Abstract
To understand how the Rhizobium leguminosarum raiI-raiR quorum-sensing system is regulated, we identified mutants with decreased levels of RaiI-made N-acyl homoserine lactones (AHLs). A LuxR-type regulator, ExpR, is required for raiR expression, and RaiR is required to induce raiI. Since raiR (and raiI) expression is also reduced in cinI and cinR quorum-sensing mutants, we thought CinI-made AHLs may activate ExpR to induce raiR. However, added CinI-made AHLs did not induce raiR expression in a cinI mutant. The reduced raiR expression in cinI and cinR mutants was due to lack of expression of cinS immediately downstream of cinI. cinS encodes a 67-residue protein, translationally coupled to CinI, and cinS acts downstream of expR for raiR induction. Cloned cinS in R. leguminosarum caused an unusual collapse of colony structure, and this was delayed by mutation of expR. The phenotype looked like a loss of exopolysaccharide (EPS) integrity; mutations in cinI, cinR, cinS, and expR all reduced expression of plyB, encoding an EPS glycanase, and mutation of plyB abolished the effect of cloned cinS on colony morphology. We conclude that CinS and ExpR act to increase PlyB levels, thereby influencing the bacterial surface. CinS is conserved in other rhizobia, including Rhizobium etli; the previously observed effect of cinI and cinR mutations decreasing swarming in that strain is primarily due to a lack of CinS rather than a lack of CinI-made AHL. We conclude that CinS mediates quorum-sensing regulation because it is coregulated with an AHL synthase and demonstrate that its regulatory effects can occur in the absence of AHLs.
Production of N-acyl homoserine lactones (AHLs) is common to many plant-associated bacteria (7), in which it is usually associated with population density-dependent regulation of genes affecting adaptive responses (49). Within the family Rhizobiaceae, population density-regulated gene expression (quorum sensing) mediated via AHLs has been identified in several agrobacteria and rhizobia (13, 51). In Agrobacterium spp., quorum-sensing regulation was initially identified as a mechanism of regulating plasmid transfer. As the bacterial population density increases, plasmid transfer genes are induced by TraR in response to AHLs made by TraI (55). In several rhizobia, traI-like AHL synthase genes are also in an operon along with plasmid transfer genes (13).
There are other quorum-sensing loci in different strains of rhizobia. In Sinorhizobium meliloti strain Rm1021, AHLs produced by SinI activate SinR and ExpR, LuxR-type regulators, to induce several genes, including those determining the production of an exopolysaccharide, exopolysaccharide II (EPS-II) (17, 23, 24, 35), that plays an important role in the symbiosis. In S. meliloti, two LuxR-type regulators, VisN and VisR, are involved in chemotaxis and motility (24, 44). Rhizobium etli has multiple AHL synthase genes (9, 39), but the functions of many of the regulated genes remain to be established. The cinR and cinI genes are required for normal symbiotic nitrogen fixation and swarming in R. etli (5, 9, 11) and for normal levels of expression of raiI, which encodes another AHL synthase. The expression of raiI in R. etli is regulated by RaiR (39).
Analysis of AHLs produced by strain A34 of Rhizobium leguminosarum bv. viciae led to the characterization of four LuxI-type AHL synthases (RhiI, CinI, RaiI, and TraI) and five LuxR-type regulators (RhiR, CinR, RaiR, TraR, and BisR) (8, 31, 50, 53). In this strain, the cinI and cinR genes are chromosomally located; CinI produces N-(3-hydroxy-7-cis-tetradecenoyl)-l-homoserine lactone (3-OH-C14:1-HSL) (20, 31), CinR induces cinI expression in response to this AHL (31), and this appears to be associated with adaptation to starvation and salt stress (47). Mutation of cinI or cinR affects the expression of the other three AHL synthase genes in R. leguminosarum bv. viciae strain A34. Thus, in a cinI mutant, the expression of raiI is reduced, resulting in very low levels of 3-OH-C8-HSL, the major AHL made by RaiI (53). Similarly, the expression levels of the traI and rhiI genes on the symbiotic plasmid pRL1JI are reduced in cinI and cinR mutants (31). RhiI-made AHLs activate RhiR to induce the expression of the rhiABC operon in R. leguminosarum bv. viciae (38), enhancing the interaction with the legume host (8).
The cinI and cinR quorum-sensing genes control induction of the traI and traR quorum-sensing regulons via CinI-made 3-OH-C14:1-HSL, which activates BisR (another LuxR-type regulator) to induce traR and hence traI (12). However, the mechanism by which cinI and/or cinR control raiI and raiR expression has not been established. In this work we demonstrate that raiI and raiR expression requires both expR and a small gene (cinS) cotranscribed with cinI. CinS also regulates the expression of plyB encoding an extracellular glycanase and is required for swarming of R. etli.
MATERIALS AND METHODS
Microbiological techniques.
Rhizobia were grown at 28°C in TY medium (4) or Y minimal medium (43) containing mannitol (0.2% [wt/vol]) (Y-mannitol medium) as the carbon source. Antibiotics were added as appropriate to maintain selection for plasmids. Bacterial growth and β-galactosidase activity were measured using a Perkin-Elmer MBA2000 spectrophotometer as described previously (53) using at least six independent cultures. To measure formation of biofilm rings, bacteria were inoculated into 100 ml of Y-mannitol medium in 250-ml Erlenmeyer flasks using 2 ml of TY cultures grown to an optical density at 600 nm (OD600) of 1.0. Flasks were shaken at 300 rpm at 28°C, and growth of biofilm rings was assayed after 5 days. The biofilm rings were harvested using cotton buds and resuspended in 1 ml H2O, and crystal violet was added to a final concentration of 0.01% from a fresh 0.4% (wt/vol) aqueous stock and left for 30 min. The bacteria were washed four times with water and then resuspended in 1 ml ethanol. After 1 h, the samples were centrifuged, and the OD575 was determined. Colony morphology changes were assayed by inoculating TY plates directly from glycerol stocks and incubating the plates for 3 days at 28°C. Swarming of R. etli was assayed in yeast extract-mannitol (YEM) swarm plates containing 0.7% agar as described previously (10) after growth at 28°C for 4 days. Nodulation tests were done using peas (Pisum sativum L.) variety Frisson as described previously (25), using a minimum of 16 matched plants per test.
Bacterial strains.
The strains and plasmids used are listed in Table 1 or in the text. R. leguminosarum strain 8400 lacks a symbiotic plasmid, and all R. leguminosarum strains were derived from strain 8400 or its streptomycin-resistant derivative, strain 8401. A34 is a derivative of strain 8401 carrying the symbiotic plasmid pRL1JI. Plasmids were mobilized into Rhizobium by triparental matings using a helper plasmid. Strain 8401 was mutagenized with Tn5-gus by using Escherichia coli strain MM294/pRK600::Tn5-gus as a donor of the suicide plasmid pRK600::Tn5-gus essentially as described previously (41). A population of about 8,000 colonies was screened for impaired AHL production by picking colonies onto a lawn of the AHL biosensor strain Chromobacterium violaceum CV026 (33) to identify mutants that did not induce the purple pigment violacein. One of these mutants (A702; expR) was severely affected for production of RaiI-made AHLs, and small-bacteriocin tests revealed that strain A702 retained the ability to produce 3-OH-C14:1-HSL. Strain A1085 was made by plating bacteriophage RL38 (6) on strain A702 and using the phage lysate to transduce strain 8400, selecting for kanamycin resistance. To make strain A1102 (cinS::Spcr), cinS with 1-kb flanking regions was first amplified by PCR using primers CTGAAGAGCGGCCGCTTCAAGCTC and GAGAAACTTAGCGGCCGCTATTGATTTC containing introduced NotI sites (bold face). The product was digested with NotI and cloned into pJQ254 (37). The cinS1::Spcr allele was then created by cloning a spectinomycin resistance cassette on a BamHI fragment from pHP45Ω (36) into the unique BamHI site in cinS. The 4-kb NotI fragment containing cinS1::Spcr was then subcloned into pJQ200 (37), and the cinS1::Spcr allele was recombined into strain 8401 by selecting for spectinomycin-resistant, sucrose-resistant transconjugants (37).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| R. leguminosarum strains | ||
| 8400 | Strain lacking a symbiotic plasmid | 29 |
| 8401 | Strr derivative of strain 8400 | 29 |
| A34 | Strain 8401 containing pRL1JI (pSym) | 14 |
| A616 | 8401 prsD1::Tn5 | 16 |
| A552 | 8401 cinR1::Tn5 | 31 |
| A702 | 8401 expR1::Tn5-gus | This work |
| A740 | 8401 cinI3::Spcr | 31 |
| A789 | 8401 raiI7::Tn5 | 53 |
| A797 | 8401 cinI3::SpcrraiI7::Tn5 | 53 |
| A802 | 8401 raiR8::Tn5 | 53 |
| A1085 | 8400 expR1::Tn5-gus | This work |
| A1102 | 8401 carrying cinS::Spcr | This work |
| R. etli CNPAF512 | WT R. etli | 10 |
| R. etli FAJ4006 | CNPAF512 cinI::Tn5-gus | 10 |
| A. tumefaciens NT1/pZLR4 | traG-lacZ-based AHL detection strain | 7 |
| C. violaceum CV026 | AHL detection strain | 33 |
| Plasmids | ||
| pIJ7516 | Cosmid carrying cinRI locus | 31 |
| pIJ7910 | cinI′-lacZ in pMP220 | 31 |
| pIJ9001 | Cosmid carrying raiIR region | 53 |
| pIJ9115 | 7-kb EcoRI fragment from strain A702 carrying part of the Tn5-gus in pBluescript SK | This work |
| pIJ9123 | Cosmid carrying expR region in pLAFR1 | This work |
| pIJ9229 | expR on 3-kb EcoRI fragment from pIJ9123 in pBluescript KS | This work |
| pIJ9252 | plyB′-lacZ fusion in pMP220 | This work |
| pIJ9263 | expR′-lacZ fusion in pMP220 | This work |
| pIJ9272 | raiR′-lacZ fusion in pMP220 | 53 |
| pIJ9276 | raiR in pBBR1-MCS5 | 53 |
| pIJ9280 | raiI′-lacZ fusion in pMP220 | 53 |
| pIJ9493 | expR in pBBR1-MCS5 | This work |
| pIJ9655 | cinI and truncated cinS in pKT230 | This work |
| pIJ9692 | cinS in pKT230 | This work |
| pIJ9769 | expR in pBBR1-MCS2 | This work |
| pMP220 | Broad-host-range lacZ expression vector; Tcr | 45 |
| pKT230 | Broad-host-range lacZ expression vector; Kanr | 2 |
| pBBR1-MCS2 | Broad-host-range plasmid; Kanr | 27 |
| pBBR1-MCS5 | Broad-host-range plasmid; Gentr | 27 |
Molecular biology techniques and plasmid construction.
DNA work was done using standard methods (40). Plasmid pIJ9115 was made by cloning the kanamycin resistance cassette from the genome of A702 as a 7-kb EcoRI fragment in pBluescript SK. Plasmid pIJ9123 was isolated from a cosmid library (28) on the basis of its ability to complement strain A702 for AHL production using C. violaceum CV026 as assayed previously (53). A 3-kb EcoRI fragment containing expR was subcloned from pIJ9123 into pBluescript to form pIJ9229, and pIJ9263 (expR′-lacZ) was made by cloning a 1.4-kb EcoRI-PstI fragment from pIJ9123 into pMP220. To construct plasmid pIJ9252 (plyB′-lacZ), a 0.5-kb EcoRI-PstI fragment from pIJ7708 (16) was cloned into the EcoRI-PstI sites of pMP220. To make plasmid pIJ9493, expR was subcloned from pIJ9229 on a 2.7-kb EcoRI-SacI fragment into pBBR1-MCS5, and expR was recloned on a 2.6-kb HindIII-SacI fragment from pIJ9493 into pBBR1-MCS2 to make pIJ9769. To make pIJ9655, cinI and a truncated cinS (encoding the first 36 residues) was amplified from pIJ7655 (31) by PCR using primers CATTCTGGGATCCACGAACTTGAAAAC and ATCTGAATCCCGATCTGCGTG which carry introduced BamHI and EcoRI sites (shown in boldface type); the product was cloned into pKT230 using BamHI and EcoRI. To make pIJ9692, cinS was amplified from plasmid pIJ7655 (31) using the forward primer CTTCGAATTCTCGGACCGCGTGCTGCGCAAGATG and the reverse primer GATATCGACCATGAATTCCCTGATGAC containing introduced EcoRI sites (boldface); the product was digested with EcoRI and cloned as a 305-bp fragment into the EcoRI site in pKT230 in the same orientation as the streptomycin resistance gene in that plasmid. All plasmid inserts made using PCR were checked by DNA sequencing.
DNA sequencing of the region containing expR (accession no. FM992852) was carried out on both strands using primer walking on pIJ9229. The location of Tn5-gus in expR was determined by sequencing pIJ9115 using a Tn5-specific primer. Database searches of the predicted protein sequences were carried out by using the BLAST and FASTA (1) programs to find related sequences in the EMBL and SwissProt protein sequence databases.
AHL assays.
R. leguminosarum cultures were grown for 48 h in TY medium to an OD600 of about 1.0, and cells were removed by centrifugation. AHLs were extracted from culture supernatants and analyzed by thin-layer chromatography (31) using Agrobacterium tumefaciens NT1/pZLR4 (7, 42). The amount loaded corresponds to the extract from 1 ml of culture supernatant. CinI-made 3-OH-C14:1-HSL was bioassayed on TY medium (50).
RESULTS
Identification of a regulatory gene (expR) required for raiI expression.
To identify genes required for raiI and raiR expression, R. leguminosarum strain 8401 (lacking a symbiotic plasmid) mutants were screened for decreased levels of RaiI-made AHLs. We used C. violaceum strain CV026, which produces a purple pigment (violacein) in response to RaiI (but not CinI)-made AHLs (33, 53). Strain A702 produced low levels of AHLs, and the mutation was not in the raiIR or cinIR region on the basis of the lack of complementation with pIJ9001 and pIJ7516, respectively. Analysis of A702-made AHLs by thin-layer chromatography using A. tumefaciens carrying traG-lacZ to detect AHLs revealed that, in contrast with the wild type (WT) (strain 8401) (Fig. 1A, lane 2), strain A702 produced no detectable C8-HSL and a very low level of 3-OH-C8-HSL (Fig. 1A, lane 3), slightly more than that made by a raiR mutant (Fig. 1A, lane 4), which lacks raiI expression (53). The A702 mutant phenotype cotransduced (100%) with kanamycin resistance, showing that the transposon mutation in A702 caused the loss of AHL production. The transductant A1085 behaved as strain A702 did.
FIG. 1.
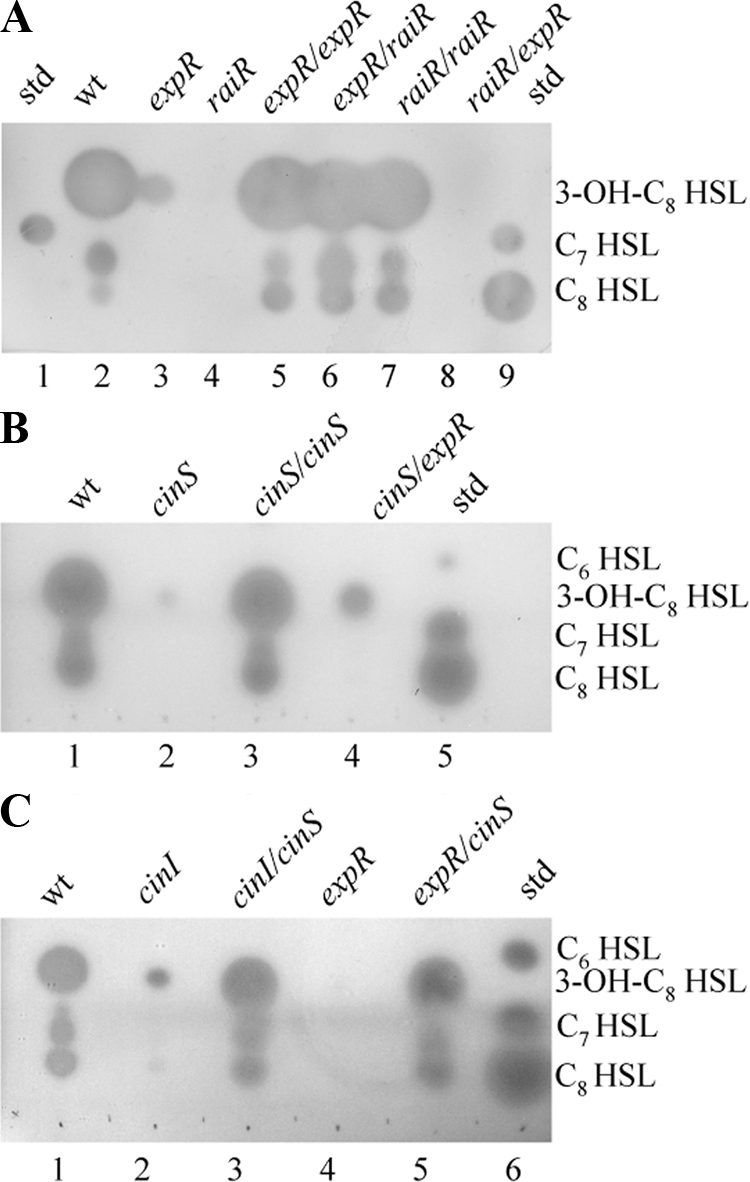
Effects of cloned expR, raiR, and cinS on production of AHLs by R. leguminosarum quorum-sensing mutants. AHLs were extracted from cell-free culture supernatants, separated by thin-layer chromatography, and detected using an overlay of agar seeded with the biosensor A. tumefaciens NT1/pZLR4. Each lane was loaded with a volume of extract equivalent to 1 ml of culture. (A) Lane 1, C7-HSL standard (std) (14 pmol); lane 2, strain 8401 (WT); lane 3, strain A702 (expR); lane 4, strain A802 (raiR); lane 5, strain A702 (expR) carrying pIJ9493 (expR); lane 6, A702 (expR) carrying pIJ9276 (raiR); lane 7, A802 (raiR) carrying pIJ9276 (raiR); lane 8, A802 (raiR) carrying pIJ9493 (expR); lane 9, standards, i.e., C8-HSL (100 pmol) and C7-HSL (14 pmol). (B) Lane 1, strain 8401 (WT); lane 2, strain A1102 (cinS); lane 3, A1102 (cinS) carrying pIJ9692 (cinS); lane 4, A1102 (cinS) carrying pIJ9493 (expR); lane 5, standards, i.e., C8-HSL (100 pmol), C7-HSL (14 pmol), and C6-HSL (5 pmol). (C) Lane 1, strain 8401 (WT), lane 2, strain A740 (cinI); lane 3, strain A740 (cinI) carrying pIJ9692 (cinS); lane 4, A1085 (expR); lane 5, A1085 (expR) carrying pIJ9692 (cinS); lane 6, standards, i.e., C8-HSL (100 pmol), C7-HSL (14 pmol), and C6-HSL (5 pmol).
A cosmid (pIJ9123) that restored AHL production to strain A702 was identified and used to probe EcoRI-digested genomic DNA from A702 and the control strain 8401, revealing that the transposon in A702 had inserted in a 3-kb EcoRI fragment. This fragment was cloned from pIJ9123 and partially sequenced, revealing the expR gene between ndvA and pyc. expR is transcribed divergently from ndvA and in the same orientation as pyc. The transposon in strain A702 was inserted 678 bp downstream of the predicted translation start of expR. The ndvA expR pyc gene arrangement is similar to that seen in R. leguminosarum 3841 (54), R. etli CFN42 (19), and S. meliloti strain 1021 (http://sequence.toulouse.inra.fr/meliloti.html), except that in strain 1021, there is an insertion element within expR (35). expR encodes a predicted LuxR-type regulator with predicted N-terminal AHL-binding and C-terminal DNA-binding domains. Cloned expR (on pIJ9493) complemented AHL production in strain A702 (expR) (Fig. 1A, lane 5) but did not suppress the defect in AHL production in the raiR mutant strain A802 (Fig. 1A, lane 8). Conversely, raiR cloned behind a vector promoter on pIJ9276 (Fig. 1A, lane 6) restored AHL production to the expR mutant A702 (as well as to the raiR mutant; Fig. 1A, lane 7), suggesting that ExpR induces raiR and hence raiI (Fig. 2).
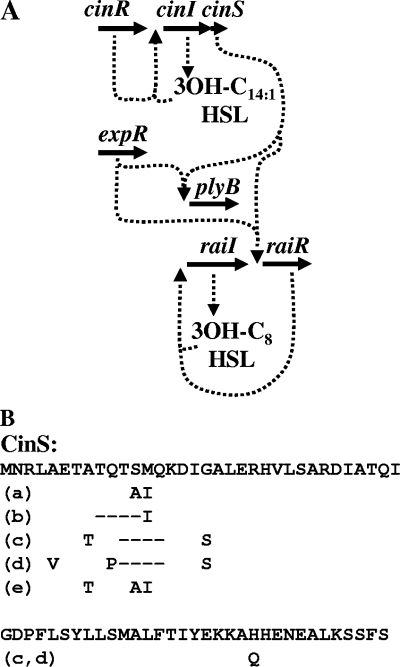
FIG. 2.
(A) Model of quorum-sensing regulation in R. leguminosarum strain 8401. The cinS gene is cotranscribed with cinI and is predicted to be translationally coupled to cinI and so is induced by CinR in response to CinI-made AHL. Both cinS and expR are required for full induction of raiR and plyB; RaiR regulates raiI in response to RaiI-made AHL. (B) Predicted sequence of CinS from strain 8401 (top line) and the differences in the conserved homologues in R. leguminosarum bv. viciae 3841 (GenBank nucleotide accession no. AM236080) and R. leguminosarum bv. trifolii strain WSM1325 (GenBank protein accession no. EDR73518) (line a), R. leguminosarum bv. trifolii strain WSM2304 (GenBank protein accession no. ACI55940) (line b), R. etli strains CNPAF512 (GenBank nucleotide accession no. AF393621) and CIAT652 (GenBank protein accession no. ACE92023) (line c), R. etli strain CFN42 (GenBank nucleotide accession no. CP000133) (line d), and Mesorhizobium tianshanense (GenBank nucleotide accession no. DQ123807) (line e). The residues different from those in CinS in strain 8401 are shown below the sequence; deletions are shown by dashes.
raiR (and hence raiI) expression is regulated by both ExpR and a novel small protein, CinS.
The expR mutation in strain A702 strongly reduced expression of both raiI′-lacZ (on pIJ9280) and raiR′-lacZ (on pIJ9272) (Table 2). These assays were done in Y medium, in which raiI and raiR expression is significantly higher than that seen previously (53) in TY medium. Mutation of cinI (Table 2) and cinR (data not shown) similarly reduced raiI and raiR expression. Mutation of raiR or raiI did not affect raiR expression, although as expected (53), they did reduce raiI expression (Table 2). Cloned raiR (pIJ9276) caused high levels of raiI expression in the expR mutant; A702/pIJ9280 (raiI′-lacZ) carrying pIJ9276 (constitutive raiR) had 15,286 (±300) units of β-galactosidase activity, similar to the high levels of raiI′-lacZ expression seen previously with cloned raiR in the cinI and cinR mutants or in strain 8401 (53).
TABLE 2.
Effect of an expR mutation on raiR′-lacZ and raiI′-lacZ expressiona
| Strain (relevant genotype) | Expressionb of:
|
|
|---|---|---|
| raiI′-lacZ (pIJ9280) | raiR′-lacZ (pIJ9272) | |
| 8401 (WT) | 5,873 ± 10 | 1,075 ± 146 |
| A702 (expR) | 340 ± 25 | 282 ± 44 |
| A740 (cinI) | 346 ± 18 | 302 ± 53 |
| A1102 (cinS) | 362 ± 9 | 318 ± 30 |
| A789 (raiI) | 462 ± 16 | 1,135 ± 99 |
| A802 (raiR) | 344 ± 13 | 1,030 ± 7 |
Bacteria were cultured for 48 h in Y-mannitol medium.
Data shown are mean values for β-galactosidase activity (in Miller units) ± standard errors, based on averages of at least six assays.
Induction of raiR requires expR, cinI, and cinR, but there is no known AHL synthase associated with ExpR. To test whether ExpR induced raiR in response to CinI-made 3-OH-C14:1-HSL, we added 1 μM 3-OH-C14:1-HSL to the growth medium of the cinI mutant A740 carrying raiR′-lacZ (pIJ9272) but saw no induction (236 ± 37 units); we could not restore raiI or raiR expression in the cinI mutant with other AHLs, including C6-HSL, 3-O-C8-HSL, and crude AHL extracts from strain 8401 (data not shown). This suggested that the effect of the cinI mutation may be due to polarity. Although there is only a short gap between the coding region of cinI and the adjacent convergently transcribed gene (31), there is a short open reading frame encoding a predicted protein of 67 residues that appears to be translationally coupled to CinI; as described below, this is indeed a gene which we named cinS (Fig. 2).
A cinS mutant (strain A1102) produced normal levels of CinI-made 3-OH-C14:1-HSL (data not shown), and A1102/pIJ7910 (cinI′-lacZ) had normal levels of β-galactosidase expression (20,628 ± 2,694 units compared with 24,692 ± 1,940 units seen with the control 8401/pIJ7910). This contrasts with the relatively low expression (986 ± 75 units) in the cinI mutant A740 carrying pIJ7910. Strain A1102 (cinS) lacked RaiI-made AHLs (Fig. 1B, lane 2); like the expR mutant (and cinI and cinR mutants), it did not induce raiI′-lacZ, apparently due to a lack of raiR expression (Table 2). Cloned cinS behind a vector promoter (pIJ9692) restored the expression of raiR′-lacZ in the cinI, cinR, and cinS mutants (Fig. 3A) and restored formation of the RaiI-made AHLs in the cinS and cinI mutants A1102 and A740 (Fig. 1B, lane 3, and Fig. 1C, lane 3), and in the cinR mutant A552 (data not shown). In contrast, pIJ9655 carrying cloned cinI and part of cinS (encoding the N-terminal 36 residues) did not complement raiR-lacZ expression in the cinI mutant (A740/pIJ9272 had 278 ± 23 units compared with over 1,000 units in the WT; Table 2); pIJ9655 (cinI) also did not restore formation of RaiI-made AHLs to the cinI mutant, although as expected, it restored production of CinI-made AHLs (data not shown). The complementation of raiR expression in the cinI mutant by cinS, but not by cinI, confirms the prediction that the cinI mutation in A740 is polar on cinS and that cinI and cinS are cotranscribed (Fig. 2A).
FIG. 3.
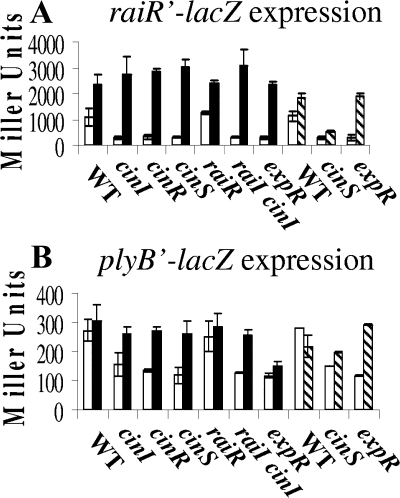
Effects of cloned cinS and expR on raiR′-lacZ and plyB′-lacZ expression in quorum-sensing mutants of R. leguminosarum. Strains were grown in Y-mannitol medium, and β-galactosidase activity was assayed after 48 h (for raiR′-lacZ) or 72 h (for plyB′-lacZ) of growth. The WT strain (strain 8401) and mutant strains were used. The mutant strains used were cinI (strain A740), cinR (A552), cinS (A1102), and raiI (A789) single mutants, the raiI cinI double mutant (A797), and expR mutant (A1085) (in this experiment A1085 was used instead of A702 to facilitate plasmid selection). (A) raiR′-lacZ activities of WT and mutants with cloned cinS on pIJ9692 (black bars) or cloned expR on pIJ9769 (hatched bars) or the strains containing the appropriate vector controls pKT230 or pBBR1-MCS2 (white bars). (B) plyB′-lacZ activities of WT and mutants with cloned cinS on pIJ9692 (black bars) or cloned expR on pIJ9769 (hatched bars) or the strains containing the appropriate vector control, pKT230 or pBBR1-MCS2 (white bars).
Taken together, these observations show that the lack of RaiI-made AHLs in cinI and cinR mutants is due to a lack of cinS expression and that CinI-made AHLs are not essential for raiI expression if cinS is expressed constitutively. The effect of CinS is on raiR expression, because cloned cinS on pIJ9692 restored raiR′-lacZ expression to normal levels in the cinI (A740) and cinR (A552) mutants (Fig. 3A). Thus, both CinS and ExpR are normally required for raiR (and hence raiI) induction (Fig. 2), and CinI-made AHL is not required if cinS is expressed constitutively. Cloned cinS also restored raiR expression in a raiI cinI double mutant, strain A797 (Fig. 3A), which makes no detectable AHLs, confirming that CinS can act independently of AHLs. Cloned cinS (pIJ9692) did not restore production of RaiI-made AHLs (data not shown) or raiI′-lacZ expression in the raiR mutant A802; raiI′-lacZ expression on pIJ9280 in the raiR mutant A802 was low (75 ± 9 units compared with 4,089 ± 347 units in WT strain 8401) and was not affected by cloned cinS (79 ± 18 units), showing that RaiR acts downstream of CinS for induction of raiI.
Cloned cinS (pIJ9692) restored both raiR expression (Fig. 3A) and the production of RaiI-made AHLs to the expR mutant A702 (Fig. 1C, lane 5), but in the cinS mutant A1102, cloned expR (pIJ9769) only very slightly increased both AHL production (Fig. 1B, lane 4) and raiR′-lacZ (pIJ9272) expression (Fig. 3A). This shows that CinS acts downstream of ExpR (the expR gene on pIJ9769 is functional because it complemented the expR mutant; Fig. 3A).
These data are not consistent with a model in which ExpR directly regulates raiR or raiI expression. The observations point toward both CinS and ExpR having an effect on the levels of raiR transcript (Fig. 2A), although high-level expression of CinS can compensate for the absence of ExpR.
expR expression and cinIS expression are not interdependent.
The expR mutant A702 carrying cinI′-lacZ (pIJ7910) expressed cinI′-lacZ normally (23,114 ± 1,753 units compared with 24,692 ± 1,940 units in strain 8401/pIJ7910) and produced normal levels of CinI-made 3-OH-C14:1-HSL (data not shown), showing that expR is not required for expression of the cinIS operon. These observations are different from those made in S. meliloti, in which expR and SinI-made AHLs are required for full induction of sinI (23, 34).
We tested whether expR expression is regulated by CinR or by CinI-made AHLs. An expR′-lacZ fusion (pIJ9263) was expressed similarly in the control strain 8401 and the cinR mutant A552 (856 ± 45 units compared with 902 ± 23 units, respectively). Therefore, expR expression is not regulated by CinR. Similar levels of expR′-lacZ expression were seen with pIJ9263 in strain A740 (cinI) (911 ± 56 units), A789 (raiI) (813 ± 119 units), A797 (raiI cinI) (865 ± 86 units), and A802 (raiR) (732 ± 164 units), showing that expR expression is independent of the Cin and Rai AHL-based regulatory systems.
cinS influences colony morphology and formation of biofilm rings.
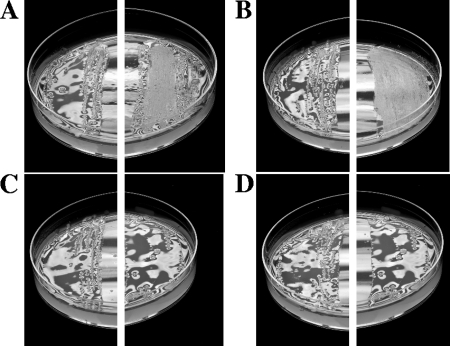
Mutation of cinS, cinR, cinI, or expR had no obvious effect on the growth rate of R. leguminosarum in minimal or complex media or on the symbiosis (nodulation and nitrogen fixation) of peas (data not shown), although the colonies on TY medium appeared slightly more mucoid than normal. Plated cultures of strain 8401 (WT) carrying cloned cinS (on pIJ9692) had an unusual phenotype, because the primary inoculum on TY plates appeared to age prematurely. Normally the primary inoculum region is quite mucoid after about 2 days of growth but appears to “collapse” or dry up after about 7 days of incubation. However, cloned cinS (pIJ9692) caused this collapse to occur in the control, strain 8401, after about 3 days (Fig. 4A). This effect of cloned cinS was independent of AHL-dependent quorum-sensing regulation, because the collapse phenotype was induced by pIJ9692 in the cinR mutant (Fig. 4B), and similar results (data not shown) were seen with the cinI, cinS, raiR, and raiI mutants as well as the raiI cinI double mutant. However, the expR mutation in strain A702 blocked the cinS-induced collapse at 3 days (Fig. 4C). This suggests that, like raiR induction, both expR and cinS are required to induce the colony collapse, but since the effect occurred in the raiI and raiR mutants, the collapse cannot be induced via an effect of ExpR and CinS on the raiI-raiR regulatory system.
FIG. 4.
Cloned cinS induced a colony collapse phenotype. The photographs show cultures of strains grown for 3 days on TY agar medium. For each strain, the photograph on the left shows the strain grown with the vector (pKT230) control and the photograph on the right shows the strain with cinS on pIJ9692. Strains 8401 (WT) (A), A552 (cinR) (B), A702 (expR) (C), and A616 (plyB) (D) are shown. After 2 days, the appearance of R. leguminosarum 8401/pIJ9692 and A552/pIJ9692 cultures looked normal (not shown) but collapsed, forming a dry appearance after 3 days. Cloned cinS had no effect on strain A616 (plyB) even after prolonged culture, but with strain A702 (expR), a delayed colony collapse was observed (not shown). pIJ9692 (cinS) induced a similar collapse to that seen in panels A and B in strains A740 (cinI), A1102 (cinS), A789 (raiI) A802 (raiR), and A797 (raiI cinI) (not shown).
Most colony mucoidicity is due to the acidic exopolysaccharide (EPS), and so we postulated that the collapse phenotype might be caused by premature degradation of the EPS. PlyA and PlyB are extracellular glycanases that can cleave the acidic EPS (16, 56) and so we tested whether they were quorum sensing regulated. A plyB′-lacZ reporter fusion (on pIJ9252) was induced during stationary phase (Fig. 5), and its expression was significantly decreased by mutations in cinI, expR, cinS, and cinR (Fig. 5). No change was seen with the plyA′-lacZ fusion (data not shown) in those mutants.
FIG. 5.
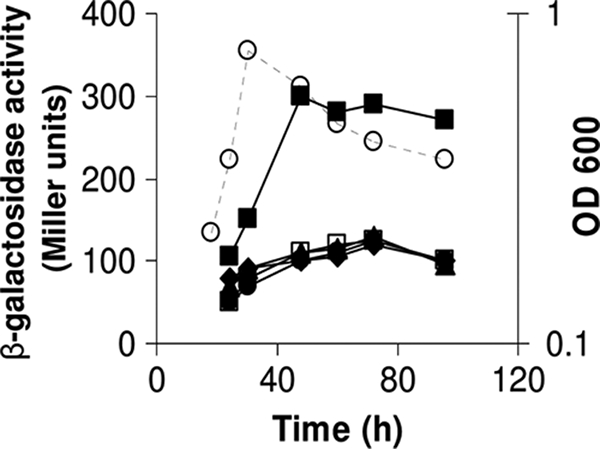
Decrease of plyB-lacZ expression in cinI, cinR, cinS, and expR mutants. Expression of plyB was assayed throughout growth in Y-mannitol medium using pIJ9252 (plyB′-lacZ) in the WT strain 8401 (solid squares), the cinI mutant A740 (solid triangles), the cinR mutant A552 (solid diamonds), the cinS mutant A1102 (open squares), and the expR mutant A702 (solid circles). The growth of strain 8401 measured at OD600 is shown as open circles and was very similar for all strains.
As with raiR′-lacZ, plyB′-lacZ expression in the cinR, cinI, and cinS mutants was restored by cloned cinS (Fig. 3B); its expression was also restored by cloned expR in the expR, but not the cinS, mutant (Fig. 3B). The correlation of plyB and raiR expression raised the possibility that plyB may be regulated by RaiR, but the expression of plyB′-lacZ in the raiR mutant showed that was not the case (Fig. 3B). Mutation of expR reduced plyB′-lacZ expression, but in this case cloned cinS did not restore plyB expression (Fig. 3B); this correlates with the observation (Fig. 4C) that the expR mutation delayed the colony collapse phenotype induced by cloned cinS. These observations are consistent with both ExpR and CinS being required for full plyB expression, but increased expression of cinS can cause an enhanced amount of PlyB, which causes degradation of EPS and a resulting collapse in colony structure. Consistent with this, cloned cinS did not induce the colony collapse in the plyB mutant A616 (Fig. 4D).
After 3 to 4 days in shaken flask cultures in Y-mannitol medium, the cinS mutant produced a biofilm ring that was enhanced compared with that of the control strain 8401 (Fig. 6A), and this was restored to normal by cloned cinS on pIJ9692 (Fig. 6B). Similar results were seen with the cinI and cinR mutants (Fig. 6). Biofilm rings produced by strain 8401 and the cinS, cinI, cinR, expR, raiI, and raiR mutants were quantified, confirming that the cinS, cinI, and cinR mutations enhanced the formation of biofilm rings (Fig. 6C). These enhanced biofilm rings were suppressed by cloned cinS (Fig. 6B, C), demonstrating that the enhanced biofilm rings in the cinI and cinR mutants are due to a lack of cinS expression, rather than a lack of CinI-made AHLs. The enhanced biofilm ring phenotype is independent of the raiIR quorum-sensing system, because raiI or raiR mutants did not produce an enhanced biofilm ring (Fig. 6C). The expR mutant A702 also produced an enhanced biofilm ring (Fig. 6), but it was less stable than that seen in the other mutants, resulting in high variation in the levels quantified (Fig. 6C); the enhanced biofilm in strain A702 (expR) was not suppressed by cloned cinS (Fig. 6C).
FIG. 6.
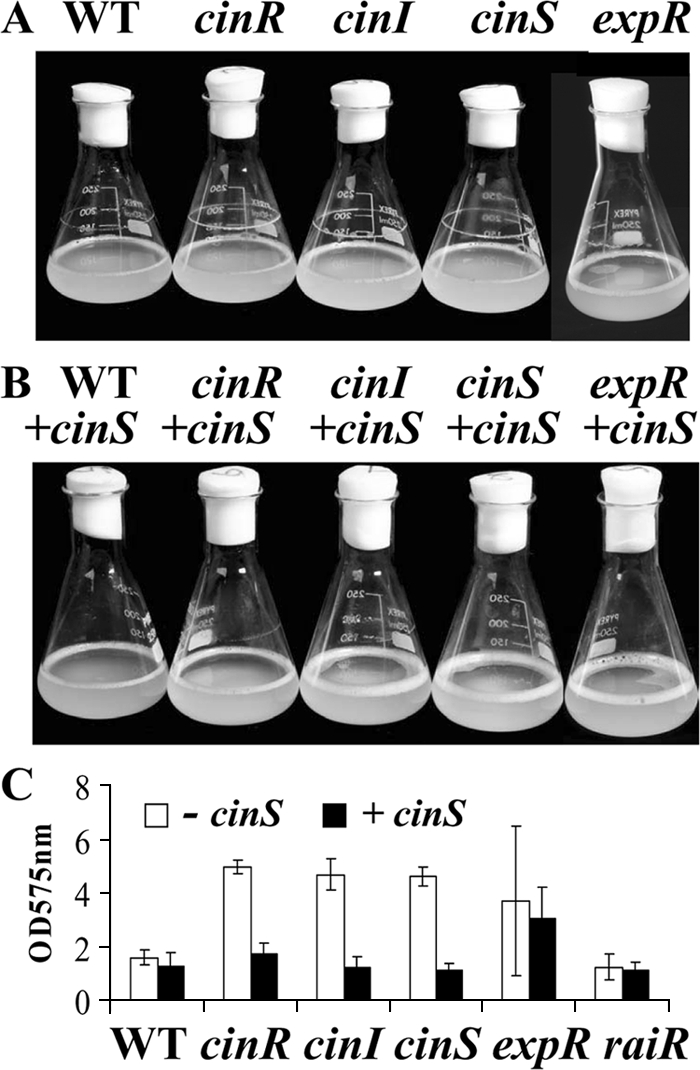
Effects of cloned cinS on biofilm rings produced by quorum-sensing mutants. (A) Biofilm rings formed at the air-liquid interface of shaking flask cultures of strains grown for 5 days in Y-mannitol medium are shown. Strains A552 (cinR), A740 (cinI), A1102 (cinS), and A702 (expR) produced enhanced biofilm rings compared to that seen with 8401 (WT), although the biofilm formed by strain A702 was less stable and tended to be shed into the growth medium. (B) Cloned cinS (on pIJ9692) suppressed the formation of the enhanced biofilm rings in the cin mutants. (C) Biofilm rings similar to those illustrated above were quantified by staining with crystal violet following growth of strain 8401 (WT), A552 (cinR), A740 (cinI), A1102 (cinS), A702 (expR), and A802 (raiR) carrying the pKT230 vector (absence of cinS [− cinS]) (open bars) or cinS on pIJ9692 (cinS present [+ cinS]) (black bars). Standard deviations (n = 3) are indicated by the error bars.
cinS regulates swarming in R. etli.
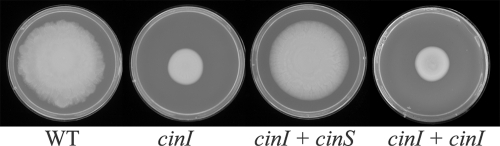
The predicted CinS sequence was used to search for related proteins using a TBLASTN search of the translated GenBank and EMBL DNA sequence databases. Closely related (several unannotated) proteins (Fig. 2B) were found in R. leguminosarum bv. viciae 3841, R. leguminosarum bv. trifolii strains WSM1325 and WSM2304, Rhizobium etli strains CFN42, CNPAF512, and CIAT652, and Mesorhizobium tianshanense (Fig. 2B). In each case, the coding region was immediately downstream from and apparently translationally coupled to cinI-like genes. In R. etli, mutation of cinR or cinI was previously shown to abolish swarming; the full swarming phenotype was not restored by the addition of the AHL made by CinI in that strain, but the AHL did induce a ruffling effect on the edge of the colony (10). On the basis of our results, it seemed likely that the cinI mutation was polar on cinS, which had not been recognized in that work as a separate gene cotranscribed with cinI. As shown (Fig. 7), cloned cinS (from R. leguminosarum strain 8401) fully restored swarming to the R. etli cinI mutant, but cloned cinI did not. This shows that the swarming must be regulated by CinS and not, as previously concluded (10) by contact with CinI-made AHLs; in addition, it shows that the mutation in cinI is polar on cinS. The R. etli cinI mutant carrying cloned cinS (pIJ9692) produced a swarm with an even edge, whereas the WT strain produced a swarm with an uneven edge (Fig. 7). Since the major difference is the presence of CinI-made AHLs in the WT, but not the mutant complemented with cinS, it is probable that the CinI-made AHLs induce the uneven edge on the swarming colony. We tested R. leguminosarum swarming under similar conditions but were unable to show cinI-dependent swarming clearly, and we did not see any difference between the control strain 8401 and the cinI, cinR, or cinS mutant derivatives of this strain, so an equivalent test of cloned cinS on swarming was not possible in that strain.
FIG. 7.
Swarming by an R. etli cinI mutant was complemented by cinS, but not cinI. R. etli CNPAF512 (WT), FAJ4006 (cinI), and FAJ4006 containing cloned cinS on pIJ9692 (cinI + cinS) or cloned cinI on pIJ9655 (cinI + cinI) were spot inoculated onto the surfaces of YEM swarm plates, revealing that cinS, but not cinI, restored swarming to FAJ4006.
DISCUSSION
In Rhizobium spp., cinI-cinR-determined quorum-sensing regulation is at the top of a hierarchy of regulation that includes the induction of the raiI-raiR genes (9, 52, 53). It is evident from the work here that the CinI-CinR-mediated effect also requires both the LuxR-type regulator ExpR and cinS, a small gene immediately downstream of, and in the same operon as, cinI. It is clearly predicted that cinS encodes a protein, based on (i) the predicted translational coupling to cinI (which is also observed with all the orthologues shown in Fig. 2B), and (ii) the highly conserved amino acid sequences downstream of the variable N-terminal region (Fig. 2B).
The effect of the hierarchical regulation is to induce the expression of raiR in a population density-dependent manner, and a consequence of this is the increased expression of raiI and of AHLs produced by RaiI. The observations that cloned cinS can restore the decreased raiR expression in an expR mutant but that cloned expR cannot restore raiR expression in the cinS mutant indicate that CinS acts downstream of ExpR. ExpR is not required for the expression of cinS, because mutation of expR did not affect the expression of the promoter of the cinIS operon.
The expR gene identified here is homologous to expR from S. meliloti. The two predicted proteins are about 58% identical, and both are LuxR-type regulators encoded by genes located between the ndvA and pyc genes. In S. meliloti strain 1021, which is the most widely used strain for genetic studies, expR is interrupted by an endogenous IS element, a mutation which prevents the formation of an exopolysaccharide called EPS-II (35). ExpR induces the expression of EPS biosynthetic genes in response to C16:1-HSL, produced by SinI (32, 34). Mutation of expR or sinI, both of which are required for the production of EPS-II, alter the colony morphology of strains, and expR is required for full sinI induction, which also requires the LuxR-type regulator SinR- and SinI-made AHLs (32, 34). Whereas expR in S. meliloti is required for full sinI expression, expR in R. leguminosarum is not required for full cinI expression. Furthermore, it appears likely that, although it appears to have an AHL-binding domain, ExpR in R. leguminosarum can act independently of AHLs because, in a cinI raiI double mutant lacking detectable AHLs, cloned cinS restored full expression of plyB, which is ExpR regulated.
R. leguminosarum produces no polysaccharide equivalent to ExpR-regulated EPS-II made by S. meliloti, and the colony morphology of the R. leguminosarum expR mutant was very similar to the WT. In addition, CinI and CinR from R. leguminosarum show only about 30% identity to SinI and SinR from S. meliloti, as might be expected from nonorthologous AHL synthases and LuxR-type regulators. R. leguminosarum produces an acidic EPS structurally very different from the succinoglycan or EPS-II produced by S. meliloti (3), so there is not a clearly homologous target that ExpR is likely to regulate. The expR and cinS genes are needed for normal induction of plyB, which encodes a secreted glycanase (15, 16) that cleaves the acidic EPS (56). Since mutations in raiI and raiR had no effect on plyB expression, the raiR and plyB genes must be regulated in parallel, rather than plyB being regulated via raiR. There is a difference between raiR and plyB regulation, because although cloned cinS can restore raiR expression in an expR mutant, it cannot restore plyB expression in the expR mutant. The lack of cinS restoration of plyB expression in the expR mutant is consistent with the observation that the colony collapse phenotype induced by cloned cinS is absent or delayed in the plyB or expR mutants, possibly implying a more direct role for ExpR in plyB expression compared with raiR expression.
Clearly, cinS can influence raiR expression and presumably other genes regulated by RaiR and surface EPS via its effects on plyB induction. Mutation of plyB or cinI in R. etli abolished swarming (5, 10) and since we have shown (Fig. 7) that the cinI mutation in R. etli is polar on cinS, this would be consistent with our proposed model of CinS playing a role in levels of plyB expression. Our data are not consistent with the proposal (10) that CinI-made AHLs have a direct effect on the extent of swarming by acting as a surfactant to reduce surface tension. It does appear that CinI-made AHLs may directly affect the characteristics of the swarm rather than its extent, because the swarming pattern in the cinI mutant carrying cloned cinS looked qualitatively different from that of the WT. It remains to be seen how many genes are regulated by CinS; in S. meliloti, ExpR and the sinIR quorum-sensing system regulate genes involved in motility and production of surface polysaccharides (24, 34).
We have not identified how CinS exerts its function. It is clear that, once expressed, CinS enhances plyB and raiR expression in the absence of CinI or RaiI-made AHLs, because cloned cinS restored plyB and raiR expression in a raiI cinI double mutant. Therefore, although CinI-made AHLs are normally required for cinS induction, they are not required for its activity, even though cinI and cinS are predicted to be translationally coupled in all strains in which they have been identified. There are no clear homologues of cinS outside the rhizobia, so it is difficult to predict how it acts. The most likely options are that CinS acts as a DNA- or RNA-binding protein that can enhance gene expression or RNA levels. There are several RNA-binding proteins that can influence quorum-sensing regulation in other bacteria (18, 22, 46, 48), sometimes circumventing the quorum-sensing regulatory system (21). However, there is also evidence for small DNA-binding proteins playing a role in quorum-sensing regulation (30) and a small quorum-sensing-induced protein (DegQ) that stimulates phosphotransfer to a transcriptional regulator that affects motility and biofilm formation (26). At this stage, we cannot easily determine whether any of these possibilities is likely, because CinS shows no similarity to any of these known regulators.
Acknowledgments
We thank our colleagues Jim Lithgow, Adam Wilkinson, Vittoria Danino, Alan Williams, Fang Xie, and Daniela Russo for helpful discussions, and we thank Paul Williams and Siri Chhabra for making AHL standards available.
This work was supported in part by the Biotechnology and Biological Sciences Research Council via a grant-in-aid, a studentship (to J.J.), a grant (208/BRE13665) under the BIRE initiative, an award from CERES, and a contract (QLK3-CT-2000-31795) and a Marie Curie EST training award (019727) from the European Union.
Footnotes
Published ahead of print on 6 March 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors: 2 broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16237-247. [DOI] [PubMed] [Google Scholar]
- 3.Becker, A., and A. Puhler. 1998. Production of exopolysaccharides, p. 97-118. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Kluwer Academic, Dordrecht, The Netherlands.
- 4.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84188-198. [DOI] [PubMed] [Google Scholar]
- 5.Braeken, K., R. Daniels, K. Vos, M. Fauvart, D. Bachaspatimayum, J. Vanderleyden, and J. Michiels. 2008. Genetic determinants of swarming in Rhizobium etli. Microb. Ecol. 5554-64. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan-Wollaston, A. V. 1979. Generalised transduction in Rhizobium leguminosarum. J. Gen. Microbiol. 112135-142. [Google Scholar]
- 7.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 111119-1129. [DOI] [PubMed] [Google Scholar]
- 8.Cubo, M. T., A. Economou, G. Murphy, A. W. Johnston, and J. A. Downie. 1992. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J. Bacteriol. 1744026-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277462-468. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, R., S. Reynaert, H. Hoekstra, C. Verreth, J. Janssens, K. Braeken, M. Fauvart, S. Beullens, C. Heusdens, I. Lambrichts, D. E. De Vos, J. Vanderleyden, J. Vermant, and J. Michiels. 2006. Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc. Natl. Acad. Sci. USA 10314965-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels, R., J. Vanderleyden, and J. Michiels. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28261-289. [DOI] [PubMed] [Google Scholar]
- 12.Danino, V. E., A. Wilkinson, A. Edwards, and J. A. Downie. 2003. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 50511-525. [DOI] [PubMed] [Google Scholar]
- 13.Downie, J. A., and J. E. Gonzalez. 2008. Cell-to-cell communication in rhizobia: quorum sensing and plant signalling, p. 213-232. In S. C. Winans and B. L. Bassler (ed.), Chemical communication among bacteria. ASM Press, Washington, DC.
- 14.Downie, J. A., C. D. Knight, A. W. B. Johnston, and L. Rossen. 1985. Identification of genes and gene-products involved in the nodulation of peas by Rhizobium leguminosarum. Mol. Gen. Genet. 198255-262. [Google Scholar]
- 15.Finnie, C., N. M. Hartley, K. C. Findlay, and J. A. Downie. 1997. The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol. Microbiol. 25135-146. [DOI] [PubMed] [Google Scholar]
- 16.Finnie, C., A. Zorreguieta, N. M. Hartley, and J. A. Downie. 1998. Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapeptide repeat motif. J. Bacteriol. 1801691-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenn, S. A., N. Gurich, M. A. Feeney, and J. E. Gonzalez. 2007. The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J. Bacteriol. 1897077-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez, N., S. Heeb, C. Valverde, E. Kay, C. Reimmann, T. Junier, and D. Haas. 2008. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, V., R. I. Santamaria, P. Bustos, I. Hernandez-Gonzalez, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramirez, V. Jimenez-Jacinto, J. Collado-Vides, and G. Davila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 1033834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray, K. M., J. P. Pearson, J. A. Downie, B. E. Boboye, and E. P. Greenberg. 1996. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of a stationary phase and rhizosphere-expressed genes. J. Bacteriol. 178372-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer, B. K., and B. L. Bassler. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA 10411145-11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heeb, S., S. A. Kuehne, M. Bycroft, S. Crivii, M. D. Allen, D. Haas, M. Camara, and P. Williams. 2006. Functional analysis of the post-transcriptional regulator RsmA reveals a novel RNA-binding site. J. Mol. Biol. 3551026-1036. [DOI] [PubMed] [Google Scholar]
- 23.Hoang, H. H., A. Becker, and J. E. González. 2004. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J. Bacteriol. 1865460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoang, H. H., N. Gurich, and J. E. González. 2008. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J. Bacteriol. 190861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight, C. D., L. Rossen, J. G. Robertson, B. Wells, and J. A. Downie. 1986. Nodulation inhibition by Rhizobium leguminosarum multicopy nodABC genes and analysis of early stages of plant infection. J. Bacteriol. 166552-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66395-409. [DOI] [PubMed] [Google Scholar]
- 27.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1mcs, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 28.Lamb, J. W., J. A. Downie, and A. W. Johnston. 1985. Cloning of the nodulation (nod) genes of Rhizobium phaseoli and their homology to R. leguminosarum nod DNA. Gene 34235-241. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, J. W., G. Hombrecher, and A. W. B. Johnston. 1982. Plasmid-determined nodulation and nitrogen-fixation abilities in Rhizobium phaseoli. Mol. Gen. Genet. 186449-452. [Google Scholar]
- 30.Lenz, D. H., and B. L. Bassler. 2007. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol. Microbiol. 63859-871. [DOI] [PubMed] [Google Scholar]
- 31.Lithgow, J. K., A. Wilkinson, A. Hardman, B. Rodelas, F. Wisniewski-Dye, P. Williams, and J. A. Downie. 2000. The regulatory locus cinRI in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 3781-97. [DOI] [PubMed] [Google Scholar]
- 32.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. González. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1433703-3711. [DOI] [PubMed] [Google Scholar]
- 34.McIntosh, M., E. Krol, and A. Becker. 2008. Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 1905308-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pellock, B. J., M. Teplitski, R. P. Boinay, W. D. Bauer, and G. C. Walker. 2002. A LuxR homolog controls production of symbiotically active extracellular polysaccharide II by Sinorhizobium meliloti. J. Bacteriol. 1845067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 37.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 12715-21. [DOI] [PubMed] [Google Scholar]
- 38.Rodelas, B., J. K. Lithgow, F. Wisniewski-Dye, A. Hardman, A. Wilkinson, A. Economou, P. Williams, and J. A. Downie. 1999. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 1813816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosemeyer, V., J. Michiels, C. Verreth, and J. Vanderleyden. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 180815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sharma, S. B., and E. R. Signer. 1990. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 4344-356. [DOI] [PubMed] [Google Scholar]
- 42.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 946036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherwood, M. T. 1970. Improved synthetic medium for the growth of Rhizobium. J. Appl. Bacteriol. 33708-713. [DOI] [PubMed] [Google Scholar]
- 44.Sourjik, V., P. Muschler, B. Scharf, and R. Schmitt. 2000. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 182782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spaink, H. P., C. A. Wijffelman, E. Pees, R. J. H. Okker, and B. J. J. Lugtenberg. 1987. Rhizobium nodulation gene nodD as a determinant of host specificity. Nature 328337-340. [Google Scholar]
- 46.Svenningsen, S. L., C. M. Waters, and B. L. Bassler. 2008. A negative feedback loop involving small RNAs accelerates Vibrio cholerae transition out of quorum-sensing mode. Genes Dev. 22226-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorne, S. H., and H. D. Williams. 1999. Cell density-dependent starvation survival of Rhizobium leguminosarum bv. phaseoli: identification of the role of an N-acyl homoserine lactone in adaptation to stationary-phase survival. J. Bacteriol. 181981-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu, K. C., and B. L. Bassler. 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 21221-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25365-404. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson, A., V. Danino, F. Wisniewski-Dye, J. K. Lithgow, and J. A. Downie. 2002. N-acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 1844510-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winans, S. C. 2008. Cell-cell signalling within crown gall tumors, p. 291-306. In S. C. Winans and B. L. Bassler (ed.), Chemical communication among bacteria. ASM Press, Washington, DC.
- 52.Wisniewski-Dye, F., and J. A. Downie. 2002. Quorum-sensing in Rhizobium. Antonie van Leeuwenhoek 81397-407. [DOI] [PubMed] [Google Scholar]
- 53.Wisniewski-Dyé, F., J. Jones, S. R. Chhabra, and J. A. Downie. 2002. raiIR genes are part of a quorum-sensing network controlled by cinI and cinR in Rhizobium leguminosarum. J. Bacteriol. 1841597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, J. P. W., L. C. Crossman, A. W. B. Johnston, N. R. Thomson, Z. F. Ghazoui, K. H. Hull, M. Wexler, A. R. J. Curson, J. D. Todd, P. S. Poole, T. H. Mauchline, A. K. East, M. A. Quail, C. Churcher, C. Arrowsmith, I. Cherevach, T. Chillingworth, K. Clarke, A. Cronin, P. Davis, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, M. Sanders, M. Simmonds, S. Whitehead, and J. Parkhill. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biology 7R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 1823885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zorreguieta, A., C. Finnie, and J. A. Downie. 2000. Extracellular glycanases of Rhizobium leguminosarum are activated on the cell surface by an exopolysaccharide-related component. J. Bacteriol. 1821304-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]