Abstract
Molecular methods are increasingly used to identify pathogens that are difficult to cultivate. We report a case of disseminated infection with “Mycobacterium tilburgii,” a proposed species that has never been cultivated. The case illustrates the diagnostic utility of sequence analysis of the 16S rRNA gene directly from clinical specimens.
Molecular methods are increasingly used to detect pathogens that are difficult to cultivate or have thus far not been cultivated. Amplification and sequencing of the 16S rRNA gene directly from clinical specimens can produce data that enable rapid detection of such organisms and their identification to the species level (2, 5). “Mycobacterium tilburgii” is a proposed species that has not been cultivable and has been reported in only four previous patients. It was first described by Buiting et al. in a previously healthy woman with acid-fast smear-positive infection of her bowels and bladder (1, 9). The organism had a unique 16S rRNA gene sequence and was named for the Dutch city in which it was first detected, Tilburg. The only other published cases are those of patients with human immunodeficiency virus (HIV)/AIDS who developed enteritis (8, 9) and pulmonary nodules (6) due to noncultured mycobacterial species. Comparison of the 16S rRNA gene sequences against those in GenBank identified the organisms in all three cases as “M. tilburgii”.
We report an additional case of disseminated “M. tilburgii” infection in an HIV-negative patient seen at the National Institutes of Health (NIH).
A 53-year-old man from New Hampshire with a history of sarcoidosis and atherosclerotic cardiovascular disease was admitted to the NIH for evaluation of abdominal pain and diarrhea.
The patient had undergone a partial right lower lobectomy 13 years earlier for pulmonary nodules diagnosed histologically as sarcoidosis. He was treated with prednisone for 1 year with resolution of the residual nodules. He also had recurrent esophageal strictures treated with endoscopic dilation.
The patient was in stable health until a year prior to admission, when he developed daily episodes of severe nonradiating periumbilical abdominal pain and intermittent watery diarrhea with occasional streaks of blood. Esophagogastroduodenoscopy and colonoscopy findings were normal, but small bowel enteroscopy demonstrated yellow duodenal and jejunal plaques. Acid-fast smear of the biopsy samples of the plaques revealed numerous acid-fast bacilli, but cultures were negative.
He was treated empirically for disseminated Mycobacterium avium complex infection with clarithromycin, ethambutol, and rifabutin with transient improvement of his symptoms. A computed tomography (CT) scan performed for recurrent abdominal pain 3 months before admission to the NIH demonstrated a mesenteric abscess. A needle biopsy yielded pus that had a positive acid-fast smear but was again culture negative. Following the biopsy, he was treated with clarithromycin, rifabutin, and amikacin. His CD4+ T-cell counts ranged from 140 to 350/μl (normal, >400/μl). He was repeatedly negative for HIV type 1 (HIV-1) and HIV-2 by enzyme-linked immunosorbent assay. He had lost more than 14 kg over a year. The patient had fatigue but no fevers, chills, or night sweats. He had never traveled outside North America and had a past history of heavy tobacco and alcohol use.
On examination at NIH, he was afebrile with normal vital signs. He appeared well nourished but mildly ill. He was barrel chested and had diffusely decreased breath sounds with scattered expiratory wheezes. There was moderate tenderness to palpation at the left periumbilical region with no palpable masses or liver edge. The spleen was palpable just below the left costal margin. Hemoccult-positive brown stool was in the rectal vault. He had mild peripheral clubbing and leukonychia. Cardiac, neurologic, and lymph node examination results were normal.
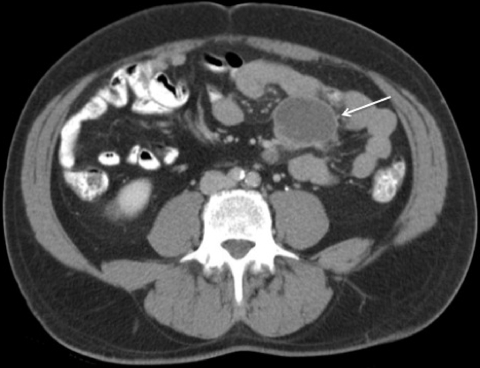
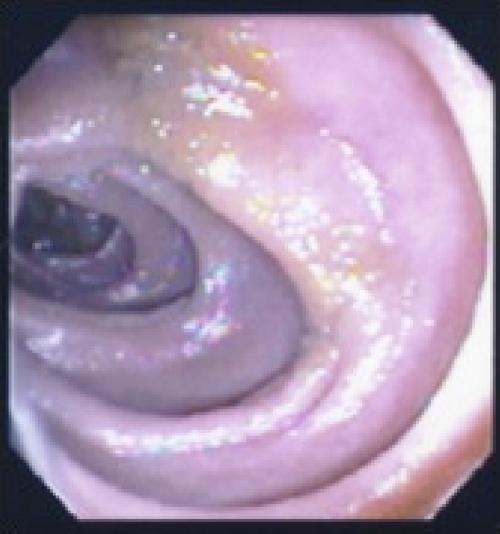
His erythrocyte sedimentation rate was 22 mm/h. His white blood cell count was 5,600/μl. He was negative for HIV-1 and -2 by enzyme-linked immunosorbent assay. A CT scan showed several large, low-density, ring-enhancing masses in the mesentery, the largest of which was 5.1 by 3.6 cm and involved the small bowel wall (Fig. 1). Upper endoscopy with push enteroscopy found scattered yellow plaques in the jejunum and duodenum (Fig. 2). After failed percutaneous drainage, the patient underwent a laparotomy. He had a thickened, nodular jejunal mucosa with matted purulent mesenteric lymph nodes, the largest of which measured 4.5 cm. A partial small bowel resection, including removal of the associated mesentery, was performed.
FIG. 1.
Abdominal CT scan of our patient, demonstrating the largest of several mesenteric masses (arrow), measuring 5.1 by 3.6 cm.
FIG. 2.
Endoscopic photograph of the jejunal wall, which had scattered yellow plaques.
Biopsy samples from the small bowel abscesses showed granulomatous changes and numerous acid-fast organisms. Cultures remained sterile. The patient was treated with clarithromycin, ethionamide, moxifloxacin, and subcutaneous gamma interferon (50 μg/m2 three times weekly). His symptoms and CT findings resolved over several months. CT scans 1, 2, and 3 years postoperatively showed continued resolution of his abscesses. After 2 years, gamma interferon was discontinued and the patient has remained well on secondary prophylaxis with moxifloxacin and clarithromycin.
A total of 14 clinical specimens were submitted for mycobacterial culture (two each of blood, sputum, and a mesenteric abscess; one each of urine, stool, and a left foot ulcer; and biopsy samples of the colon, small bowel, duodenum, skin, and a mesenteric lymph node). Variable numbers of acid-fast bacilli were seen, by auramine-rhodamine staining (Becton Dickinson, Sparks, MD), on the direct smears of the colonic, jejunal, and duodenal biopsy samples and of the lymph node and mesenteric abscess materials.
All samples were plated on BACTEC 12B, Middlebrook 7H11 agar, and Mitchison 7H11 agar (Middlebrook 7H11 with carbenicillin, polymyxin B, amphotericin B, and trimethoprim; Remel, Lenexa, KS). In an attempt to isolate fastidious mycobacterial species known to have specific nutritional requirements, material from some specimens was inoculated onto one or more of the following media: Lowenstein-Jensen slants, Herrold's egg yolk agar with mycobactin J, sheep blood agar, horse blood agar, and charcoal yeast extract agar (Remel). Multiple BACTEC 12B vials were also inoculated for each specimen; various supplements were added to individual vials, including reconstituting fluid (BD), egg yolk (BD), mycobactin J (Allied Monitor, Fayette, MO), and hemin solution (Sigma, St. Louis, MO). Solid media were incubated either at 30°C in ambient air or at 35°C in 5 to 8% CO2; BACTEC 12B vials were incubated at both 30 and 35°C. Cultures were incubated for 10 to 16 weeks and remained negative, except for single colonies of Mycobacterium mucogenicum, Mycobacterium gordonae, and Acremonium species from the colonic biopsy sample. These organisms were not considered pathogens in this patient, given the source, the species, and the fact that only a single colony of each was isolated.
For DNA extraction, amplification, and sequence analysis, organismal DNA was extracted from mesenteric abscess fluid by a modification of a previously described procedure (4). Briefly, the fluid sample was lysed in guanidinium thiocyanate buffer. The DNA was extracted with phenol-chloroform-isoamyl alcohol, purified with the Gene Clean II kit (MP Biomedicals, Solon, OH), and eluted with Tris-EDTA buffer.
A 1,236-bp fragment of the 16S rRNA gene was amplified and sequenced as previously described (3, 4). Sequences were assembled by SeqMan II software (DNAstar, Madison, WI). Related sequences were identified by the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information, Bethesda, MD). No material other than the abscess fluid was available for molecular analysis.
Numerous specimens from this patient were positive for acid-fast bacilli, but no pathogens could be isolated despite the use of a variety of culture methodologies. The sequence of the 1,236-bp region of the 16S rRNA gene amplified from the abscess material contained no ambiguous bases and showed 100% similarity to the sequence of “M. tilburgii,” a proposed species submitted to GenBank in 1995 (GenBank accession no. Z50172). Given the perfect sequence match and the failure of any likely pathogen to grow from this patient's specimens, it is most probable that “M. tilburgii” was the cause of his infection.
“M. tilburgii” has been reported four times previously as a human pathogen. The original report described a patient with cutaneous anergy but no other evidence of immune compromise (1). The three subsequent cases occurred in profoundly immunosuppressed patients with HIV/AIDS. Our patient is similar to the one in the original report; he had no known immune defect, the infection was visceral, and therapy was successful. It is possible that his earlier diagnosis of sarcoidosis was due to an abnormal immune reaction to this organism. However, his improvement with steroids makes this unlikely. Steroid therapy could have put him at risk for acquiring this infection, but that seems improbable, as it preceded the infection by many years.
The molecular study of organisms difficult or impossible to cultivate, or difficult to identify by phenotypic testing, has led to the recognition of many new microbial species, including new species of mycobacteria (7). “M. tilburgii” is representative of organisms currently identifiable only through molecular techniques.
There are at least two cogent reasons for thinking that “M. tilburgii” was, in fact, a pathogen in this case. First, numerous acid-fast organisms were seen in the biopsy samples and abscess material, so one would have predicted the pathogen to be a species of Mycobacterium. Second, the “M. tilburgii” sequence has only been reported four times previously and only from human patient material, not from animal or environmental sources.
In addition to providing a fifth report of “M. tilburgii” infection, this case illustrates the diagnostic utility of organismal DNA extraction from clinical lesions, particularly those in which organisms can be visualized but from which no likely pathogen can be cultivated, followed by amplification and sequencing of an appropriate genetic target.
Acknowledgments
This research was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases and the National Institutes of Health Clinical Center.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Buiting, A., M. Vos, A. M. Bergmans, and L. Schhouls. 1995. A new mycobacterial species causing a disseminated infection, abstr. K45. In Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 2.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conville, P. S., J. M. Brown, A. G. Steigerwalt, J. W. Lee, D. E. Byrer, V. L. Anderson, S. E. Dorman, S. M. Holland, B. Cahill, K. C. Carroll, and F. G. Witebsky. 2003. Nocardia veterana as a pathogen in North American patients. J. Clin. Microbiol. 412560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiratisin, P., L. Li, P. R. Murray, and S. H. Fischer. 2003. Identification of bacteria recovered from clinical specimens by 16S rRNA gene sequencing. Eur. J. Clin. Microbiol. Infect. Dis. 22628-631. [DOI] [PubMed] [Google Scholar]
- 6.Kolditz, M., M. Halank, P. Spornraft-Ragaller, H. Schmidt, and G. Hoffken. 2005. Localized pulmonary infection associated with Mycobacterium tilburgii in an HIV-infected patient. Infection 33278-281. [DOI] [PubMed] [Google Scholar]
- 7.Pauls, R. J., C. Y. Turenne, J. N. Wolfe, and A. Kabani. 2003. A high proportion of novel mycobacteria species identified by 16S rDNA analysis among slowly growing AccuProbe-negative strains in a clinical setting. Am. J. Clin. Pathol. 120560-566. [DOI] [PubMed] [Google Scholar]
- 8.Richter, E., S. Rusch-Gerdes, S. Niemann, A. Stoehr, and A. Plettenberg. 2000. Detection, identification, and treatment of a novel, non-cultivable Mycobacterium species in an HIV patient. AIDS 141667-1668. [DOI] [PubMed] [Google Scholar]
- 9.Wagner, D., M. Vos, A. G. Buiting, A. Serr, A. M. Bergmans, W. Kern, and L. M. Schouls. 2006. “Mycobacterium tilburgii” infections. Emerg. Infect. Dis. 12532-534. [DOI] [PMC free article] [PubMed] [Google Scholar]