligated with NheI-HindIII-digested plasmid pET28a+ (Novagen), yielding pET28a-bgaC. For expression of recombinant BgaC, an overnight culture of E. coli BL21(DE3) cells transformed with pET28a-bgaC was reinoculated into LB broth supplemented with kanamycin (40 μg/ml) and grown at 37°C. At the mid-exponential phase (A600, ∼0.7), the expression of the His6-tagged BgaC protein was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were then incubated at 18°C for 24 h after induction to avoid inclusion body formation. Cells were harvested by centrifugation, and the cell pellet was resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 500 mM NaCl) for disruption by sonication. All purification steps were carried out at 4°C, and BgaC was purified by Ni-nitrilotriacetic acid affinity chromatography with the ÄKTA prime fast protein liquid chromatography system (Amersham Pharmacia Biotech, Sweden).
β-Galactosidase activity assay.
The β-galactosidase activity assay was initiated by adding BgaC (0.33 nmol) into the reaction mixture (50 mM NaPO4 [pH 6.5], 10 mM p-nitrophenyl-β-galactopyranoside [PNPG], 10 mM MgCl2, 45 mM β-mercaptoethanol [BME]), and the mixture was incubated for 30 min at 30°C. Reactions were stopped by adding 500 μl of 1 M sodium carbonate solution, and the amount of p-nitrophenol (PNP) released was determined by measuring the increase in absorbance at 420 nm. To determine the pH dependence of the enzymatic release of PNP from PNPG, enzyme activity was measured between pH 5 and 8 by using acetic acid (50 mM, pH 5.0 and 5.5) and sodium phosphate (50 mM, pH 6.0 to 8.0) buffers. The temperature dependency of the enzyme activity was measured by assaying the enzymatic release of PNP over the temperature range of 10 to 70°C. To evaluate the effects of various divalent cations on BgaC activity, enzyme reactions in the reaction mixture (90 mM NaPO4 [pH 6.5], 10 mM PNPG, 45 mM BME) were carried out in the presence of a 10 mM final concentration of various cations (MgSO4, MnCl2, CaCl2, ZnCl2, FeSO4, NiSO4, or CuSO4) or EDTA.
Linkage specificity assay.
To determine the linkage specificities of both BgaC and BgaA, enzyme activities were assayed with the sugar chains listed in Table 2. Each 50-pmol 2-pyridylamine(PA)-labeled glycan was reacted with 2 mU BgaC or BgaA in a 50-μl reaction mixture (50 mM NaPO4 [pH 6.5], 10 mM MgCl2, 45 mM BME) for 20 h at 30°C. Release of terminal galactose from specific sugar was detected by high-performance liquid chromatography (HPLC; Waters) or matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS; Bruker, Germany). After the reaction, protein was removed with Microcon YM-10 (Millipore) and the product was analyzed using an HPLC device connected to a model 2475 multi-λ fluorescence detector (Waters) with an Asahipak amine NH2P-50 4E column (4.5 by 250 mm; Shodex, Japan). Product sugar chains were separated by an isocratic mobile phase (200 mM acetic acid-triethylamine [pH 7.3]-acetonitrile [25:75, vol/vol]) with a 1-ml/min flow rate. MALDI-TOF MS was performed in the reflector-positive and linear negative-ion mode, using 2,5-dihydroxybenzoic acid (DHB) and 6-aza-2-thiothymine (6ATT) as a matrix. DHB and 6ATT were prepared as a saturated solution in 25% acetonitrile-0.1% trifluoroacetic acid, equally mixed. All samples were irradiated with UV light (337 nm) from an N2 laser at a 20-kV accelerating voltage.
TABLE 2.
Hydrolysis of terminal galactose from various sugar chains by BgaC and BgaAa
Fifty picomoles of each substrate was incubated with 2 mU BgaC or BgaA in a 50-μl reaction mixture (50 mM NaPO4 [pH 6.5], 10 mM MgCl2, 45 mM β-mercaptoethanol) for 20 h at 30°C. Release of the terminal galactose from substrates was analyzed by HPLC or MALDI-TOF. The sugar chains 025, 028, 042, 043, and 044 were purchased from Takara (Japan). The sugar chain NA2 was from Sigma.
Localization of BgaC activity of S. pneumoniae R6.
To investigate the localization of BgaC in S. pneumoniae R6, enzyme activity against the sugar chain in growth medium, soluble cell lysates, and intact cells was measured. Cells were inoculated into 5 ml brain heart infusion (Becton Dickinson) broth and incubated for 24 h up to stationary phase under anaerobic conditions. Cells were harvested by centrifugation at 2,000 × g, washed once with 50 mM sodium phosphate buffer (pH 6.5), and then resuspended in 500 μl sodium phosphate buffer (pH 6.5). The culture supernatant was concentrated 10-fold by using an Amicon Ultra-15 device with a molecular weight cutoff of 10,000 (Millipore). The soluble cell lysates were obtained by centrifugation after treatment of cells with 200 μl of 1 mg/ml lysozyme solution for 1 h and sonication. The BgaC activity for the sugar chain was determined according to linkage specificity assay methods. Twenty-five microliters of the culture supernatant, the cell suspension, or the cell lysates was added into the 55-μl reaction mixture (50 mM NaPO4[pH 6.5], 10 mM MgCl2, 45 mM BME, and 20 μg lacto-N-tetraose [LNT]) and incubated for 120 h at 30°C. Cleavage of galactose was analyzed by thin-layer chromatography (TLC). A small volume of product (4 to 5 μl) was spotted in a tight band on a model 60 silica gel aluminum-backed TLC plate (Merck). The bands were completely dried in dry oven at 60°C for 15 min. The plate was developed in an ethylacetate-acetic acid-H2O (2:1:1, vol/vol/vol) solution. The spots were visualized with 2% orcinol reagent dissolved in 25% sulfuric acid.
Western blot and immunofluorescence analyses.
The cell wall fraction of S. pneumoniae was obtained as previously described (38), but with a slight modification. Briefly, the wild-type strain of S. pneumoniae R6 grown overnight in THY broth was harvested, disrupted by lysozyme treatment and sonication, and separated by centrifugation to obtain the soluble cell lysate. The remaining cell debris pellet was then dissolved in 4% sodium dodecyl sulfate (SDS) solution, boiled for 15 min, and centrifuged to obtain the cell wall fraction. Each fractionated sample was separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting with a 1:500 dilution of rabbit antiserum, which was raised against the purified recombinant BgaC protein, as a primary antibody. The goat anti-rabbit immunoglobulin G (IgG) conjugated with alkaline phosphatases was used at a 1:2,000 dilution as a secondary antibody, and then BgaC was detected by color development of a 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium substrate. For immunofluorescence microscopy analysis, pneumococci were reacted with anti-BgaC antiserum, which was preadsorbed to heat-killed S. pneumoniae R6 ΔbgaC mutant cells to eliminate cross-reactivity, counterstained with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG antibodies (Sigma), and inspected by confocal microscopy (Carl Zeiss LSM 510 META). To examine the reassociation of soluble BgaC protein to the surfaces of streptococci, purified BgaC was labeled by using an FITC antibody-labeling kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. The wild-type strain of S. pneumoniae R6 was grown overnight in THY broth and harvested by centrifugation at 1,500 × g for 5 min. Cell pellets were washed with phosphate-buffered saline (PBS; pH 7.4) twice and resuspended in PBS at a concentration of approximately 5 × 108 bacteria/ml. The bacterial suspension (100 μl) was mixed with 33 μg of FITC-labeled BgaC and incubated at room temperature for 1 h. For the competition experiment, the bacterial suspension was mixed with 120 μg of nonlabeled BgaC and incubated at room temperature for 30 min prior to treatment of FITC-labeled BgaC. Unbound FITC-labeled BgaC was removed by washing it in PBS with centrifugation before being inspected by confocal microscopy.
Tissue culture assays.
Invasion assays were performed as described previously (35). Briefly, A549 or HEp-2 cells were grown to confluence in 12-well tissue culture plates and washed three times with PBS (140 mM NaCl, 3 mM KCl, 10 mM NaH2P04, 1.5 mM KH2P04, pH 7.2), after which 1 ml of culture medium (without antibiotics) was added per well. Exponential-phase cultures of D39 and its isogenic ΔbgaC mutant derivatives were harvested by centrifugation, washed with PBS, and resuspended in Dulbecco's modified Eagle's medium. Monolayers were infected with 2 × 107 bacteria (bacterium/cell ratio [multiplicity of infection {MOI}], 100:1), followed by 1, 2, and 3 h of incubation at 37°C. Fresh medium containing 10 μg/ml penicillin and 200 μg/ml gentamicin was added to each well to kill extracellular bacteria. After an additional 1 h of incubation, the monolayers were washed with PBS, and the cells were detached from the plates by treatment with 0.25% trypsin-0.02% EDTA and then lysed with Triton X-100 (0.025% in H2O). Appropriate dilutions were plated on blood agar to determine the numbers of viable bacteria. To determine the total numbers of adherent and intracellular bacteria, infected monolayers were washed as described above and then trypsinized, lysed, and plated quantitatively without antibiotic treatment. All samples were assayed in triplicate, and each assay was repeated at least three times.
Intranasal challenge.
The intranasal challenge was carried out essentially as described previously (20). Before the challenge, bacteria were cultured at 37°C overnight on blood agar (supplemented with erythromycin where appropriate) and then grown in THY broth for approximately 4 h at 37°C to give ca. 4 × 107 CFU/ml (A600, 0.1). Each bacterial culture then was adjusted in THY broth to ca. 109 CFU/ml. Groups of five CD1 mice (5 weeks old) were infected intranasally with 10 μl of either D39 or D39 ΔbgaC at ca. 2 × 107 CFU/mouse. Survival of mice was monitored four times daily for the first 5 days, twice daily for the next 5 days, and then daily until 21 days after challenge. To enumerate bacteria in different organs after intranasal challenge, mice were sacrificed at 6, 12, and 24 h postinfection, and blood samples, nasopharynxes, and lungs were collected aseptically and then washed three times with PBS (pH 7.3). Samples were then homogenized in PBS with a tissue homogenizer (model 200, double insulated; PRO Scientific, Inc., Oxford, CT) on ice, serially diluted as appropriate in sterile PBS, and plated in duplicate on blood agar containing the appropriate antibiotic(s). Subsequently, plates were incubated for approximately 16 h at 37°C in an atmosphere of 95% air-5% CO2, after which colonies were counted and averaged between replicates.
Statistics.
Statistical analysis was performed using paired or unpaired Student t tests. Data presented are means ± standard deviations from the mean for two to four independent experiments. Differences in median survival times between groups were analyzed by the Mann-Whitney U test (two tailed), and differences in overall survival rate between groups were analyzed by the Fisher exact test.
RESULTS
Sequence analysis and comparison of S. pneumoniae BgaC.
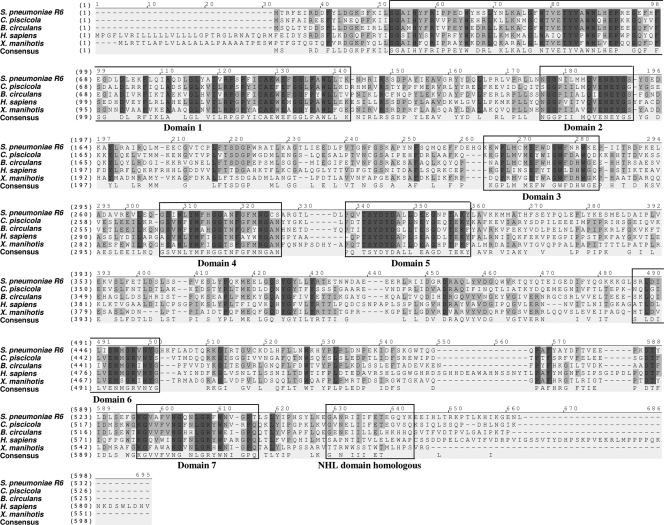
S. pneumoniae bgaC was predicted to encode a protein showing sequence homology with β-galactosidases of glycosyl hydrolase family 35, which mainly includes enzymes from higher eukaryotes (12). Thus, S. pneumoniae BgaC more closely resembles β-galactosidases of higher eukaryotes (such as mammalian lysosomal β-galactosidases) and those of microbial pathogens than typical prokaryotic β-galactosidases in sequence and structural organization. The BgaC protein of S. pneumoniae shows 47%, 43%, 36%, and 41% identity and 64%, 63%, 54%, and 57% similarity to those of Carnobacterium piscicola, Bacillus circulans, Homo sapiens, and Xanthomonas axonopodis pv. manihotis, respectively (Fig. 2). The sequence homology comparison between BgaC and other homologs reveals that S. pneumoniae BgaC has seven highly conserved domains, comprising two amino-terminal domains (domains 1 and 2), a cluster of three small central domains (domains 3, 4, and 5), and two small carboxy-terminal domains (domains 6 and 7), similar to those reported in previous studies of the X. axonopodis pv. manihotis β-galactosidase (31) and of the NHL (NCL-1, HT2A, and LIN-41) repeat homologous domain (29) (Fig. 2). Interestingly, to date, the known prokaryotic NHL domain-containing proteins have all been found in prokaryotes that are human pathogens (28).
FIG. 2.
Sequence comparison of BgaC homologs of various organisms. Identical and similar amino acids are shaded in dark and faint gray. The BgaC protein of S. pneumoniae (NP_357763) shows 47%, 43%, 36%, and 41% identity and 64%, 63%, 54%, and 57% similarity to those of Carnobacterium piscicola (AAL27306), Bacillus circulans (BAA21669), Homo sapiens (P16278), and Xanthomonas axonopodis pv. manihotis (AAC41485), respectively. S. pneumoniae BgaC has seven highly conserved domains, two amino-terminal domains (domains 1 and 2), a cluster of three small central domains (domains 3, 4, and 5), two small carboxy-terminal domains (domains 6 and 7), and the NHL repeat homologous domain. Alignment of amino acid sequences was carried out by Vector NTI (version 9.0).
Expression and biochemical characterization of recombinant BgaC.
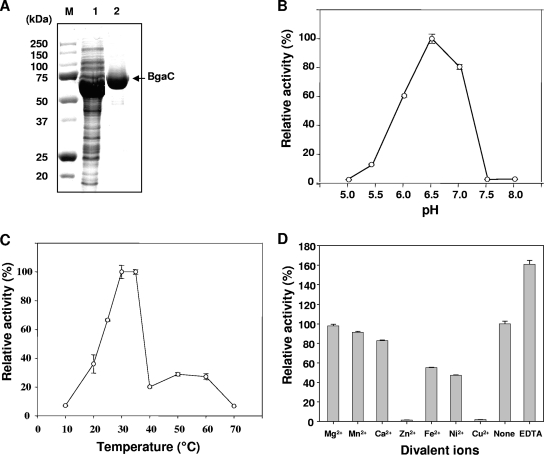
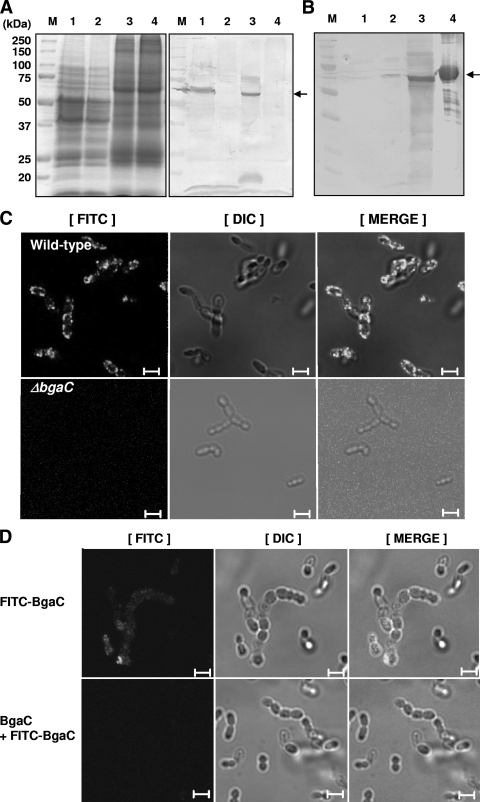
In order to investigate its biochemical properties, S. pneumoniae BgaC was overexpressed as a His6-tagged protein in E. coli and purified by Ni-nitrilotriacetic acid affinity chromatography (Fig. 3A). The molecular mass of the denatured recombinant BgaC protein was estimated to be ∼69 kDa by SDS-PAGE analysis, which is in good agreement with the molecular mass deduced from the amino acid sequence. The native molecular mass of the enzyme, as determined by size exclusion chromatography, was also ∼69 kDa, suggesting that BgaC is a monomeric enzyme (data not shown).
FIG. 3.
Purification and biochemical characterization of recombinant BgaC protein. (A) Overexpression and purification of BgaC. BgaC protein was overexpressed and purified as His6-tagged protein from E. coli as described in Materials and Methods. M, molecular mass markers; lane 1, crude extracts of E. coli transformed with pET28-bgaC; lane 2, purified BgaC. Results are shown for optimal pH (B) and temperature (C) analyses. β-Galactosidase activity was measured at various pHs (5.0 to 8.0) and temperatures (10 to 70°C) as described in Materials and Methods. (D) Analysis of effects of divalent cations on BgaC activity. The respective divalent cations or EDTA (10 mM final concentration) was added to the BgaC reaction mixture, and the enzyme activity was determined as described in Materials and Methods.
The β-galactosidase activity of the purified recombinant BgaC protein was analyzed by using PNPG as a chromogenic substrate. The pH optimum for the BgaC reaction was determined over the range of pH 5.0 to 8.0. The enzyme exhibited maximum activity at pH 6.5 (Fig. 3B). The enzyme activity decreased sharply at pH 5.5 or 7.5 to less than 20% of that observed at the optimum pH. The optimum temperature for the reaction was determined by incubating the assay mixture at various temperatures, ranging from 10 to 70°C. The enzyme showed the highest activity at 30°C, while at 40°C, the activity decreased sharply to about 20% of that at 30°C (Fig. 3C). The effects of various divalent cations on BgaC activity were tested at final concentrations of 10 mM. In the presence of Zn2+ and Cu2+, the enzyme exhibited less than 5% of the activity observed in the absence of any added metal ion. In the presence of Fe2+ and Ni2+, the enzyme showed about 50% activity (Fig. 3D). In contrast, addition of Mg2+, Mn2+, and Ca2+ did not affect the enzyme activity and resulted in no significant change compared with the activity in the control, implying that BgaC activity does not require divalent cations, in contrast to E. coli LacZ (14). Furthermore, the enzyme activity was rather increased, about 60%, in the presence of EDTA, compared to that observed in the absence of any metal ion.
Linkage specificity and localization of BgaC activity at the cell surface.
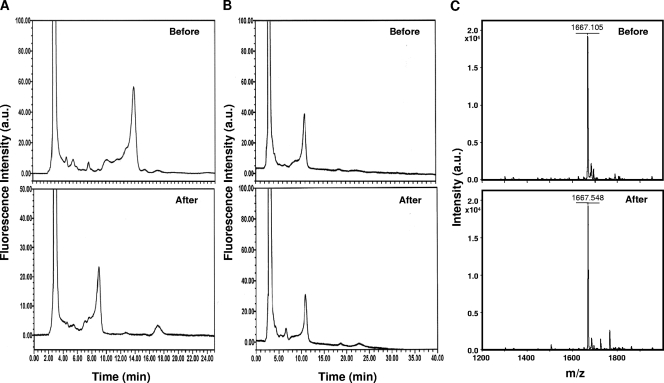
To identify the linkage specificities of BgaC, sugar chains 028 (asialo GM1-tetrasaccharide), 025 (N-acetyllactosamine type, tetrasialylated triantennary), 042 (lacto-N-tetraose), 043 (lacto-N-fucopentaose I), 044 (lacto-N-fucopentaose II), and NA2 (asialo galactosyl biantennary) were used as substrates (Table 2). Each sugar chain was treated with BgaC and analyzed by HPLC or MALDI-TOF MS to determine whether BgaC was able to liberate galactose moiety from the substrates. BgaC was shown to catalyze the hydrolysis of galactose from sugar chain 042, which contains Galβ1-3GlcNAc. However, BgaC was not able to release galactose from sugar chains 028 and NA2, which contain Galβ1-3GalNAc and Galβ1-4GlcNAc, respectively (Fig. 4). Moreover, BgaC was not able to hydrolyze the Galβ1-3GlcNAc linkage of sugar chain substrates 043, 044, and 025, which contain galactose or N-acetylglucosamine residues modified by fucosylation or sialylation. In contrast, when these sugar chains were treated with recombinant BgaA, expressed and purified from E. coli, only NA2 was hydrolyzed (data not shown). Our data indicate that BgaC is highly specific for the terminal Gal-GlcNAc moiety with a β1,3-glycosidic bond without any modification.
FIG. 4.
Linkage hydrolysis specificity of BgaC. To determine the linkage specificity of BgaC-catalyzed hydrolysis, sugar chains 042, 028, and NA2 were treated with purified recombinant BgaC at 30°C for 20 h and analyzed by HPLC or MALDI-TOF MS. (A) HPLC chromatogram of PA-labeled sugar chain 042 before and after BgaC treatment; (B) HPLC chromatogram of PA-labeled sugar chain 028 before and after BgaC treatment; (C) MALDI-TOF mass spectrum of sugar chain NA2 before and after BgaC treatment. a.u., arbitrary units.
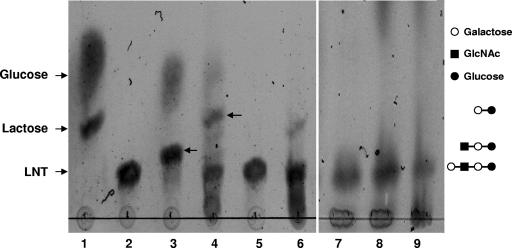
Because BgaC hydrolyzed the galactose from LNT, which is a lacto structure in the host glycosphingolipid (1, 25), we speculated that BgaC might be secreted or exposed on the S. pneumoniae surface to act on the host oligosaccharides. To test this hypothesis, LNT was treated with the culture supernatant, intact cells, or soluble cell lysates of the wild-type strain of S. pneumoniae R6, and the resultant products were analyzed by TLC (Fig. 5). While purified BgaC completely released terminal β(1,3)-galactose from LNT to produce trisaccharide (lane 3), other fractions did not. Interestingly, the intact cells and culture supernatant converted LNT to disaccharide, which had a migration pattern similar to that of lactose (lanes 4 and 6), but the soluble cell lysate did not cleave LNT to tri- or disaccharide (lane 5). Presumably, the generation of disaccharide as a final product by the intact cells and culture supernatant is due to the action of another active glycosidase, hexosaminidase, which is a product of strH. The cell surface-associated hexosaminidase of S. pneumoniae is involved in sequential deglycosylation of human glycoconjugates (19). In contrast, when LNT was treated with the fractions of a ΔbgaC mutant strain of S. pneumoniae R6, no cleavage was observed (Fig. 5, lanes 7 to 9). These results suggest that BgaC might be located on the cell surface and thus involved in the cleavage of terminal galactose in the lacto structure of host glycosphingolipids.
FIG. 5.
Localization of BgaC activity in S. pneumoniae R6. To localize the BgaC activity, culture broths of wild-type (lanes 4 to 6) and ΔbgaC mutant (lanes 7 to 9) strains of S. pneumoniae R6 were fractionated into culture supernatant, intact cell pellets, and soluble cell lysates, and then each fraction was incubated with lactose-N-tetraose (LNT) for 120 h at 30°C. The reaction mixtures were separated by TLC and visualized as described in Materials and Methods. The arrows in lanes 3 and 4 indicate trisaccharide and disaccharide, respectively. Lanes: 1, lactose plus glucose; 2, LNT; 3, LNT plus purified recombinant BgaC; 4, LNT plus intact cell pellets; 5, LNT plus soluble cell lysates; 6, LNT plus culture supernatant; 7, LNT plus intact cell pellets; 8, LNT plus soluble cell lysates; 9, LNT plus culture supernatant.
Construction of bgaC deletion mutants and analysis of BgaC expression.
To further study the expression and function of BgaC, bgaC deletion (ΔbgaC) mutations in both nonencapsulated R6 and encapsulated D39 strains were generated by replacing a 365-bp internal fragment of bgaC with an ermB erythromycin resistance cassette by homologous recombination (Fig. 1A). The correct deletion of bgaC was confirmed by PCR (Fig. 1B). Moreover, the absence of BgaC protein expression was confirmed by immunoblot analysis using anti-BgaC antibody. No signal was detected in the ΔbgaC strains in either the R6 or the D39 background, whereas a single 69-kDa protein was clearly detected in the total cell lysate from both wild-type strains (Fig. 6A). This result also indicates that BgaC is expressed not only in the nonencapsulated avirulent R6 strain but also in the encapsulated virulent D39 strain when cultivated under normal laboratory conditions. However, the ΔbgaC mutant strains did not exhibit detectable changes in growth or morphology (data not shown).
FIG. 6.
Expression of BgaC on the surface of S. pneumoniae R6. (A) Total cell lysate analysis. Wild-type and ΔbgaC mutant S. pneumoniae R6 and D39 strains grown overnight at 37°C in THY broth were harvested by centrifugation, lysed by lysozyme treatment, boiled for 15 min in 4% SDS solution, and then analyzed by SDS-PAGE (left) and Western blotting (right). M, molecular mass markers; lane 1, R6 wild type; lane 2, R6 ΔbgaC mutant; lane 3, D39 wild type; lane 4, D39 ΔbgaC mutant. An arrow indicates a signal corresponding to BgaC. (B) Cell wall fractionation analysis by Western blotting. The wild-type strain of S. pneumoniae R6 grown overnight in THY broth was fractionated into the soluble cell lysate and the cell wall fraction, separated by SDS-PAGE, and then analyzed by Western blotting as described in Materials and Methods. M, molecular weight markers; lane 1, culture supernatant; lane 2, soluble cell lysates; lane 3, cell wall fraction; lane 4, purified recombinant BgaC. (C) Immunofluorescence microscopy analysis for localization of BgaC on the surface of S. pneumoniae R6. Wild-type (top) and ΔbgaC mutant (bottom) strains were incubated with anti-BgaC antibodies, counterstained with FITC-conjugated anti-rabbit IgG antibodies, and inspected by confocal microscopy. Images of binding of anti-BgaC antibody to bacteria (left), differential interference contrast (DIC) microscopy images of same bacteria (middle), and merged pictures of the two (right) are shown. Bars, 5 μm. (D) Fluorescence microscopy analysis for reassociation of the recombinant BgaC protein on the surface of S. pneumoniae R6. The wild-type strain was incubated with FITC-labeled BgaC protein (top) or preincubated with unlabeled BgaC prior to incubation with FITC-labeled BgaC protein (bottom). Bacterial suspensions were washed in PBS by centrifugation to remove unbound proteins and inspected by confocal microscopy. Images of binding of FITC-labeled BgaC to bacteria (left), differential interference contrast microscopy images of same bacteria (middle), and merged pictures of the two (right) are shown. Bars, 5 μm.
Since the linkage specificity analysis of BgaC-mediated sugar chain hydrolysis suggested that BgaC is located on the surface of S. pneumoniae, the localization of BgaC was further analyzed by fractionation experiments and immunofluorescence microscopy. Since it is difficult to fractionate the cell wall from the encapsulated D39 strain because of the thick capsule, the nonencapsulated R6 strain was used for the fractionation study. Pneumococci were fractionated into cell wall and cytosol fractions and subjected to immunoblotting analysis using anti-BgaC polyclonal rabbit IgG (Fig. 6B). The appearance of an immunoblotting band in the cell wall fraction strongly indicated that BgaC was located on the cell surface; this was further supported by immunofluorescence microscopy analysis (Fig. 6C). The S. pneumoniae R6 wild-type and bgaC-deleted mutant strains were treated with anti-BgaC rabbit polyclonal antibody and FITC-conjugated anti-rabbit secondary antibody. Fluorescence signals surrounding cells were detected in the S. pneumoniae R6 wild-type strain treated with primary and secondary antibodies, whereas no signal was detected in the ΔbgaC mutant strain. In order to investigate whether soluble BgaC protein could bind back to the cell surfaces of streptococci, FITC-labeled BgaC was treated with a suspension of S. pneumoniae R6 and inspected by confocal microscopy. As shown in Fig. 6D, FITC-labeled BgaC protein was able to reassociate to the surfaces of bacteria (top panels). When unlabeled BgaC was challenged as a competitor prior to treatment of FITC-labeled BgaC, no fluorescence signal was detected (bottom panels). Along with the biochemical fractionation analysis, the immunofluorescence microscopy analysis and reassociation assay strongly indicate that BgaC is expressed on the outer surface of S. pneumoniae.
Effect of bgaC deletion mutation on virulence and adherence.
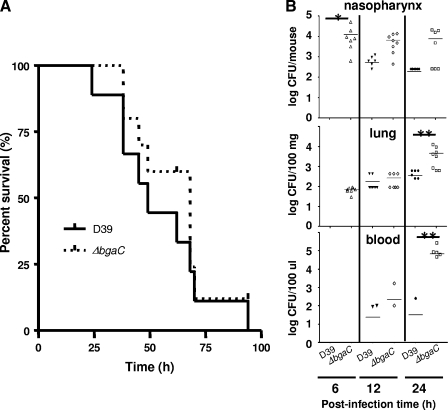
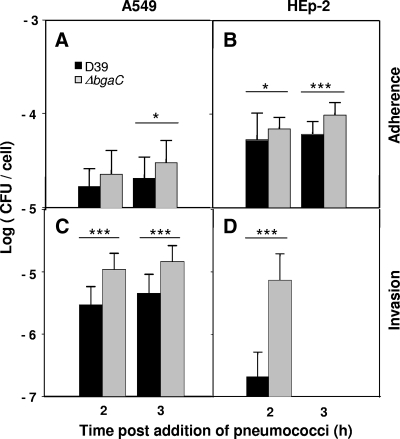
If BgaC degrades galactosides of the host cells upon contact with the pneumococci, adherence of pneumococci to the host cells could be affected by the absence of BgaC protein. Therefore, the effect of the bgaC deletion on virulence was investigated by determining survival time for mice after infection with pneumococci via the intranasal route. However, the encapsulated ΔbgaC mutant did not significantly attenuate the virulence, compared to the wild-type D39 strain (Fig. 7A), indicating that the expression of the bgaC gene, which could be induced during invasion from the nasopharynx to the lungs and blood, may not play a critical role in host damage. To assess the effect of the bgaC deletion on colonization after intranasal infection, numbers of viable cells of the encapsulated ΔbgaC mutant in mice were determined. At 6 h postinfection, the numbers of viable cells of the ΔbgaC mutant in the nasopharynxes (P < 0.05) were significantly higher than those of the parental strain. Consistent with this, at 12 and 24 h postinfection, the numbers of viable cells of the ΔbgaC mutant in the nasopharynxes, lungs, and blood samples were always higher than those of the parental strain (Fig. 7B), indicating that the ΔbgaC mutant could colonize more efficiently than the wild type at the early phase of infection. Moreover, when the nonencapsulated R-type ΔbgaC mutant was used for the colonization experiment, the bgaC mutant could colonize more efficiently than the encapsulated ΔbgaC mutant (data not shown). To test the possibility that the ΔbgaC mutant might compete out the wild type at earlier time points, thus showing the higher colonization level, an in vitro coinfection experiment was performed. When A549 cells were infected with 2 × 107 CFU (MOI = 100) of the wild type alone or in combination with 2 × 107 CFU of the ΔbgaC mutant for up to 3 h, the number of viable cells of the wild type was not decreased in the presence of the ΔbgaC mutant (data not shown), demonstrating that the higher adherence and colonization levels of the ΔbgaC mutant were not due to the slower growth of the wild-type strain. To investigate the mechanism underlying the higher numbers of viable cells in the ΔbgaC mutant at the early stage of infection in vivo, human lung epithelial carcinoma A549 cells and laryngeal Hep-2 cells were infected in vitro with the ΔbgaC mutant for 2 and 3 h and, numbers of viable cells were determined. Consistent with the in vivo results, the ΔbgaC mutant showed significantly higher adherence and invasion than the wild-type D39 strain in both cell types (Fig. 8). These results clearly demonstrate that the ΔbgaC mutant can adhere and subsequently invade the host cells more efficiently in the early phase but that it becomes subsequently more vulnerable to the host immune system, resulting in lower viability than the wild-type.
FIG. 7.
No attenuated virulence but higher colonization levels of the bgaC deletion mutant at the early phase of infection in vivo. (A) Effect of the bgaC deletion mutation on virulence. Groups of 10 CD1 mice were challenged intranasally with approximately 4 × 107 CFU of D39 or 3.5 × 107 CFU of the isogenic bgaC mutant. Each datum point represents one mouse. Solid line, wild-type D39; dotted line, ΔbgaC mutant. (B) Effect of the bgaC deletion mutation on bacterial recovery from nasopharynxes, lungs, and blood samples of CD1 mice after intranasal challenge. Twenty-four CD1 mice/group were challenged intranasally with either wild-type D39 or the ΔbgaC mutant at 1 × 107 CFU/mouse. At 6, 12, and 24 h postinfection, eight mice from each group were sacrificed and the number of recovered bacteria was determined by plating on blood agar. The figure shows the standard deviations for three independent experiments. Asterisks denote values significantly different from that for the wild type (*, P < 0.05; **, P < 0.01).
FIG. 8.
Increased adherence and invasion of the ΔbgaC mutant into host cells in vitro. Adherence to and invasion of A549 cells (A and C) or HEp-2 cells (B and D) were analyzed by infecting cells with 2 × 107 CFU (MOI = 100) of wild-type D39 or the ΔbgaC mutant. For adherence, the monolayer was washed after infection and the total number of bacteria in each well was determined by a viable-cell count. For invasion, extracellular bacteria were removed by treatment with penicillin and gentamicin after infection. The monolayer was then washed extensively and the number of intracellular bacteria was determined by a viable-cell count. *, P values of <0.05; ***, P values of <0.001 for comparison with the wild-type-infected group. The figure shows the standard deviations for three independent experiments. Black bars, wild-type D39 strain; gray bars, ΔbgaC mutant strain.
Adherence of pneumococci could be affected by the polysaccharide capsule, and nonencapsulated strains adhere more efficiently than encapsulated strains (30). Therefore, the higher adherence level of the ΔbgaC mutant could be ascribed to the lower capsular polysaccharide level. However, measuring polysaccharide in the ΔbgaC mutant by Western blotting using type 2 antiserum revealed almost the same amount as that in the wild type (data not shown), demonstrating that the bgaC deletion did not decrease capsular polysaccharide.
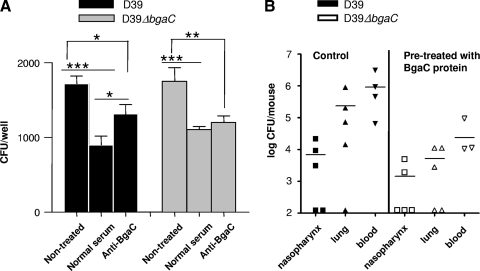
In an effort to confirm that BgaC is directly involved in the adherence of pneumococci to A549 cells, BgaC antibody was used to block BgaC on the surface of wild-type D39 for 30 min, and adherence of the pneumococci on A549 cells was then determined. Adherence of the wild type treated with anti-BgaC antibody was significantly increased compared with that of the wild type treated with normal serum (before immunization); however, adherence of the ΔbgaC mutant treated with anti-BgaC antibody was not increased, and there was no apparent difference from the treatment with normal serum (Fig. 9A). We determined the effect of anti-BgaC antibody on the enzyme activity of recombinant BgaC and observed that the enzyme activity was decreased in the presence of anti-BgaC antibody (data not shown). These results support our hypothesis that BgaC is present on the pneumococci surface and that the enzyme activity of BgaC is important in determining adherence.
FIG. 9.
Adherence of pneumococci altered by availability of BgaC on the surface. (A) Increased adherence of the wild type to the host cells by pretreatment with anti-BgaC antibody in vitro. Wild-type D39 or the ΔbgaC mutant was preincubated with anti-BgaC antibody for 30 min at 37°C. Subsequently, A549 cells were infected with 2 × 107 CFU (MOI = 100) of the wild type or the ΔbgaC mutant. For adherence, the monolayer was washed after infection, and the total number of bacteria in each well was determined by a viable-cell count. *, P values of <0.05; **, P values of <0.01; ***, P values of <0.001 for comparison with the control group. The figure shows the standard deviations for three independent experiments. Black bars, wild-type D39 strain; gray bars, ΔbgaC mutant strain. (B) Colonization of the ΔbgaC mutant decreased by pretreatment with BgaC protein in vivo. Mice (five mice/group) were treated with BgaC enzyme (225 μg/50 μl) intranasally for 30 min and infected with the ΔbgaC mutant (5 × 108 CFU/10 μl). The colonization level was determined at 12 h postinfection. The mice pretreated with BgaC protein showed lower colonization levels than the control in vivo.
To corroborate further the possibility that surface-associated exoglycosidase BgaC can hydrolyze and remove the host cell ligand, purified BgaC protein was added onto the nasopharynxes of mice and incubated for 30 min, and then each mouse was infected with the bgaC mutant and the colonization level was determined. Consistently, when mice were pretreated with the BgaC protein, the colonization of the mutant was decreased in comparison to that of the nontreated control (Fig. 9B), indicating that surface-associated exoglycosidase BgaC can hydrolyze and remove the host cell ligand so that binding of pneumococci to the host would be decreased in vitro.
DISCUSSION
Exoglycosidases in pathogenic bacteria are important enzymes for infection and colonization of host cells. The essential role of BgaA, a surface-associated β1,4-galactosidase of S. pneumoniae, was demonstrated in degalactosylation of human glycoconjugates for colonization and/or pathogenesis (19). In contrast to typical β-galactosidases that comprise approximately 1,000 amino acids and are cytoplasmic proteins, S. pneumoniae bgaA encodes a 2,235-amino-acid polypeptide with a putative signal sequence at the N terminus (41). In S. pneumoniae BgaA, 365 residues located in the N-terminal half of the protein show homology to the E. coli and Streptococcus thermophilus β-galactosidases, but the remainder of the protein displays no homology to the other proteins. Interestingly, the complete genome sequencing of S. pneumoniae (13) reveals that S. pneumoniae has another β-galactosidase of 595 amino acids, BgaC, which shows relatively high homology to eukaryotic β-galactosidases as well as to pathogenic microbial enzymes.
In the present study, we showed that the β-galactosidases encoded by bgaA and bgaC of S. pneumoniae R6 differ in their substrate specificities. While S. pneumoniae BgaA releases a terminal galactose β1,4 linked to GlcNAc in sugar chains of glycoproteins, BgaC shows high substrate specificity only for Galβ1-3GlcNAc (Table 2). It was previously reported that BgaC proteins of B. circulans and X. axonopodis pv. manihotis could cleave terminal galactoses of Galβ1-3GlcNAc or Galβ1-4GlcNAc, but the specific hydrolysis of Galβ1-3GlcNAc was 1,000-fold more efficient than that of Galβ1-4GlcNAc (11, 15, 40). Moreover, BgaC proteins from C. piscicola and B. circulans had specificity for galactose linked to both GalNAc and GlcNAc with a β1,3-glycosidic bond (15, 40). In addition, BgaC of B. circulans also has activity for galactose linked to GlcNAc modified with Neu5Ac or Fuc (11). In contrast, BgaC of S. pneumoniae cleaved only the terminal galactose linked to GlcNAc that was not modified with Fuc or Neu5Ac (Table 2). The analysis of BgaC activity with various sugar chains indicates that S. pneumoniae BgaC is a β-galactosidase with high oligosaccharide specificity for the β1,3-glycosidic bond rather than the β1,4-glycosidic bond with GlcNAc.
Localization analysis of BgaC indicated that it is expressed on the cell surface, even though BgaC does not have a typical signal peptide and LPXTG motif or choline-binding repeats on its amino or carboxyl terminus, which are required for anchorage of proteins on the cell surface. Recently, it was reported that there was a new class of virulence factors that did not have anchors but rather underwent surface-located adhesion and invasions (7). Expression of BgaC on the cell surface may belong to this new class of virulence factors and play a role in adherence to the host cell by exposing GlcNAc in glycolipid. Amino acid sequence alignment and motif scanning of BgaC revealed an interesting motif, homologous to the NHL repeat sequence (28), located at amino acids 562 to 573. NHL is defined by amino acid sequence homologies among Ncl-1, HT2A, and Lin-41 proteins (28) and is a conserved structural motif present in a large family of growth regulators. According to structural model analysis, the NHL domain is expected to be involved in protein-protein interaction (28). Bacterial NHL repeat domains are homologues of the YWTD repeat family of proposed β-propeller domains, which are widespread in eukaryotic extracellular proteins (29).
Adherence is an initial stage of the pathogen invasion into the host cells and involves a number of ligands, such as oligosaccharides and protein adhesins. In S. pneumoniae, NanA, BgaA, and StrH act sequentially to remove sialic acid, galactose, and N-acetylglucosamine and expose mannose on human glycoproteins for binding by pneumococci, suggesting that S. pneumoniae deglycosylates host airway defense glycoproteins, thereby enhancing adherence of S. pneumoniae to airway components (18, 19, 33, 34). Interestingly, although adherence of nonencapsulated nanA and bgaA mutants to epithelial cells was decreased in vitro, in vitro results revealed no decrease in the colonization of bgaA nanA strH triple exoglycosidase mutants (19). The surface-anchored pullulanases of S. pneumoniae recognize and bind multivalently to host glycogen, thus increasing interaction with alveolar type II cells in mouse lung tissue (36). Moreover, sugar moieties such as lacto-N-neotetraose and asialoganglioside GM1 can contribute to the adherence of pneumococci to host cells (33). These results suggest that exoglycosidases can unmask receptors upon hydrolysis of targets.
It was reported that the disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells was Galβ1-3GlcNAc (1, 2). Adherence inhibition tests with neolactotetraose and lactotetraose indicated that pneumococci prefer binding to the lactotetraose structure (1). This lactotetraose structure is one of the major core structures of vertebrate glycosphingolipids, suggesting that BgaC can remove galactose from β1,3-linked GlcNAc in lacto-N-tetraose of the glycolipid that could serve as a possible binding site for S. pneumoniae infection. If that is the case, absence of BgaC protein caused by gene deletion or by antibody treatment would increase adherence rather than decrease adherence, as observed in Fig. 7 and 9.
Notably, we report for the first time that surface-associated exoglycosidase BgaC can hydrolyze and remove the host cell ligand so that binding of pneumococci to the host is decreased in vivo and in vitro. This appears to be analogous to the function of influenza virus sialidase, which causes release from host cells by cleaving sialic acid, the sugar residue important for binding to host receptor (8). On the other hand, the bgaC deletion mutant could adhere more efficiently than the wild-type by some other factors. For example, galactoside hydrolysis by BgaC might trigger mucin synthesis; this would inhibit binding of pneumococci to the epithelial cells since infection with bacteria induces mucin synthesis in epithelial cells as an antimicrobial response of the innate immune system to protect the host (10, 16, 22, 23, 24). More work is required for investigation of key factors that might play a role in BgaC-mediated adherence.
During the progression from colonization to invasive disease, adaptation to different environmental niches in the host is mediated by changes in the expression of key virulence factors. Modulation of gene expression for a few virulence factors in S. pneumoniae has been reported, although the exact mechanisms involved in S. pneumoniae are not well characterized (17, 39). Through microarray analysis, we also observed that the bgaC gene was induced 2.54-fold and slightly decreased to 0.91-fold in A549 human lung cells infected with wild-type D39 after 10 min and 2 h of infection, respectively (data not shown). This suggests that BgaC is immediately induced upon contact with the host cells, indicating the involvement of BgaC during host cell invasion.
The present study showed that BgaC could hydrolyze the host galactoside moiety and thus affect adherence to the host cells and viability in phagocytes. The results indicate that BgaC is localized on the outer surface to hydrolyze β-galactosides on the surfaces of host cells and subsequently to remove ligands responsible for pneumococcal binding to the host cells. The underlying mechanism of BgaC expression and its role in pathogenesis should be investigated. This study will provide further insight into one of the diverse microbial strategies employed during pathogenesis.
Acknowledgments
This work was supported by grants from the Next Generation New Technology Development Program of the Korean Ministry of Knowledge and Economy (to H. A. Kang) and from the Korean Center for Disease Control (to D.-K. Rhee). This research was also partially supported by the Chung-Ang University Excellent Researcher Grant in 2008.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Andersson, B., J. Dahmen, T. Frejd, H. Leffler, G. Magnusson, G. Noori, and C. S. Eden. 1983. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J. Exp. Med. 158559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, B., B. Gray, and C. S. Eden. 1988. Role of attachment for the virulence of Streptococcus pneumoniae. Acta Otolaryngol. Suppl. 454163-166. [DOI] [PubMed] [Google Scholar]
- 3.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobrov, A. G., and R. D. Perry. 2006. Yersinia pestis lacZ expresses a beta-galactosidase with low enzymatic activity. FEMS Microbiol. Lett. 25543-51. [DOI] [PubMed] [Google Scholar]
- 5.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cámara, M., G. J. Boulnois, P. W. Andrew, and T. J. Mitchell. 1994. A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infect. Immun. 623688-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhatwal, G. S. 2002. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 10205-208. [DOI] [PubMed] [Google Scholar]
- 8.Colman, P. M. 1999. A novel approach to antiviral therapy for influenza. J. Antimicrob. Chemother. 44(Suppl. B)17-22. [DOI] [PubMed] [Google Scholar]
- 9.Conzelmann, E., and K. Sandhoff. 1987. Glycolipid and glycoprotein degradation. Adv. Enzymol. Relat. Areas Mol. Biol. 6089-216. [DOI] [PubMed] [Google Scholar]
- 10.Dohrman, A., S. Miyata, M. Gallup, J. D. Li, C. Chapelin, A. Coste, E. Escudier, J. Nadel, and C. Basbaum. 1998. Mucin (MUC 2 and MUC 5 AC) transcriptional regulation in response to Gram-positive and -negative bacteria. Biochim. Biophys. Acta 1406251-259. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto, H., M. Miyasato, Y. Ito, T. Sasaki, and K. Ajisaka. 1998. Purification and properties of recombinant beta-galactosidase from Bacillus circulans. Glycoconj. J. 15155-160. [DOI] [PubMed] [Google Scholar]
- 12.Henrissat, B., and G. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7637-644. [DOI] [PubMed] [Google Scholar]
- 13.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber, R. E., C. Parfett, H. Woulfe-Flanagan, and D. J. Thompson. 1979. Interaction of divalent cations with beta-galactosidase (Escherichia coli). Biochemistry 184090-4095. [DOI] [PubMed] [Google Scholar]
- 15.Ito, Y., and T. Sasaki. 1997. Cloning and characterization of the gene encoding a novel beta-galactosidase from Bacillus circulans. Biosci. Biotechnol. Biochem. 611270-1276. [DOI] [PubMed] [Google Scholar]
- 16.Kadioglu, A., J. N. Weiser, J. C. Paton, and P. W. Andrew. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6288-301. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, G. E., and J. Yother. 2007. CcpA-dependent and -independent control of beta-galactosidase expression in Streptococcus pneumoniae occurs via regulation of an upstream phosphotransferase system-encoding operon. J. Bacteriol. 1895183-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King, S. J., K. R. Hippe, J. M. Gould, D. Bae, S. Peterson, R. Cline, C. Fasching, E. N. Janoff, and J. N. Weiser. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol. Microbiol. 54159-171. [DOI] [PubMed] [Google Scholar]
- 19.King, S. J., K. R. Hippe, and J. N. Weiser. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59961-974. [DOI] [PubMed] [Google Scholar]
- 20.Kwon, H. Y., S. W. Kim, M. H. Choi, A. D. Ogunniyi, J. C. Paton, S. H. Park, S. N. Pyo, and D. K. Rhee. 2003. Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae. Infect. Immun. 713757-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D 39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 18938-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemjabbar, H., and C. Basbaum. 2002. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat. Med. 841-46. [DOI] [PubMed] [Google Scholar]
- 23.Li, J. D., A. F. Dohrman, M. Gallup, S. Miyata, J. R. Gum, Y. S. Kim, J. A. Nadel, A. Prince, and C. B. Basbaum. 1997. Transcriptional activation of mucin by Pseudomonas aeruginosa lipopolysaccharide in the pathogenesis of cystic fibrosis lung disease. Proc. Natl. Acad. Sci. USA 94967-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J. D., W. Feng, M. Gallup, J. H. Kim, J. Gum, Y. Kim, and C. Basbaum. 1998. Activation of NFkB via a Src-dependent Ras-MAPK-pp90rsk pathway is required for P. aeruginosa-induced mucin overproduction in epithelial cells. Proc. Natl. Acad. Sci. USA 955718-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrill, A. H., Jr., M. D. Wang, M. Park, and M. C. Sullards. 2007. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 32457-468. [DOI] [PubMed] [Google Scholar]
- 26.Poch, O., H. L'Hote, V. Dallery, F. Debeaux, R. Fleer, and R. Sodoyer. 1992. Sequence of the Kluyveromyces lactis beta-galactosidase: comparison with prokaryotic enzymes and secondary structure analysis. Gene 11855-63. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, B. F., R. M. Adams, C. Requadt, S. Power, and S. E. Mainzer. 1989. Expression and nucleotide sequence of the Lactobacillus bulgaricus beta-galactosidase gene cloned in Escherichia coli. J. Bacteriol. 171625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slack, F. J., and G. Ruvkun. 1998. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem. Sci. 23474-475. [DOI] [PubMed] [Google Scholar]
- 29.Springer, T. A. 1998. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 283837-862. [DOI] [PubMed] [Google Scholar]
- 30.Talbot, U. M., A. W. Paton, and J. C. Paton. 1996. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. Infect. Immun. 643772-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taron, C. H., J. S. Benner, L. J. Hornstra, and E. P. Guthrie. 1995. A novel beta-galactosidase gene isolated from the bacterium Xanthomonas manihotis exhibits strong homology to several eukaryotic beta-galactosidases. Glycobiology 5603-610. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 33.Tong, H. H., M. A. McIver, L. M. Fisher, and T. F. DeMaria. 1999. Effect of lacto-N-neotetraose, asialoganglioside-GM1 and neuraminidase on adherence of otitis media-associated serotypes of Streptococcus pneumoniae to chinchilla tracheal epithelium. Microb. Pathog. 26111-119. [DOI] [PubMed] [Google Scholar]
- 34.Tong, H. H., X. Liu, Y. Chen, M. James, and T. Demaria. 2002. Effect of neuraminidase on receptor-mediated adherence of Streptococcus pneumoniae to chinchilla tracheal epithelium. Acta Otolaryngol. 122413-419. [DOI] [PubMed] [Google Scholar]
- 35.Tu, L. N., H. Y. Jeong, H. Y. Kwon, A. D. Ogunniyi, J. C. Paton, S. N. Pyo, and D. K. Rhee. 2007. Modulation of adherence, invasion, and tumor necrosis factor alpha secretion during the early stages of infection by Streptococcus pneumoniae ClpL. Infect. Immun. 752996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Bueren, A. L., M. Higgins, D. Wang, R. D. Burke, and A. B. Boraston. 2007. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat. Struct. Mol. Biol. 1476-84. [DOI] [PubMed] [Google Scholar]
- 37.Vasseghi, H., and J. P. Claverys. 1983. Amplification of a chimeric plasmid carrying an erythromycin-resistance determinant introduced into the genome of Streptococcus pneumoniae. Gene 21285-292. [DOI] [PubMed] [Google Scholar]
- 38.Vijayakumar, M. N., and D. A. Morrison. 1986. Localization of competence-induced proteins in Streptococcus pneumoniae. J. Bacteriol. 165689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiser, J. N., D. Bae, H. Epino, S. B. Gordon, M. Kapoor, L. A. Zenewicz, and M. Shchepetov. 2001. Changes in availability of oxygen accentuate differences in capsular polysaccharide expression by phenotypic variants and clinical isolates of Streptococcus pneumoniae. Infect. Immun. 695430-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong-Madden, S. T., and D. Landry. 1995. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology 519-28. [DOI] [PubMed] [Google Scholar]
- 41.Zähner, D., and R. Hakenbeck. 2000. The Streptococcus pneumoniae beta-galactosidase is a surface protein. J. Bacteriol. 1825919-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]