Abstract
Heat shock response in Corynebacterium glutamicum was characterized by whole-genome expression analysis using a DNA microarray. It was indicated that heat shock response of C. glutamicum included not only upregulation of heat shock protein (HSP) genes encoding molecular chaperones and ATP-dependent proteases, but it also increased and decreased expression of more than 300 genes related to disparate physiological functions. An extracytoplasmic-function sigma factor, SigH, was upregulated by heat shock. The SigH regulon was defined by gene expression profiling using sigH-disrupted and overexpressing strains in conjunction with mapping of transcription initiation sites. A total of 45 genes, including HSP genes and genes involved in oxidative stress response, were identified as the SigH regulon. Expression of some HSP genes was also upregulated by deletion of the transcriptional regulators HspR and HrcA. HspR represses expression of the clpB and dnaK operons, and HrcA represses expression of groESL1 and groEL2. SigH was shown to play an important role in regulation of heat shock response in concert with HspR and HrcA, but its role is likely restricted to only a part of the regulation of C. glutamicum heat shock response. Upregulation of 18 genes encoding transcriptional regulators by heat shock suggests a complex regulatory network of heat shock response in C. glutamicum.
Corynebacterium glutamicum is a nonpathogenic, highly G+C gram-positive bacterium that belongs to the order Actinomycetales, which includes the genera Mycobacterium and Streptomyces. C. glutamicum has been widely used for the industrial production of various amino acids and nucleic acids (19, 20, 47). We previously demonstrated that growth-arrested C. glutamicum cells under conditions of oxygen deprivation exhibit high productivity of ethanol, d-lactic acid and succinic acid (22, 36, 37). Meanwhile, this species is of increasing interest as a model organism for closely related pathogenic species, such as Corynebacterium diphtheriae, Mycobacterium tuberculosis, and Mycobacterium leprae (4, 32, 33).
The heat shock response is the most frequently used model system for studying the impact of stress on biological systems. All organisms respond to a sudden increase in temperature with elevated expression of a set of highly conserved proteins, termed heat shock proteins (HSPs), which include molecular chaperones that assist folding nascent proteins and repairing damaged proteins and ATP-dependent proteases that degrade misfolded proteins (9, 49). Genes with various physiological functions in addition to HSPs have been identified as being part of a heat shock stimulon by whole-genome expression analysis using DNA microarrays (6, 18, 41).
The heat shock response is controlled by a combination of alternative sigma factors that recognize specific heat shock promoters upstream of heat shock genes and transcriptional regulators that act as the repressors or activators by binding to the promoter regions of certain heat shock genes (48, 52). In Mycobacterium tuberculosis, the extracytoplasmic-function (ECF) sigma factor SigH plays a central role in a regulatory network of heat and oxidative stress responses (31, 39). Two transcriptional repressors, HrcA and HspR, are responsible for control of heat shock genes of M. tuberculosis in association with DNA elements called CIRCE (for controlling inverted repeat of chaperone expression) and HAIR (for HspR-associated inverted repeat), respectively (45). In Streptomyces albus G, another transcriptional repressor, RheA, in addition to HrcA and HspR, is involved in heat shock regulation (15, 16, 43).
In C. glutamicum, expression of some HSPs has been shown to be upregulated by heat shock (1, 2, 12). Detailed analysis of transcriptional regulation of the clpP1-clpP2 operon and the clpC gene, each encoding a subunit of ATP-dependent protease, revealed involvement of the ECF sigma factor SigH of C. glutamicum in their expression (12). In addition, sequence analyses of promoter regions of groES, groEL2, and dnaK allow the identification of CIRCE and HAIR elements close to the −10 and −35 regions of the promoters, suggesting involvement of HrcA and HspR in regulation of heat shock response in C. glutamicum (2). Although our knowledge of C. glutamicum heat shock response has increased during the past decade, transcriptional responses to heat shock and its regulatory systems have not been analyzed at the whole-genome level.
In the present study, we defined the heat shock stimulon of C. glutamicum using DNA microarray. Expression of about 10% of the genes on the chromosome related to disparate cellular functions was shown to respond to heat shock. We elucidated the regulatory roles of SigH, HrcA, and HspR in response to heat shock by combining whole-genome expression analyses with targeted mutagenesis. In identifying the 45 genes that comprise the SigH regulon, we showed that in addition to ClpP1, ClpP2, and ClpC, expression of most HSPs, including DnaK, GrpE, DnaJ, ClpB, DnaJ2, GroES, GroEL1, and GroEL2, was regulated by SigH in concert with HrcA and HspR. Finally, we identified 18 genes encoding transcriptional regulators that were upregulated by heat shock, suggesting complex regulatory systems of C. glutamicum heat shock response.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
C. glutamicum strain R (51) and its derivatives were grown at 33°C in medium A with 4% (wt/vol) glucose as described previously (11). C. glutamicum cells were grown at 33°C until they reached an optical density at 610 nm of 2 to 3. The culture temperature was raised to 45°C in a water bath, and then cells were incubated on a rotary shaker at 45°C. The culture was mixed with an equal amount of the RNAprotect bacteria reagent (Qiagen, Hilden, Germany) 15 min after heat shock and settled at room temperature for 10 min. The cells were harvested by centrifugation and stored at −80°C until they were used to prepare RNA.
Mutant construction.
sigH, sigM, and hrcA disruptants were constructed by the transposon-mediated mutagenesis method as described previously (46). Transposon insertion locations were identified based on the sequences of thermal asymmetric interlaced-PCR products of mutant cells. In each disruptant, a transposon was inserted at 426, 162, and 476 bases downstream of the 5′ end of the genes sigH, sigM, and hrcA, respectively. Construction of an hspR disruptant was carried out as follows. The hspR coding region was amplified by PCR using the primer pair 2687-F and 2687-R (see Table S1 in the supplemental material) and cloned between the EcoRI and PstI sites of pHSG398 (Takara Bio, Shiga, Japan). A kanamycin resistance cassette was inserted into the unique EcoO65I site that lies at 144 bases downstream of the 5′ end of the hspR gene. The resultant plasmid was transferred by electroporation into C. glutamicum, and a strain resistant to kanamycin and sensitive to chloramphenicol was selected. Disruption of the hspR gene was confirmed by PCR. Construction of sigH-overexpressing shuttle vector pCRD601 was carried out as follows. A DNA fragment containing the sigH promoter and coding regions was amplified by PCR using the primer pair sigH-F and sigH-R (see Table S1 in the supplemental material) and was cloned between the PstI and EcoRI sites of pCRB1 (23). pCRD601 was transferred by electroporation into C. glutamicum.
RNA isolation and DNA microarray analysis.
Total RNA was extracted from C. glutamicum cells by using the RNeasy mini kit (Qiagen) and treated with DNase I (Takara Bio) as described previously (11). Global gene expression analysis was performed with the C. glutamicum R DNA microarray as described previously (11). Microarray analyses were carried out using two sets of RNA samples isolated from independently grown cultures with different combinations of Cy dyes (a dye swap strategy). Since the C. glutamicum R DNA microarray contains two replicates per gene, a total of four replicates per gene were available to determine changes in gene expression. Genes with significantly differential transcript levels (P < 0.05; Student's t test) by at least a factor of two were determined.
Mapping of transcription initiation sites by RACE-PCR.
Transcription initiation sites were determined by using the SMART RACE cDNA amplification kit (Clontech, CA) with RNA prepared from cells subjected to heat shock. 5′ Rapid amplification of cDNA ends (RACE)-PCR analyses were carried out as recommended by the supplier with 1 μg of total RNA and gene-specific primers (see Table S1 in the supplemental material). Resulting PCR products were cloned into a pGEM-T Easy vector by using the pGEM-T Easy vector system (Promega Corporation, WI). At least 10 clones for each 5′ RACE-PCR product were sequenced by the dye terminator method using the BigDye Terminator v3.1 cycle sequencing kit and the 3130xl Genetic Analyzer (Applied BioSystems, CA).
Real-time qRT-PCR.
A one-step real-time quantitative reverse transcription-PCR (qRT-PCR) analysis was performed with the Power SYBR green PCR master mix (Applied Biosystems) and a pair of gene-specific forward and reverse primers (see Table S1 in the supplemental material) by using the 7500 Fast real-time PCR system (Applied Biosystems) as described previously (11). Relative ratios were normalized with the value for 16S rRNA and are represented as means of triplicate measurements.
A two-step real-time qRT-PCR was performed to determine the transcript levels of cgR_2620 and cgR_2674. cDNA was synthesized from total RNA with reverse primers, RT16S-R, RT2620-R, and RT2674-R, as follows. An aliquot (1 μg) of total RNA and 1 pmol each of reverse primers were denatured at 90°C for 5 min and then gradually cooled to 55°C. The RT reaction was performed in 20 μl of first-strand buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2) containing total RNA, reverse primers, 5 mM dithiothreitol, 0.5 mM each of dATP, dGTP, dCTP, and dTTP, and 200 U of SuperScript III RNase H− reverse transcriptase (Invitrogen, CA). The reaction mixture was incubated at 55°C for 1 h, and then the reaction was stopped by the addition of 80 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). An aliquot (1 μl) of the cDNA solution was taken for real-time qPCR analysis. For the experiment with 16S rRNA, the cDNA solution was further diluted 10-fold.
RESULTS
Global changes in gene expression in response to heat shock.
There are 13 genes encoding HSPs in the genome of C. glutamicum. Present are a single gene each for groES (cgR_0715), grpE (cgR_2689), clpX (cgR_2269), clpB (cgR_2676), and clpC (cgR_2580) and two paralogues each for the genes groEL, dnaK, clpP, and dnaJ, designated groEL1 (cgR_0716) and groEL2 (cgR_2619), dnaK (cgR_2690) and dnaK2 (cgR_2260), clpP1 (cgR_2308) and clpP2 (cgR_2307), and dnaJ (cgR_2688) and dnaJ2 (cgR_2162), respectively. Some of these genes constitute operons, such as groES-groEL1, dnaK-grpE-dnaJ, and clpP1-clpP2. The genes hspR and hrcA encoding transcriptional regulators are located next to the dnaK-grpE-dnaJ operon and the dnaJ2 gene, respectively.
A gene expression profile of C. glutamicum cells subjected to heat shock (45°C) for 15 min was compared to that of cells grown at 33°C. The transcript levels of 157 genes were increased, while those of another 147 genes were decreased at least twofold by heat shock (see Tables S2 and S3 in the supplemental material). In addition to genes such as clpC, clpP1, clpP2, groES, groEL1, groEL2, and the dnaK operon that have been shown to be heat inducible in previous studies (2, 12), most of the HSP genes except dnaK2 and clpX were upregulated by heat shock. Genes such as msrA, mtr, trxB, msrB, and trxC, involved in oxidative stress response, as well as the cys genes that are involved in sulfate reduction and the suf gene cluster that is involved in iron-sulfur cluster formation were also upregulated. Elevated expression of the genes sigB and sigH encoding RNA polymerase sigma factors indicates that these sigma factors should take part in regulation of gene expression in response to heat shock.
The downregulated genes included genes of glycolysis (ptsG, gapA, glgP1, fba, eno, pyk, pfkA, and ptsF), the tricarboxylic acid cycle (sdhCAB, acn, lpd, fum, sucB, gltA, icd, and aceE), and the pentose phosphate pathway (gnd). In addition, the transcript levels of genes involved in transcription and translation (rpl and rps genes, infC, tig, fusA, rpoA, nusB, and efp) and respiration (qcrBAC, ctaEFC, ctaD, narKG, and ccsA) and atp genes encoding F1F0-ATP synthase were decreased. Cell growth was arrested after heat shock (data not shown), resulting in downregulation of these genes.
Regulatory roles of SigH in expression of heat shock-responsive genes.
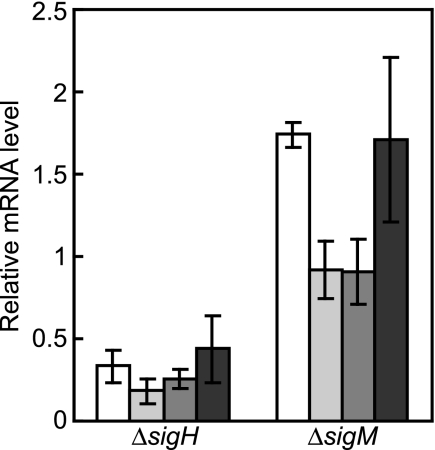
The ECF sigma factor SigH has been shown to control expression of clpP1-clpP2 and clpC in response to heat shock (12). Global regulation of the heat shock response by SigH was elucidated using a DNA microarray. Gene expression profiles of cells subjected to heat shock were compared between the wild type and the sigH disruptant. The transcript levels of 19 genes upregulated by heat shock were decreased by sigH disruption (Table 1). In addition to clpP1-clpP2 and clpC, expression of the clpB gene and the dnaK operon is downregulated in the sigH disruptant. Transcription from the promoters of clpB, dnaK, trxB, and sufR has been shown to depend on the other ECF sigma factor, SigM (34). In the sigM disruptant, the transcript levels of these genes at 45°C were comparable with or higher than those in the wild type, while they were significantly reduced in the sigH disruptant (Fig. 1). Moreover, DNA microarray analyses revealed that sigM disruption did not affect the transcript levels of heat-inducible genes (data not shown). These results indicate that SigM is not involved in regulation of gene expression in response to heat shock. Genes regulated by SigH were further identified using the sigH-overexpressing strain, in which the sigH gene was expressed from a multicopy plasmid, pCRD601. Global changes in gene expression during exponential growth by sigH overexpression were examined by a comparison with the strain harboring the control plasmid pCRB1. The transcript levels of 34 genes upregulated by heat shock were increased by the sigH overexpression even at 33°C (Table 1). A total of 37 genes that were downregulated in the sigH disruptant and/or upregulated in the sigH-overexpressing strain were considered to be under the control of SigH.
TABLE 1.
Heat shock-inducible genes affected by sigH disruption or sigH overexpressionf
| Genec | Product | ΔsigHa | Pb | SigHoxa | Pb | Heat shockd | Pb |
|---|---|---|---|---|---|---|---|
| Downregulated in the sigH disruptant and upregulated in the sigH-overexpressing strain | |||||||
| cgR_0627 | Hypothetical protein | −1.34 | 3.9E−02 | 2.29 | 3.2E−04 | 1.48 | 1.6E−05 |
| cgR_1297 | Thioredoxin domain-containing protein | −1.04 | 5.4E−03 | 1.29 | 2.9E−03 | 1.02 | 4.6E−04 |
| cgR_1317*e | tRNA-methyltransferase | −1.35 | 2.1E−02 | 1.52 | 4.9E−02 | 2.64 | 1.6E−02 |
| cgR_1318* | Hypothetical protein | −0.66 | 3.4E−03 | 1.29 | 1.3E−02 | 1.21 | 1.3E−02 |
| cgR_1554 | Hypothetical protein | −1.37 | 2.6E−03 | 1.21 | 2.6E−03 | 1.23 | 1.2E−03 |
| cgR_1727 (msrB) | Peptide methionine sulfoxide reductase | −2.78 | 1.1E−03 | 2.43 | 9.1E−04 | 2.08 | 1.5E−03 |
| cgR_2078 | Hypothetical protein | −1.91 | 5.4E−03 | 2.30 | 6.3E−03 | 1.22 | 4.7E−02 |
| cgR_2183 | Alkanal monooxygenase α chain | −3.05 | 1.4E−04 | 3.47 | 2.2E−05 | 2.72 | 4.7E−03 |
| cgR_2308 (clpP1)†e | ATP-dependent Clp protease, proteolytic subunit | −1.78 | 4.9E−02 | 1.85 | 3.0E−03 | 1.32 | 3.3E−03 |
| cgR_2307 (clpP2)† | ATP-dependent Clp protease, proteolytic subunit | −1.55 | 4.3E−02 | 1.76 | 8.6E−04 | 1.22 | 1.1E−03 |
| cgR_2320 | Putative dithiol-disulfide isomerase | −1.41 | 2.6E−03 | 2.29 | 1.7E−04 | 1.11 | 1.9E−03 |
| cgR_2580 (clpC) | ATP-dependent Clp protease, ATP-binding subunit | −1.46 | 2.1E−02 | 1.13 | 1.2E−04 | 2.25 | 4.9E−03 |
| cgR_2676 (clpB)‡e | Molecular chaperone | −1.78 | 3.3E−02 | 1.82 | 2.6E−04 | 4.92 | 5.8E−04 |
| cgR_2675‡ | Hypothetical protein | −1.79 | 3.4E−02 | 1.49 | 1.4E−04 | 4.16 | 2.8E−03 |
| cgR_2690 (dnaK)§e | Molecular chaperone | −1.29 | 1.2E−01 | 1.57 | 7.6E−03 | 4.20 | 4.0E−06 |
| cgR_2689 (grpE)§ | Molecular chaperone | −1.17 | 6.9E−02 | 1.23 | 7.2E−03 | 4.29 | 3.2E−06 |
| cgR_2688 (dnaJ)§ | Molecular chaperone | −1.15 | 7.7E−03 | 0.95 | 2.8E−02 | 4.20 | 2.6E−05 |
| cgR_2687 (hspR)§ | Transcriptional regulator, MerR family | −0.87 | 2.7E−02 | 0.35 | 9.3E−02 | 4.33 | 1.6E−05 |
| cgR_2964 | Putative oxidoreductase/dehydrogenase | −1.80 | 6.1E−03 | 3.33 | 1.7E−04 | 1.66 | 4.3E−02 |
| cgR_2980 (trxB)¶e | Thioredoxin reductase | −1.76 | 2.2E−02 | 2.88 | 1.0E−03 | 2.30 | 2.1E−03 |
| cgR_2981 (trxC)¶ | Thioredoxin | −1.26 | 2.2E−03 | 1.59 | 1.3E−02 | 1.79 | 5.6E−05 |
| cgR_2982 (cwlM)¶ | N-Acetylmuramoyl-l-alanine amidase | −0.95 | 1.8E−02 | 1.33 | 2.2E−02 | 1.74 | 3.0E−04 |
| Downregulated in the sigH disruptant | |||||||
| cgR_1616 (sufR)‖e | Transcriptional regulator, ArsR family | NA | 0.77 | 3.9E−02 | 2.34 | 3.6E−02 | |
| cgR_1615 (sufB)‖ | Fe-S cluster assembly protein | −1.62 | 5.3E−02 | 0.75 | 1.5E−01 | 2.52 | 2.1E−02 |
| cgR_1614 (sufD)‖ | Fe-S cluster assembly protein | −1.21 | 3.0E−02 | 0.65 | 1.4E−01 | 2.06 | 1.7E−02 |
| cgR_1613 (sufC)‖ | Fe-S cluster assembly ATPase | −1.15 | 2.4E−02 | 0.78 | 1.1E−01 | 1.94 | 2.3E−02 |
| cgR_1612 (sufS)‖ | Cysteine desulfurase | −0.78 | 1.9E−02 | 0.64 | 1.1E−01 | 1.47 | 4.5E−02 |
| cgR_1611 (sufU)‖ | Fe-S cluster assembly protein | −0.77 | 1.2E−01 | 0.63 | 2.7E−01 | 1.57 | 4.6E−03 |
| cgR_1610‖ | Hypothetical protein | NA | 0.61 | 2.0E−01 | 1.75 | 1.2E−02 | |
| Upregulated in the sigH-overexpressing strain | |||||||
| cgR_0488 (hemA)∧e | Glutamyl-tRNA reductase | −0.17 | 6.4E−01 | 1.15 | 1.0E−03 | 2.06 | 9.1E−06 |
| cgR_0489 (hemC)∧ | Porphobilinogen deaminase | −0.20 | 4.4E−01 | 0.74 | 1.3E−04 | 1.09 | 3.2E−03 |
| cgR_0715 (groES)◊e | Molecular chaperone | 0.00 | 9.9E−01 | 1.14 | 4.8E−04 | 1.89 | 9.7E−03 |
| cgR_0716 (groEL1)◊ | Molecular chaperone | −0.19 | 2.6E−01 | 1.32 | 7.2E−03 | 1.92 | 1.6E−03 |
| cgR_1749 (sigB) | RNA polymerase sigma factor | −0.53 | 2.6E−01 | 1.00 | 4.6E−03 | 1.34 | 3.7E−02 |
| cgR_1753 | Hypothetical protein | NA | 3.66 | 2.7E−05 | 3.50 | 4.4E−03 | |
| cgR_1832 (mtr) | Mycothione reductase | NA | 3.31 | 8.4E−03 | 2.66 | 1.3E−03 | |
| cgR_1848 | rRNA large subunit methyltransferase | −1.06 | 6.0E−02 | 1.29 | 2.0E−04 | 1.19 | 4.0E−03 |
| cgR_2162 (dnaJ2) | Molecular chaperone | −0.67 | 1.7E−01 | 1.69 | 1.5E−03 | 1.19 | 5.3E−04 |
| cgR_2417 | Glutamate racemase | NA | 1.75 | 5.7E−03 | 1.86 | 2.3E−02 | |
| cgR_2451 | Na+/H+-dicarboxylate symporter | −1.15 | 1.7E−01 | 2.65 | 6.7E−03 | 4.02 | 1.8E−03 |
| cgR_2470 | Dithiol-disulfide isomerase | NA | 2.66 | 4.3E−04 | 1.45 | 5.0E−03 | |
| cgR_2619 (groEL2) | Molecular chaperone | NA | 1.13 | 9.3E−04 | 3.09 | 1.2E−04 | |
| cgR_2830 (msrA)⧫e | Peptide methionine sulfoxide reductase | NA | 2.07 | 1.2E−03 | 3.39 | 1.2E−03 | |
| cgR_2829⧫ | Amidohydrolase | NA | NA | 1.89 | 1.1E−03 | ||
| cgR_2903 | NADH dehydrogenase/NAD(P)H nitroreductase | −0.35 | 8.1E−02 | 1.45 | 4.5E−04 | 1.96 | 1.3E−02 |
Relative ratios of the transcript levels in the sigH disruptant to the wild type 15 min after heat shock (ΔsigH) and those in the sigH-overexpressing strain to the wild type at 33°C (SigHox) determined by DNA microarray analyses are shown in the base 2 logorithm.
P values for the relative ratio were determined by Student's t test.
Genes that constitute a putative operon are indicated with the same character.
Relative ratios of transcript levels 15 min after heat shock to 33°C in the wild type.
First gene of the operon.
Data determined to be significantly changed using the criteria described in Materials and Methods are bold. NA, data not available.
FIG. 1.
Transcript levels of heat-inducible genes in the sigH and sigM disruptants. The transcript levels of the genes clpB (open bars), dnaK (light gray bars), trxB (gray bars), and sufR (dark gray bars) 15 min after heat shock in the sigH (left) and sigM (right) disruptants were compared to those in the wild type by qRT-PCR. Experiments were repeated three times. The transcript levels of the wild type were taken as 1.
The sigB gene was also upregulated by heat shock (see Table S2 in the supplemental material). Gene expression profiles of cells subjected to heat shock were compared between the wild type and the sigB disruptant. However, disruption of the sigB gene did not affect the transcript levels of heat-inducible genes, while genes that have been previously shown to be under the control of SigB (11) were downregulated in the sigB disruptant (data not shown). SigB is not involved in regulation of the heat shock response.
Determination of a consensus sequence of SigH-dependent promoters.
To determine the SigH-dependent promoter sequence, the operon structures of the genes under the control of SigH were analyzed. Analysis of the genetic organization of the respective genomic regions as well as searches for putative rho-independent transcription terminators was done using the TransTerm program (13), and nine putative operons were discovered among the candidate genes (Table 1). Taking into account these putative operon structures, 45 genes that constitute 29 transcriptional units were regarded as the SigH regulon (Table 1).
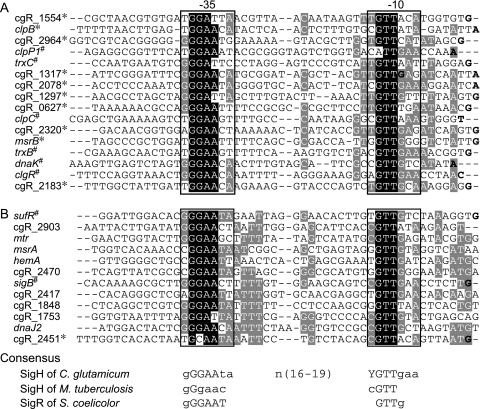
The sequence of a SigH-dependent promoter was previously estimated from promoter sequences of three genes, clpP1, clpC, and clgR (12). We tested whether promoter sequences of the newly identified members of the SigH regulon were comparable with the previously defined one by mapping the transcription initiation sites (TISs) using RACE-PCR. TISs of 10 genes that were downregulated by sigH disruption and also upregulated by sigH overexpression (see genes listed in Table 1) were determined, and promoter sequences were aligned with previously identified promoters (2, 12, 34) (Fig. 2A). Highly conserved −10 and −35 regions were observed within promoter regions of all the genes. Furthermore, these conserved sequences were analyzed for the presence of the upstream regions of genes downregulated by sigH disruption or upregulated by sigH overexpression (see genes listed in Table 1). Putative promoter sequences with the conserved −10 and −35 regions were found within the upstream regions of most genes, except for groES-groEL1 and groEL2 (Fig. 2B). A TIS of cgR_2451 was determined by RACE-PCR, confirming transcription from the predicted promoter (Fig. 2B). The consensus sequences of the SigH-dependent promoter were defined to be YGTTGAA for the −10 region and GGGAATA for the −35 region.
FIG. 2.
Promoter sequences of SigH-regulated genes. (A) Promoter sequences of the genes downregulated by sigH disruption and also upregulated by sigH overexpression. The sequences of 50 nucleotides upstream of the TISs that were determined in this study by RACE-PCR (*) and in the previous studies (#) are shown (2, 12, 34). Two TISs for cgR_1317 were detected. (B) Putative promoter sequences of the genes downregulated by sigH disruption or upregulated by sigH overexpression. A TIS of cgR_2451 was determined by RACE-PCR, and TISs of sufR and sigB have been determined previously (34, 35). The TISs are bold. The nucleotides conserved among more than 80% and 50% of the sequences are shaded in black and gray, respectively. The consensus sequences of SigH-dependent promoters were determined by the BioProspector program (30). The −10 and −35 regions are indicated by open boxes, and the consensus sequences are shown at the bottom with those recognized by SigH of M. tuberculosis and SigR of S. coelicolor. Nucleotides of the consensus sequence conserved in more than 80% of the sequences are in capital letters. Y indicates T or C.
Identification of the HspR and HrcA regulons.
The hspR gene constitutes an operon with dnaK-grpE-dnaJ, which was induced by heat shock (Table 1). The hrcA gene is located immediately upstream of the dnaJ2 gene, but it was not upregulated by heat shock. The regulons of HspR and HrcA were determined by comparing gene expression profiles at 33°C between the wild type and gene disruptants using a DNA microarray. Expression of the dnaK and clpB operons was upregulated by hspR disruption (Table 2). The HAIR elements are present in the dnaK and clpB promoter regions (Fig. 3A), indicating that these two operons are directly repressed by HspR. cgR_2674 was identified as belonging to an upregulated gene in the hspR disruptant by DNA microarray analysis, but qRT-PCR analysis showed that its transcript level was not affected by hspR disruption (data not shown).
TABLE 2.
Genes under the control of HspR and HrcA
| Gened | Product | Ratioa | Pb |
|---|---|---|---|
| Differentially expressed between the wild type and the hspR disruptant | |||
| cgR_2674c | Major facilitator superfamily permease | 1.30 | 3.5E−05 |
| cgR_2676 (clpB)*e | Molecular chaperone | 2.92 | 4.0E−05 |
| cgR_2675* | Hypothetical protein | 2.65 | 2.5E−05 |
| cgR_2690 (dnaK)†e | Molecular chaperone | 3.25 | 2.1E−04 |
| cgR_2689 (grpE)† | Molecular chaperone | 2.90 | 1.4E−03 |
| cgR_2688 (dnaJ)† | Molecular chaperone | 2.24 | 2.2E−04 |
| Differentially expressed between the wild type and the hrcA disruptant | |||
| cgR_0715 (groES)‡e | Molecular chaperone | 2.70 | 1.9E−06 |
| cgR_0716 (groEL1)‡ | Molecular chaperone | 2.90 | 2.3E−05 |
| cgR_1436 | NADPH-dependent quinone oxidoreductase | 2.11 | 3.0E−03 |
| cgR_2619 (groEL2) | Molecular chaperone | 3.13 | 3.3E−06 |
| cgR_2620c | Hypothetical protein | 1.73 | 2.8E−04 |
| cgR_2888 (dps) | Starvation-induced DNA-protecting protein | −1.49 | 2.5E−03 |
Relative ratios of the transcript levels in the hspR and hrcA disruptants to the wild type at 33°C determined by DNA microarray analyses are shown in the base 2 logarithm.
P values for the relative ratio were determined by Student's t test.
qRT-PCR analyses showed that the transcript levels of cgR_2674 and cgR_2620 were not affected by hspR and hrcA disruption, respectively.
Genes that constitute a putative operon are indicated with the same character.
First gene of the operon.
FIG. 3.
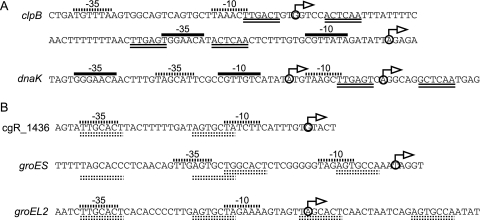
Regulatory regions of genes under the control of HspR (A) and HrcA (B). Transcription initiation sites are indicated with bent arrows. Solid and dotted lines over the sequences correspond to the SigA- and SigH-dependent promoters, respectively. Double solid and dotted lines under the sequences correspond to the HAIR and CIRCE elements, respectively.
In the hrcA disruptant, the transcript levels of groES, groEL1, cgR_1436, and groEL2 were increased, while the dps transcript level was decreased (Table 2). In the promoter regions of groES-groEL1 and groEL2, two CIRCE elements exist (Fig. 3B). A TIS of cgR_1436 was mapped by using RACE-PCR experiments (Fig. 3B). The promoter sequence of cgR_1436 was similar to that of the SigA-dependent promoter, TA(T/C)NNT for the −10 region and TTGACA for the −35 region (11), and a CIRCE element was found within its promoter region. These results indicate that HrcA acts as a transcriptional repressor for groES-groEL1, cgR_1436, and groEL2. No CIRCE element was found upstream of the dps-coding region. The dps gene was considered to be subject of an indirect regulation provoked by hrcA disruption. By qRT-PCR, cgR_2620 was indicated not to be regulated by HrcA.
Dual control of expression of clpB and dnaK by SigH and HspR.
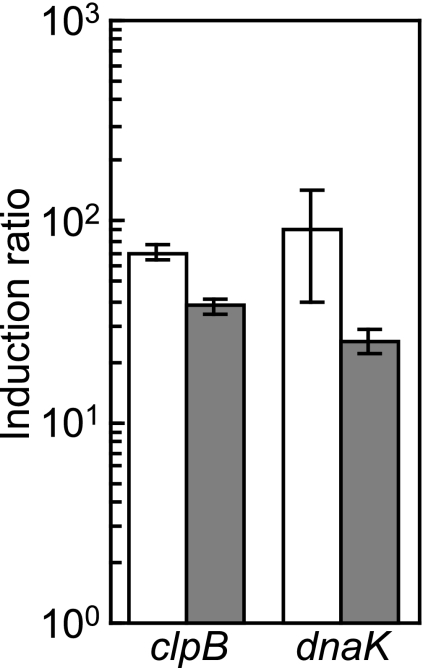
Expression of the clpB and dnaK operons was positively regulated by SigH (Table 1) and negatively regulated by HspR (Table 2). Transcriptional regulation of these genes was further investigated. The transcript levels of clpB and dnaK after heat shock in the wild type and the sigH disruptant were determined by qRT-PCR (Fig. 4). Both genes were induced by heat shock, even in the sigH disruptant, while the induction levels were reduced compared to that of the wild type. By using RACE-PCR experiments using RNA prepared from cells subjected to heat shock, two TISs were detected for clpB (Fig. 3A). The promoter proximal to the 5′ end of the gene was the SigH-dependent promoter, and the distal promoter was similar to the SigA-dependent promoter. The dnaK operon is transcribed from two promoters, of which one is SigH dependent and the other is SigA dependent (Fig. 3A). The HAIR element is located at the promoter regions of clpB and dnaK to repress transcription from the SigA-dependent promoter (Fig. 3A). Thus, it is likely that induction of the clpB and dnaK operons by heat shock is accomplished by derepression of the SigA-dependent promoter and activation of the SigH-dependent promoter.
FIG. 4.
SigH-independent induction of clpB and dnaK by heat shock. The transcript levels of clpB and dnaK 15 min after heat shock were compared to those at 33°C in the wild type (white bars) and the sigH disruptant (gray bars) by qRT-PCR. Experiments were repeated three times. The transcript levels at 33°C were taken as 1.
DISCUSSION
In the present study, the heat shock stimulon of C. glutamicum was identified using a DNA microarray. A total of 157 genes, including genes encoding HSPs, were upregulated by heat shock, and induction of most genes by heat shock was shown here for the first time (see Table S2 in the supplemental material). Drastic changes in gene expression in response to heat shock are documented in various bacteria (6, 18, 41), while its regulation is currently not completely understood (17). This study adds to the growing body of heat shock studies in bacterial systems.
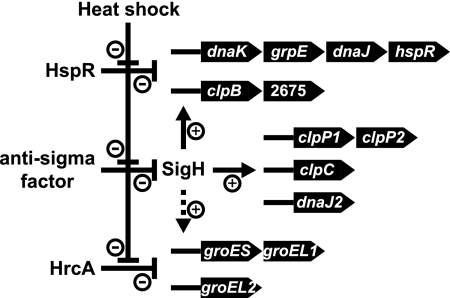
The ECF sigma factor SigH of C. glutamicum plays an important role in regulation of a total of 45 genes in response to heat shock (Table 1). It regulates expression of most HSP genes except for clpX and dnaK2, which are not upregulated by heat shock (Fig. 5). The thioredoxin (trxB and trxC) and mycothiol (mtr) systems, which are major antioxidant systems in actinobacteria (8), are also under the control of SigH. These findings are in good agreement with a previous study in which SigH function was shown to be related to heat and oxidative stress tolerance (27). The SigH-dependent promoter sequence of C. glutamicum is similar to those recognized by SigH of M. tuberculosis (26, 31) and SigR of Streptomyces coelicolor (38), which are orthologs of C. glutamicum SigH (Fig. 2). The activity of SigH of M. tuberculosis and SigR of S. coelicolor is regulated by the anti-sigma factors RshA and RsrA, respectively (25, 44). RshA has been demonstrated to bind to SigH to inhibit SigH-dependent transcription, and this interaction is disrupted by elevated temperatures as well as oxidizing conditions (44). An open reading frame encoding a putative anti-sigma factor was identified downstream of the sigH gene in the published C. glutamicum genome sequences, although this open reading frame is defined only in the genome of C. glutamicum ATCC 13032, reported by Kalinowski et al. (24), but not in C. glutamicum ATCC 13032, reported by Ikeda and Nakagawa (21), and C. glutamicum strain R, reported by Yukawa et al. (51). The activity of SigH of C. glutamicum is probably regulated by an anti-sigma factor in a mechanism similar to those of M. tuberculosis and S. coelicolor (Fig. 5).
FIG. 5.
A model of the regulatory network controlling expression of the HSP genes in response to heat shock. + and − indicate positive regulation and negative regulation, respectively. Indirect regulation of groES-groEL1 and groEL2 by SigH is shown with a dotted arrow. Involvement of an anti-sigma factor in regulation of the SigH activity has not been revealed in C. glutamicum. See text for details.
In the hspR disruptant, the transcript levels of the clpB and dnaK operons were upregulated (Table 2). The HAIR elements are present within the promoter regions of clpB and dnaK (Fig. 3A), suggesting that HspR directly represses expression of these operons. The HspR-HAIR system regulates expression of the dnaK, acr2, and clpB operons in M. tuberculosis (45), and the dnaK and clpB operons and the lon gene in S. coelicolor (5). The acr2 operon and the lon gene are found in certain strains of actinobacteria, but C. glutamicum does not contain these genes (48). The dnaK and clpB operons are the ubiquitous regulon of the HspR-HAIR system in actinobacteria. Expression of the dnaK and clpB operons was positively regulated by SigH and subjected to negative regulation by HspR. These operons were upregulated by heat shock even in the sigH disruptant (Fig. 4) and also by the sigH overexpression (Table 1). Disruption of hspR resulted in upregulation of these operons without affecting expression of other members of the SigH regulon (Table 2). These results indicate that SigH and HspR independently regulate expression of the dnaK and clpB operons (Fig. 5). In M. tuberculosis, the DNA-binding activity of HspR is modulated by chaperones (7). Expression of the dnaK and clpB operons might be inducible by various stimuli, which are sensed by an anti-sigma factor and chaperones.
Expression of groES-groEL1, cgR_1436, and groEL2 was upregulated by hrcA disruption (Table 2). CIRCE elements were observed within those promoter regions, suggesting direct regulation by HrcA. The HrcA-CIRCE system regulates expression of the groES-groEL1 operon and groEL2 and Rv0991c in M. tuberculosis (45). The groES-groEL1 operon and the groEL2 gene are also regulated by the HrcA-CIRCE system in Streptomyces spp. (10, 15), indicating that these genes constitute the core regulon of the HrcA-CIRCE system in actinobacteria. The transcript levels of groES-groEL1 and groEL2 were increased by overexpression of the sigH gene (Table 1). As the groES-groEL1 and groEL2 genes are transcribed from the SigA-dependent promoters (2), the SigH-associated control over groES-groEL1 and groEL2 expression is supposed to be indirect (Fig. 5). In M. tuberculosis, the transcript level of groES is increased by hspR disruption, in which some HSP genes under the control of HspR are derepressed (45). Upregulation of HSP genes by sigH overexpression might affect expression of groES-groEL1 and groEL2.
In the gram-positive model bacterium Bacillus subtilis, HrcA represses transcription of the hrcA-grpE-dnaK-dnaJ and groEL-groES operons from SigA-dependent promoters (42, 50). A general stress response sigma factor, SigB, positively regulates expression of the genes clpP, clpC, and clpX, which are also subjected to negative control by CtsR (29). In C. glutamicum, there is no sigma factor homologous to SigB of B. subtilis (14), but the ECF sigma factor SigH plays a regulatory role in expression of HSP genes in concert with HspR. On the other hand, the HrcA-CIRCE systems of C. glutamicum and B. subtilis negatively regulate the genes groES and groEL, suggesting its ancient origin.
Out of 157 genes upregulated by heat shock, 45 genes were defined to constitute the SigH regulon (Table 1). However, regulation of the remaining 112 genes is not unraveled. We found that some of these genes were transcribed from the SigA-dependent promoter (data not shown), suggesting that hitherto unidentified transcriptional regulators are involved in response to heat shock. Eighteen genes encoding transcriptional regulators were upregulated by heat shock (Table 3). In addition to the hspR gene, the transcript levels of the genes clgR, sufR, amtR, phoR, and mcbR were increased. ClgR activates transcription of clpP1-clpP2 and clpC (12), and SufR is the repressor of the suf gene cluster (34). McbR regulates genes involved in sulfur metabolism (40), and AmtR is the master regulator of nitrogen control (3). Some genes under the control of McbR and AmtR were upregulated by heat shock (see Table S2 in the supplemental material). A phoRS operon, which is positively regulated by PhoR (28), was included among the upregulated genes (see Table S2 in the supplemental material). Characterization of transcriptional regulators that are upregulated by heat shock would lead to further understanding of the regulatory network of gene expression in response to heat shock.
TABLE 3.
Transcriptional regulators upregulated by heat shock
| Gene | Product(s) | Ratioa | Pb |
|---|---|---|---|
| cgR_2687 (hspR) | Transcriptional regulator, MerR family | 4.33 | 1.6E−05 |
| cgR_2930 | Transcriptional regulator, ArsR family | 3.04 | 1.3E−02 |
| cgR_1792 (clgR) | Transcriptional regulator | 3.02 | 2.4E−02 |
| cgR_1435 | Transcriptional regulator, HxlR family | 2.70 | 3.5E−03 |
| cgR_1616 (sufR) | Transcriptional regulator, ArsR family | 2.34 | 3.6E−02 |
| cgR_2613 | Transcriptional regulator, MarR family | 1.94 | 7.5E−03 |
| cgR_2858 | Transcriptional regulator, MarR family | 1.82 | 6.7E−02 |
| cgR_1024 | Transcriptional regulator, TetR family | 1.73 | 1.8E−02 |
| cgR_0978 (amtR) | Transcriptional regulator, TetR family | 1.51 | 2.3E−04 |
| cgR_1106 | Transcriptional regulator, MarR family | 1.42 | 2.3E−02 |
| cgR_2511 (phoR) | Two-component response regulator | 1.40 | 1.4E−02 |
| cgR_2866 | Transcriptional regulator, PadR family | 1.27 | 1.4E−03 |
| cgR_0024 | Transcriptional regulator | 1.25 | 5.3E−04 |
| cgR_2029 | Transcriptional regulator, MerR family | 1.25 | 4.5E−02 |
| cgR_0818 | Transcriptional regulator, XRE family | 1.22 | 2.9E−03 |
| cgR_0746 | Transcriptional regulator, MarR family | 1.12 | 1.5E−03 |
| cgR_0453 | Transcriptional regulator, TetR family | 1.07 | 2.2E−04 |
| cgR_2850 (mcbR) | Transcriptional regulator, TetR family | 1.02 | 7.4E−03 |
Relative ratios of the transcript levels 15 min after heat shock to 33°C in the wild type determined by DNA microarray analyses are shown in the base 2 logarithm.
P values for the relative ratio were determined by Student's t test.
Supplementary Material
Acknowledgments
We thank Crispinus A. Omumasaba (RITE) for critical reading of the manuscript.
This study was partially supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO), Japan.
Footnotes
Published ahead of print on 6 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barreiro, C., E. Gonzalez-Lavado, S. Brand, A. Tauch, and J. F. Martin. 2005. Heat shock proteome analysis of wild-type Corynebacterium glutamicum ATCC 13032 and a spontaneous mutant lacking GroEL1, a dispensable chaperone. J. Bacteriol. 187884-889. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Barreiro, C., E. Gonzalez-Lavado, M. Patek, and J. F. Martin. 2004. Transcriptional analysis of the groES-groEL1, groEL2, and dnaK genes in Corynebacterium glutamicum: characterization of heat shock-induced promoters. J. Bacteriol. 1864813-4817. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Beckers, G., J. Strösser, U. Hildebrandt, J. Kalinowski, M. Farwick, R. Krämer, and A. Burkovski. 2005. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Mol. Microbiol. 58580-595. [DOI] [PubMed] [Google Scholar]
- 4.Brune, I., K. Brinkrolf, J. Kalinowski, A. Pühler, and A. Tauch. 2005. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucca, G., A. M. Brassington, G. Hotchkiss, V. Mersinias, and C. P. Smith. 2003. Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol. Microbiol. 50153-166. [DOI] [PubMed] [Google Scholar]
- 6.Chhabra, S. R., Q. He, K. H. Huang, S. P. Gaucher, E. J. Alm, Z. He, M. Z. Hadi, T. C. Hazen, J. D. Wall, J. Zhou, A. P. Arkin, and A. K. Singh. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 1881817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das Gupta, T., B. Bandyopadhyay, and S. K. Das Gupta. 2008. Modulation of DNA-binding activity of Mycobacterium tuberculosis HspR by chaperones. Microbiology 154484-490. [DOI] [PubMed] [Google Scholar]
- 8.den Hengst, C. D., and M. J. Buttner. 2008. Redox control in actinobacteria. Biochim. Biophys. Acta 17801201-1216. [DOI] [PubMed] [Google Scholar]
- 9.Dougan, D. A., A. Mogk, and B. Bukau. 2002. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell. Mol. Life Sci. 591607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duchêne, A. M., C. J. Thompson, and P. Mazodier. 1994. Transcriptional analysis of groEL genes in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 24561-68. [DOI] [PubMed] [Google Scholar]
- 11.Ehira, S., T. Shirai, H. Teramoto, M. Inui, and H. Yukawa. 2008. Group 2 sigma factor SigB of Corynebacterium glutamicum positively regulates glucose metabolism under conditions of oxygen deprivation. Appl. Environ. Microbiol. 745146-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor σH. Mol. Microbiol. 52285-302. [DOI] [PubMed] [Google Scholar]
- 13.Ermolaeva, M. D., H. G. Khalak, O. White, H. O. Smith, and S. L. Salzberg. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 30127-33. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira, A., M. Gray, M. Wiedmann, and K. J. Boor. 2004. Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other Gram-positive bacteria. Curr. Microbiol. 4839-46. [DOI] [PubMed] [Google Scholar]
- 15.Grandvalet, C., G. Rapoport, and P. Mazodier. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 1805129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandvalet, C., P. Servant, and P. Mazodier. 1997. Disruption of hspR, the repressor gene of the dnaK operon in Streptomyces albus G. Mol. Microbiol. 2377-84. [DOI] [PubMed] [Google Scholar]
- 17.Guisbert, E., T. Yura, V. A. Rhodius, and C. A. Gross. 2008. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol. Mol. Biol. Rev. 72545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 1837318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann, T. 2003. Industrial production of amino acids by coryneform bacteria. J. Biotechnol. 104155-172. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda, M. 2003. Amino acid production processes. Adv. Biochem. Eng. Biotechnol. 791-35. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 6299-109. [DOI] [PubMed] [Google Scholar]
- 22.Inui, M., H. Kawaguchi, S. Murakami, A. A. Vertès, and H. Yukawa. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8243-254. [DOI] [PubMed] [Google Scholar]
- 23.Inui, M., S. Murakami, S. Okino, H. Kawaguchi, A. A. Vertès, and H. Yukawa. 2004. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J. Mol. Microbiol. Biotechnol. 7182-196. [DOI] [PubMed] [Google Scholar]
- 24.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Pühler, D. A. Rey, C. Rückert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegräbe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 1045-25. [DOI] [PubMed] [Google Scholar]
- 25.Kang, J. G., M. S. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner, and J. H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 184292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative σ factor, SigH. Proc. Natl. Acad. Sci. USA 998330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, T. H., H. J. Kim, J. S. Park, Y. Kim, P. Kim, and H. S. Lee. 2005. Functional analysis of sigH expression in Corynebacterium glutamicum. Biochem. Biophys. Res. Commun. 3311542-1547. [DOI] [PubMed] [Google Scholar]
- 28.Kočan, M., S. Schaffer, T. Ishige, U. Sorger-Herrmann, V. Wendisch, and M. Bott. 2006. Two-component systems of Corynebacterium glutamicum: deletion analysis and involvement of the PhoS-PhoR system in the phosphate starvation response. J. Bacteriol. 188724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krüger, E., and M. Hecker. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 1806681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, X., D. L. Brutlag, and J. S. Liu. 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 6127-138. [PubMed] [Google Scholar]
- 31.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45365-374. [DOI] [PubMed] [Google Scholar]
- 32.Mishra, A. K., L. J. Alderwick, D. Rittmann, R. V. Tatituri, J. Nigou, M. Gilleron, L. Eggeling, and G. S. Besra. 2007. Identification of an alpha(1->6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol. Microbiol. 651503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Möker, N., M. Brocker, S. Schaffer, R. Krämer, S. Morbach, and M. Bott. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54420-438. [DOI] [PubMed] [Google Scholar]
- 34.Nakunst, D., C. Larisch, A. T. Hüser, A. Tauch, A. Pühler, and J. Kalinowski. 2007. The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes. J. Bacteriol. 1894696-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oguiza, J. A., A. T. Marcos, and J. F. Martín. 1997. Transcriptional analysis of the sigA and sigB genes of Brevibacterium lactofermentum. FEMS Microbiol. Lett. 153111-117. [DOI] [PubMed] [Google Scholar]
- 36.Okino, S., R. Noburyu, M. Suda, T. Jojima, M. Inui, and H. Yukawa. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl. Microbiol. Biotechnol. 81459-464. [DOI] [PubMed] [Google Scholar]
- 37.Okino, S., M. Suda, K. Fujikura, M. Inui, and H. Yukawa. 2008. Production of D-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 78449-454. [DOI] [PubMed] [Google Scholar]
- 38.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 421007-1020. [DOI] [PubMed] [Google Scholar]
- 39.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 1836119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey, D. A., S. S. Nentwich, D. J. Koch, C. Rückert, A. Pühler, A. Tauch, and J. Kalinowski. 2005. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol. Microbiol. 56871-887. [DOI] [PubMed] [Google Scholar]
- 41.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 273821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 1781088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Servant, P., G. Rapoport, and P. Mazodier. 1999. RheA, the repressor of hsp18 in Streptomyces albus G. Microbiology 1452385-2391. [DOI] [PubMed] [Google Scholar]
- 44.Song, T., S. L. Dove, K. H. Lee, and R. N. Husson. 2003. RshA, an anti-sigma factor that regulates the activity of the mycobacterial stress response sigma factor SigH. Mol. Microbiol. 50949-959. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 1483129-3138. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, N., N. Okai, H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2006. High-throughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl. Environ. Microbiol. 723750-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terasawa, M., and H. Yukawa. 1993. Industrial production of biochemicals by native immobilization, p. 37-52. In A. Tanaka, O. Tosaka, and T. Kobayashi (ed.), Industrial application of immobilized biocatalysts. Marcel Dekker, Inc., New York, NY. [PubMed]
- 48.Ventura, M., C. Canchaya, Z. Zhang, V. Bernini, G. F. Fitzgerald, and D. van Sinderen. 2006. How high G+C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30734-759. [DOI] [PubMed] [Google Scholar]
- 49.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 2861888-1893. [DOI] [PubMed] [Google Scholar]
- 50.Yuan, G., and S. L. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 1776462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yukawa, H., C. A. Omumasaba, H. Nonaka, P. Kos, N. Okai, N. Suzuki, M. Suda, Y. Tsuge, J. Watanabe, Y. Ikeda, A. A. Vertès, and M. Inui. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 1531042-1058. [DOI] [PubMed] [Google Scholar]
- 52.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat-shock response. Curr. Opin. Microbiol. 2153-158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.