Abstract
The presence and assortment of 16 known virulence/resistance genetic determinants carried by prophages or prophage-like elements were tested in 212 clinical group A Streptococcus (GAS) strains and related to available data from SmaI macrorestriction/pulsed-field gel electrophoresis analysis and emm typing. A strong correlation existed among the three analyses. This finding supports the substantial contribution to the evolution and diversification of the GAS genome attributed to phages.
Group A Streptococcus (GAS) (Streptococcus pyogenes) is a gram-positive bacterial pathogen that can either asymptomatically colonize the upper respiratory tract and skin or cause a wide variety of diseases ranging from relatively mild infections such as pharyngitis, tonsillitis, and impetigo to more severe forms of invasive diseases, like streptococcal toxic shock syndrome, necrotizing fasciitis, and myositis, and the postinfection sequelae glomerulonephritis and rheumatic fever (6).
Many differences between GAS strains are dependent on the presence of mobile genetic elements such as plasmids, bacteriophages, transposons, and insertion sequences (1). Although prophages, prophage-like elements, and other exogenous sequences comprise a small fraction of the GAS genome, they may account for up to 70% of the variation in the gene makeup of GAS strains. Lysogenic bacteriophages are very common in the genomes of sequenced GAS strains, and many of these strains are polylysogenic (2). Streptococcal phages are widely considered to be potential contributors to virulence and resistance. Moreover, the conversion from nonpathogenic to toxigenic strains of Streptococcus pyogenes can be mediated by bacteriophage infection (3, 10).
Many prophages encode one or two proven or putative extracellular virulence factors and antibiotic resistance determinants and may contribute to the expression and horizontal transfer of these genes (2). Prophage-encoded virulence factors can be divided into two groups (1). The first group, the pyrogenic toxin superantigens, consists of the streptococcal superantigen and the streptococcal pyrogenic exotoxins (SpeA, SpeC, SpeK, SpeH, SpeI, SpeM, and SpeL). The other group is composed of DNases (Spd1, Spd3, Spd4, Sda, and Sdn) and phospholipase A2 (18).
Additionally, antimicrobial resistance genes are frequently carried by mobile genetic elements, such as transposons, conjugative plasmids, and prophages (2). As a typical example, in S. pyogenes, efflux-mediated macrolide resistance is encoded by the mef(A) gene, which is carried by DNA chimeric elements composed of a prophage with an inserted transposon.
In this work, the presence of 16 “prophage-associated virulence/resistance genes” (“φ vir”) was determined in a population of 212 pharyngeal S. pyogenes strains obtained from throat cultures of children (up to 14 years old) with symptomatic pharyngotonsillitis and isolated in the context of a nationwide survey conducted in 1997 to 1998 (22). Only strains for which data concerning both the emm type and the SmaI macrorestriction analysis by PFGE were already available (19, 23) were included. The study was designed to obtain (i) data on the molecular epidemiology of φ vir patterns in S. pyogenes strains isolated from the oropharynx, (ii) additional insights into the contribution of prophages to the global diversification of the GAS population, and (iii) evidence on the correlation between the distribution pattern of φ vir genes and other classification schemes based on emm typing and SmaI macrorestriction profiling. The particular interest on pharyngeal isolates comes from recent findings on the lysogenic exchange of toxin genes among GAS strains in both cultured pharyngeal cells and an animal model of throat infection (3, 10). This niche would be favorable to genetic exchange and to the evolution of new assortment of virulence and resistance genetic determinants.
Moreover, 165 out of the 212 strains were also erythromycin resistant, as already determined by a broth microdilution method (19). During the last two decades, an overall increasing rate of erythromycin resistance among Streptococcus pyogenes clinical isolates has been reported in many countries (5). In Italy, a high prevalence of mef(A) and the associated efflux-mediated erythromycin resistance has been reported (17), which supports our interest in gaining data on the diffusion of prophage-associated resistance genes in this bacterial population.
Hence, all 212 strains were screened by a PCR-based method for the presence of the 16 known prophage-associated φ vir genes: speA, speC, speH, speI, speK, speL, speM, ssa, spd1, spd3, spd4, sdn, sda, sla, mef(A), and Tet O. Primer pairs for virulence genes were from Matsumoto et al. (16), those for Tet O were from Giovanetti et al. (11), and those used to detect mef(A) were from Zampaloni et al. (23).
The PCR conditions were the same for all amplification reactions. Briefly, each 25-μl test tube contained 1 μg of chromosomal DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 250 μM deoxynucleotide triphosphates (dNTPs), 1 μM oligonucleotide primer, and 0.5 U Taq polymerase (AmpliTaq Gold; Applied Biosystems). Thermal cycling conditions for amplification reactions were as follows: 94°C for 2 min; followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min; followed by 72°C for 7 min; and finally a 4°C hold. Two percent agarose gel electrophoresis was used to analyze the length and quality of the amplification products.
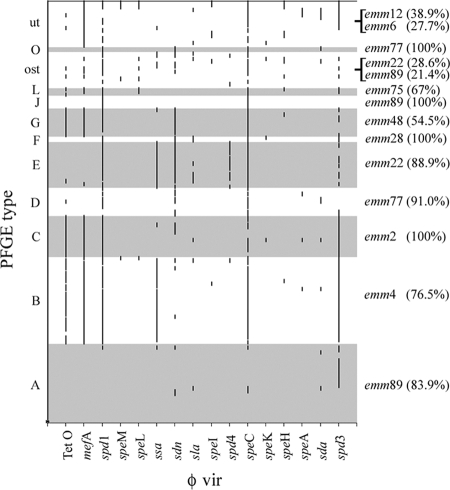
The relative distribution of φ vir genes was greatly variable, with a high prevalence of spd1 (55%), spd3 (53%), and sdn (28%) among DNases. The most represented streptococcal superantigens were ssa (30%) and speC (62%) (Fig. 1). These latter genes were also frequently detected in previous works (13, 20, 21). The incidence of the other φ vir genes ranged between 3.7 and 14.4%. In the particular case of speA, its low prevalence contrasts with the significantly higher values (30 to 50%) reported in other works (13, 15, 20, 21). The most prevalent resistance gene was mef(A) (73%) (Fig. 1). The overall mean number of phage-associated virulence/resistance genes per isolate (mφ vir) was 3.8 (± 2.1), and the analysis of their assortment led to the identification of 67 different profiles (Fig. 1). With respect to these variables, the work of Green et al., which is the only study directly comparable to the present one, even if restricted to emm28 GAS, reported an mφ vir equal to 3.6 and an overall number of 46 profiles (12). Only 50 strains analyzed here could be classified in just 11 profiles represented in the scheme of Green et al.
FIG. 1.
Schematic representation of the correlation between the distribution of φ vir genes and PFGE analysis. The positive samples are represented by dots. A continuous vertical line results from the occurrence of several contiguous strains (reported on the PFGE type axis); these are positive for the same virulence/resistance gene. On the right-hand axis, are reported the most prevalent emm types within each PFGE cluster together with their corresponding percentages.
PFGE analysis of macrorestriction patterns was possible for 189 strains (89.2%). The chromosomal DNA extracted from the remaining 23 isolates was not restricted by SmaI (untypeable). This latter subgroup was positive for mef(A) and negative for Tet O, a genotype that is associated with the presence of a modification system methylating cytosine residues in SmaI restriction sites (8, 9). Altogether, 24 different PFGE types (out of a total of 48 possible types and subtypes) were recognized (Fig. 1), of which almost 60% were recorded in only one isolate (one-strain type). Among typeable isolates, about 50% fell into three types (A, B, and E).
The comparison between the PFGE analysis and the assortment of φ vir genes showed that the second analysis had the highest discriminatory ability, at least in the set of strains examined in this study. On the other hand, Fig. 1 clearly shows the nonrandom distribution of the number and type of φ vir genes within each PFGE type. Cluster type A was composed of 31 strains, of which 14 (45.1%) were negative for all virulence genes considered. Moreover, the other 14 strains belonging to the same cluster had one virulence gene (spd3). Another example is the distribution within type cluster B with 29 strains (75%) being positive for the same set of six φ vir genes. Therefore, we uncovered a strong association between the profile of φ vir and SmaI macrorestriction PFGE clustering. This is the first observation indicating that the structure of the GAS genome as seen throughout SmaI macrorestriction depends upon polylysogeny (i.e., the presence of prophages).
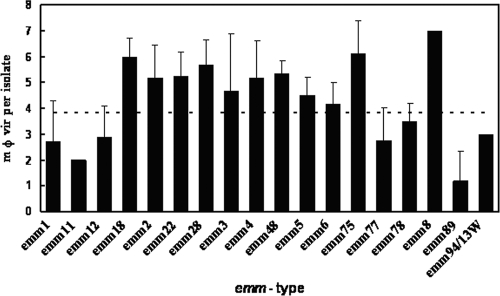
The occurrence of the more frequent emm types is reported in Fig. 2. The most represented types were emm89, emm4, emm22, emm2, and emm77. Again, in this case the association of the PFGE analysis with emm typing was evident and confirmed similar observations reported by others (4, 7), even in the case of invasive strains (14).
FIG. 2.
Mean absolute number of φ vir genes per emm type. Error bars indicate the value of the standard deviation, and the horizontal dotted line shows the mφ vir per isolate for the entire population studied.
Regarding the correlation between φ vir content/distribution and emm type, we found that the emm2, emm3, emm4, emm8, emm18, emm22, emm28, emm48, and emm75 group of strains showed high mφ vir values, ranging from 4.7 to 7.0 (Fig. 1). Notably, the emm89 group had an mφ vir of 1.2, and nearly one-third of the strains did not harbor any of the φ vir genes assayed (Fig. 2). In the set of strains studied, emm types frequently associated with invasive diseases (e.g., emm3 and emm18) showed the highest load in terms of phage-associated virulence genes. This clearly supports the hypothesis that phages may be the major factor responsible for determining the plasticity of the GAS genome as a whole and that they may also play a significant role in the evolution of the pathogenicity (1).
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Banks, D. J., S. B. Beres, and J. M. Musser. 2002. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10515-521. [DOI] [PubMed] [Google Scholar]
- 2.Beres, S. B., and J. M. Musser. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS ONE 2e800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broudy, T. B., and V. A. Fischetti. 2003. In vivo lysogenic conversion of Tox− Streptococcus pyogenes to Tox+ with lysogenic streptococci or free phage. Infect. Immun. 713782-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrico, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 442524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornaglia, G., and A. Bryskier. 2004. Macrolide resistance of Streptococcus pyogenes, vol. 3. Karger, Basel, Switzerland.
- 6.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Descheemaeker, P., F. Van Loock, M. Hauchecorne, P. Vandamme, and H. Goossens. 2000. Molecular characterisation of group A streptococci from invasive and non-invasive disease episodes in Belgium during 1993-1994. J. Med. Microbiol. 49467-471. [DOI] [PubMed] [Google Scholar]
- 8.Euler, C. W., P. A. Ryan, J. M. Martin, and V. A. Fischetti. 2007. M.SpyI, a DNA methyltransferase encoded on a mefA chimeric element, modifies the genome of Streptococcus pyogenes. J. Bacteriol. 1891044-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueiredo, T. A., S. I. Aguiar, J. Melo-Cristino, and M. Ramirez. 2006. DNA methylase activity as a marker for the presence of a family of phage-like elements conferring efflux-mediated macrolide resistance in streptococci. Antimicrob. Agents Chemother. 503689-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti, V. A. 2007. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 15297-300. [DOI] [PubMed] [Google Scholar]
- 11.Giovanetti, E., A. Brenciani, R. Lupidi, M. C. Roberts, and P. E. Varaldo. 2003. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Chemother. 472844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, N. M., S. B. Beres, E. A. Graviss, J. E. Allison, A. J. McGeer, J. Vuopio-Varkila, R. B. LeFebvre, and J. M. Musser. 2005. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J. Clin. Microbiol. 434083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jing, H.-B., B.-A. Ning, H.-J. Hao, Y.-L. Zheng, D. Chang, W. Jiang, and Y.-Q. Jiang. 2006. Epidemiological analysis of group A streptococci recovered from patients in China. J. Med. Microbiol. 551101-1107. [DOI] [PubMed] [Google Scholar]
- 14.Krucsó, B., M. Gacs, B. Libisch, Z. Hunyadi, K. Molnár, M. Füzi, and J. Pászti. 2007. Molecular characterisation of invasive Streptococcus pyogenes isolates from Hungary obtained in 2004 and 2005. Eur. J. Clin. Microbiol. Infect. Dis. 26807-811. [DOI] [PubMed] [Google Scholar]
- 15.Lorino, G., G. Gherardi, S. Angeletti, M. De Cesaris, N. Graziano, S. Maringhini, F. Merlino, F. Di Bernardo, and G. Dicuonzo. 2006. Molecular characterisation and clonal analysis of group A streptococci causing pharyngitis among paediatric patients in Palermo, Italy. Clin. Microbiol. Infect. 12189-192. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto, M., N. P. Hoe, M. Liu, S. B. Beres, G. L. Sylva, C. M. Brandt, G. Haase, and J. M. Musser. 2003. Intrahost sequence variation in the streptococcal inhibitor of complement gene in patients with human pharyngitis. J. Infect. Dis. 187604-612. [DOI] [PubMed] [Google Scholar]
- 17.Montagnani, F., L. Stolzuoli, L. Croci, C. Rizzuti, F. Arena, A. Zanchi, and C. Cellesi. 5 December 2008. Erythromycin resistance in Streptococcus pyogenes and macrolide consumption in a central Italian region. Infection. [Epub ahead of print.] doi: 10.1007/s15010-008-8023-1. [DOI] [PubMed]
- 18.Nagiec, M. J., B. Lei, S. K. Parker, M. L. Vasil, M. Matsumoto, R. M. Ireland, S. B. Beres, N. P. Hoe, and J. M. Musser. 2004. Analysis of a novel prophage-encoded group A Streptococcus extracellular phospholipase A2. J. Biol. Chem. 27945909-45918. [DOI] [PubMed] [Google Scholar]
- 19.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 765-71. [DOI] [PubMed] [Google Scholar]
- 20.Rivera, A., M. Rebollo, E. Miro, M. Mateo, F. Navarro, M. Gurgui, B. Mirelis, and P. Coll. 2006. Superantigen gene profile, emm type and antibiotic resistance genes among group A streptococcal isolates from Barcelona, Spain. J. Med. Microbiol. 551115-1123. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz, F.-J., A. Beyer, E. Charpentier, B. H. Normark, M. Schade, A. C. Fluit, D. Hafner, and R. Novak. 2003. Toxin-gene profile heterogeneity among endemic invasive European group A streptococcal isolates. J. Infect. Dis. 1881578-1586. [DOI] [PubMed] [Google Scholar]
- 22.Varaldo, P. E., E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. C. Schito, and G. Tempera. 1999. Nationwide survey in Italy of treatment of Streptococcus pyogenes pharyngitis in children: influence of macrolide resistance on clinical and microbiological outcomes. Clin. Infect. Dis. 29869-873. [DOI] [PubMed] [Google Scholar]
- 23.Zampaloni, C., P. Cappelletti, M. Prenna, L. A. Vitali, and S. Ripa. 2003. emm gene distribution among erythromycin-resistant and -susceptible Italian isolates of Streptococcus pyogenes. J. Clin. Microbiol. 411307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]