Abstract
This study reports the occurrence of “Bartonella rochalimae” in Europe and the presence of Bartonella vinsonii subsp. berkhoffii genotypes II and III in dogs in southern Italy and provides DNA sequencing evidence of a potentially new Bartonella sp. infecting dogs in Greece and Italy.
Throughout the world, Bartonella species are considered emerging pathogens in veterinary and human medicine (13, 14). Among the 11 human pathogenic Bartonella species, 7 have been isolated from dogs: B. vinsonii subsp. berkhoffii (8), B. henselae, B. clarridgeiae (4, 5), B. washoensis (3), B. elizabethae (10), B. quintana, and “B. rochalimae” (6, 7). Also, an increasing number of arthropods have been implicated in Bartonella transmission (1a, 9). Currently, there is no information about Bartonella spp. infecting dogs in Greece or Italy, countries in which ectoparasite activity is intense (11, 12). The aim of the present study was to detect and genetically characterize Bartonella spp. from naturally infected dogs in Greece and Italy.
In Italy, blood samples were collected from shelter dogs from the Apulian region—26 dogs from Bari (41°5′N, 16°5′E) and 20 dogs from Ginosa (40°3′N, 16°4′E). Additionally, blood samples and attached ticks were collected from 14 pet dogs from the Basilicata region (40°31′N, 16°06′E). In Greece, blood samples and bone marrow aspirates were collected from 50 clinically ill dogs at the Veterinary Teaching Hospital of the Aristotle University of Thessaloniki (40°38′N, 22°57′E). DNA from dog and tick samples was extracted (1, 4) and initially screened by PCR targeting a fragment of the 16S-23S intergenic transcribed spacer (ITS) region of Bartonella spp. (4). Bartonella-positive canine samples were further characterized by PCR amplification and sequencing of three different genes (16S rRNA, pap31, and rpoB genes) (4). For amplification of the 16S rRNA gene and the initial portion of the ITS region, universal forward primer 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) or 12F (5′-GCG GCA AGC YTA ACA CAT GCA AGT CGA ACG-3′), targeting the beginning of the 16S rRNA, and reverse primer 325as (5′-CGC CAG AAG GCT TGG GAT CAT CAT CTG AAG-3′), targeting the initial segment of the ITS region, were used to amplify approximately 1,700 bp. PCR was performed as described previously (2). DNA from a healthy, specific pathogen-free dog was used as a PCR negative control. B. henselae Houston-1 (GenBank accession number L35101) was used as a positive control. Amplicons were sequenced by direct sequencing or after cloning into pGem-T easy vectors (Promega, Madison, WI). Consensual DNA segments were generated (Vector NTI version 10; Invitrogen, Carlsbad, CA). The occurrence of chimeras among 16S rRNA gene clones was ruled out using the online Chimera Tool Detection program from the Ribosomal Database Project II (http://35.8.164.52/cgis/chimera.cgi?su=SSU). Final sequences were compared with others deposited in GenBank, using the BLASTN tool version 2.
The microimmunofluorescence test for the detection of anti-B. vinsonii subsp. berkhoffii (isolate 93-CO-1; ATCC 51672) and anti-B. henselae (similar to strain Houston-1) antibodies in canine sera from the Basilicata region of Italy was performed as described previously (12a). The starting dilution was 1:16, and the cutoff titer for seroreactivity was defined at 1:64.
Bartonella DNA was amplified from 2 of 50 dogs in Greece and from 7 of 60 clinically healthy dogs in Italy (Table 1). One dog from Greece (dog no. 1) was infected with B. rochalimae (ITS sequences were 100% similar [520 bp] to sequence DQ683199). Six Italian dogs and one Greek dog were infected with a novel, uncultured Bartonella sp. strain (hereafter designated strain HMD [GenBank accession number EF614393]), and of those dogs, one dog was coinfected with B. vinsonii subsp. berkhoffii genotype III (99.7% similar [677/679 bp] to sequence EU295657). Another Italian dog was infected with B. vinsonii subsp. berkhoffii genotype II (100% similar [565 bp] to sequence DQ059763). Bartonella DNA was amplified by PCR from three tick pools, as follows: one salivary gland pool from female ticks (from dog no. 3 [Table 1]), one salivary gland pool from male ticks, and one gut content pool from male ticks (both from dog no. 8). Alignment of ITS DNA sequences obtained from all three PCR-positive Italian Rhipicephalus sanguineus pools resulted in retrieval of the same sequence (GenBank accession number FJ177635), which had one base pair difference (99.8% similar [458/459 bp]) from Bartonella sp. strain HMD. Of the six Italian dogs infected with Bartonella sp. strain HMD, two had the same polymorphism (a cytosine) that was detected in ticks from the same region, whereas four dogs had the same polymorphism (a thymine) that was detected in the Greek dog.
TABLE 1.
Serology results and genetic characterization of Bartonella spp. detected in two Greek dogs and seven Italian dogs, based upon DNA amplification and sequencing of three distinct portions of the genomea
| Dog no. | Country/region | Serology titer by IFA
|
Bartonella species/strain DNA amplification by PCR of the:
|
|||
|---|---|---|---|---|---|---|
| B. henselae | B. vinsonii subsp. berkhoffii | ITS region | 16S rRNA gene | rpoB gene | ||
| 1 | Greece/Thessaloniki | NP | NP | B. rochalimae | B. rochalimae | B. rochalimae |
| 2 | Greece/Thessaloniki | NP | NP | HMD | HMD-1/HMD-2 | HMD |
| 3 | Italy/Basilicata | 64 | <16 | HMD | HMD-3 | HMD |
| 4 | Italy/Basilicata | 128 | 32 | HMD | HMD-3 | HMD |
| 5 | Italy/Basilicata | 256 | <16 | HMD and B. vinsonii subsp. berkhoffii genotype III | B. vinsonii subsp. berkhoffii genotype III | HMD and B. vinsonii subsp. berkhoffii genotype III |
| 6 | Italy/Basilicata | 64 | 32 | HMD | NP | HMD |
| 7 | Italy/Basilicata | <16 | <16 | HMD | NP | HMD |
| 8 | Italy/Basilicata | <16 | 64 | HMD | HMD-3 | HMD |
| 9 | Italy/Ginosa | NP | NP | B. vinsonii subsp. berkhoffii genotype II | NP | NP |
IFA, microimmunofluorescence test; NP, not performed; HMD, Bartonella sp. strain HMD.
ITS PCR results were confirmed by PCR amplification and sequencing of the rpoB and 16S rRNA genes (Table 1). Identical (100% similar [593/593 bp]) but unique Bartonella strain HMD rpoB gene sequences were obtained from the Italian and Greek dogs. Despite multiple attempts, pap31 gene amplification was not successful for any dog infected with Bartonella strain HMD. The pap31 gene was amplified from Italian dog no. 5, infected with B. vinsonii subsp. berkhoffii genotype III, but could not be amplified for dog no. 10 due to insufficient genomic DNA.
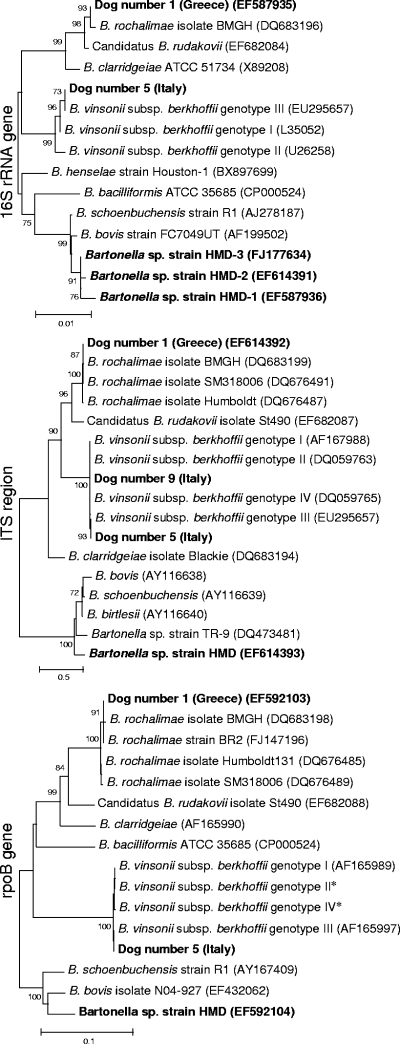
Phylogenetic analysis is presented in Fig. 1. Sequencing of multiple 16S rRNA/ITS clones of Bartonella strain HMD from Greek dog no. 2 produced two different DNA sequences (deposited as HMD-1 and HMD-2), which contained 13-bp differences within the 1,665-bp (99.2% similarity) sequence. From three Italian dogs, a third unique sequence was obtained, differing by three base pairs from strain HMD-1 and by seven base pairs from strain HMD-2 and with a deletion of tandem-repeated sequence TCTCAAGTTAATAATAAACTTG located at the beginning of the ITS region (starting at nucleotide 69); thus, these sequences were deposited as Bartonella sp. strain HMD-3 (GenBank accession number FJ177634). Despite these differences in the 16S rRNA and the initial portion of the ITS region, identical sequences were consistently obtained from clones of partial sequences of the ITS region (459 bp) and rpoB (593 bp) gene derived from all Greek and Italian dogs.
FIG. 1.
Neighbor-joining trees based on 16S rRNA gene, 16S-23S ITS region, and rpoB gene sequences of selected Bartonella spp. Sequences represented by boldface type were generated in this study from seven Italian dogs and two Greek dogs (see Table 1 for details). Values above the tree branches represent the bootstrap values calculated from 2,500 replicates. Only values of >70% are indicated. Scale bar represents the genetic distance calculated by the Kimura two-parameter method. *, sequences generated in our laboratory.
The results of this study indicate that dogs in Italy and Greece are exposed to multiple Bartonella species. This report describes the occurrence of Bartonella rochalimae in Europe and the existence of Bartonella vinsonii subsp. berkhoffii genotypes II and III in dogs from southern Italy. Importantly, a potentially new strain or species of Bartonella was identified in salivary glands of R. sanguineus and in dog blood or bone marrow samples from both countries. These results suggest that an unrecognized tick-transmitted organism may exist in this region, which could complicate the diagnosis and management of other vector-borne infections in dogs and people.
Acknowledgments
We thank Julie M. Bradley and Barbara C. Hegarty of the Intracellular Pathogens Research Laboratory, North Carolina State University, for serological testing and antigen growth and preparation and Tonya Lee for editorial assistance.
This study was supported by the State of North Carolina and in part by Bayer Animal Health and IDEXX Laboratories.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Billeter, S. A., M. K. Miller, E. B. Breitschwerdt, and M. G. Levy. 2008. Detection of two Bartonella tamiae-like sequences in Amblyomma americanum (Acari: Ixodidae) using 16S-23S intergenic spacer region-specific primers. J. Med. Entomol. 45176-179. [DOI] [PubMed] [Google Scholar]
- 1a.Billeter, S. A., M. G. Levy, B. B. Chomel, and E. B. Breitschwerdt. 2008. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med. Vet. Entomol. 221-15. [DOI] [PubMed] [Google Scholar]
- 2.Cadenas, M. B., R. G. Maggi, P. P. Diniz, K. T. Breitschwerdt, S. Sontakke, and E. B. Breitschwerdt. 2007. Identification of bacteria from clinical samples using Bartonella alpha-Proteobacteria growth medium. J. Microbiol. Methods 71147-155. [DOI] [PubMed] [Google Scholar]
- 3.Chomel, B. B., A. C. Wey, and R. W. Kasten. 2003. Isolation of Bartonella washoensis from a dog with mitral valve endocarditis. J. Clin. Microbiol. 415327-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diniz, P. P., R. G. Maggi, D. S. Schwartz, M. B. Cadenas, J. M. Bradley, B. Hegarty, and E. B. Breitschwerdt. 2007. Canine bartonellosis: serological and molecular prevalence in Brazil and evidence of coinfection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii. Vet. Res. 38697-710. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie, T. N., R. J. Washabau, M. H. Goldschmidt, J. M. Cullen, A. R. Rogala, and E. B. Breitschwerdt. 2003. Detection of Bartonella henselae and Bartonella clarridgeiae DNA in hepatic specimens from two dogs with hepatic disease. J. Am. Vet. Med. Assoc. 22247-51. [DOI] [PubMed] [Google Scholar]
- 6.Henn, J. B., M. W. Gabriel, R. W. Kasten, R. N. Brown, J. E. Koehler, K. A. MacDonald, M. D. Kittleson, W. P. Thomas, and B. B. Chomel. 2009. Infective endocarditis in a dog and the phylogenetic relationship of the associated “Bartonella rochalimae” strain with isolates from dogs, gray foxes, and a human. J. Clin. Microbiol. 47787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henn, J. B., M. W. Gabriel, R. W. Kasten, R. N. Brown, J. H. Theis, J. E. Foley, and B. B. Chomel. 2007. Gray foxes (Urocyon cinereoargenteus) as a potential reservoir of a Bartonella clarridgeiae-like bacterium and domestic dogs as part of a sentinel system for surveillance of zoonotic arthropod-borne pathogens in northern California. J. Clin. Microbiol. 452411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kordick, D. L., B. Swaminathan, C. E. Greene, K. H. Wilson, A. M. Whitney, S. O'Connor, D. G. Hollis, G. M. Matar, A. G. Steigerwalt, G. B. Malcolm, P. S. Hayes, T. L. Hadfield, E. B. Breitschwerdt, and D. J. Brenner. 1996. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int. J. Syst. Bacteriol. 46704-709. [DOI] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Mexas, A. M., S. I. Hancock, and E. B. Breitschwerdt. 2002. Bartonella henselae and Bartonella elizabethae as potential canine pathogens. J. Clin. Microbiol. 404670-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papadopoulos, B., P. C. Morel, and A. Aeschlimann. 1996. Ticks of domestic animals in the Macedonia region of Greece. Vet. Parasitol. 6325-40. [DOI] [PubMed] [Google Scholar]
- 12.Rinaldi, L., G. Spera, V. Musella, S. Carbone, V. Veneziano, A. Iori, and G. Cringoli. 2007. A survey of fleas on dogs in southern Italy. Vet. Parasitol. 148375-378. [DOI] [PubMed] [Google Scholar]
- 12a.Solano-Gallego, L., J. Bradley, B. Hegarty, B. Sigmon, and E. Breitschwerdt. 2004. Bartonella henselae IgG antibodies are prevalent in dogs from southeastern USA. Vet. Res. 35585-595. [DOI] [PubMed] [Google Scholar]
- 13.Vorou, R. M., V. G. Papavassiliou, and S. Tsiodras. 2007. Emerging zoonoses and vector-borne infections affecting humans in Europe. Epidemiol. Infect. 1351231-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wormser, G. P. 2007. Discovery of new infectious diseases—Bartonella species. N. Engl. J. Med. 3562346-2347. [DOI] [PubMed] [Google Scholar]