Abstract
Enumerating Aspergillus fumigatus CFU can be challenging since CFU determination by plate count can be difficult. CFU determination by quantitative real-time PCR (qPCR), however, is becoming increasingly common and usually relies on detecting one of the subunits of the multicopy rRNA genes. This study was undertaken to determine if ribosomal DNA (rDNA) copy number was constant or variable among different A. fumigatus isolates. FKS1 was used as a single-copy control gene and was validated against single-copy (pyrG and ARG4) and multicopy (arsC) controls. The copy numbers of the 18S rDNA subunit were then determined for a variety of isolates and were found to vary with the strain, from 38 to 91 copies per genome. Investigation of the stability of the 18S rDNA copy number after exposure to a number of different environmental and growth conditions revealed that the copy number was stable, varying less than one copy across all conditions, including in isolates recovered from an animal model. These results suggest that while the ribosomal genes are excellent targets for enumeration by qPCR, the copy number should be determined prior to using them as targets for quantitative analysis.
Aspergillosis is caused by pathogenic fungi in the genus Aspergillus and includes allergic, superficial, saprophytic, and invasive disease (12). The frequency of invasive aspergillosis (IA) continues to increase due to a growing population of immunosuppressed individuals. In fact, Aspergillus fumigatus, the most frequent Aspergillus species in IA cases (19), is now the most common airborne human fungal pathogen (25). The mortality rate for IA can be unacceptably high for some patient populations, once infected, ranging from 70 to 90%, depending on the patient type (7, 13, 31). However, in spite of the severity of disease, the ubiquitous nature of Aspergillus in the environment makes exposure difficult to avoid; consequently, susceptible patients will almost always be at risk for infection.
The life-threatening nature of IA makes accurate diagnosis and early detection crucial. Quantitative real-time PCR (qPCR) is emerging as a sensitive and cost-efficient technique for detecting Aspergillus spp. from a diverse variety of sources, including clinical specimens. Investigators studying IA with animal models routinely use qPCR to measure fungal load (17, 27), including response to drug treatment (6, 42). Bioaerosol quantitation of Aspergillus spp., particularly in the hospital environment, is also amenable to qPCR (32). Finally, even though qPCR is not the first choice for clinical diagnosis of IA, it has proven useful for quantitating Aspergillus spp. from a variety of patient specimens (2, 26, 38) and has proven extremely useful as a secondary assay for comparative purposes during assay development (8, 23).
One of the drawbacks of PCR-based detection methods is a lack of standardization (5), and one of the first areas to standardize is selection of an appropriate target for amplification. The quantitative nature of qPCR allows an estimation of the number of CFU by equating the copy number of the target sequence with the genome number through a simple ratio, provided the ratio remains invariant. With fungi, the ribosomal genes have proven to be useful PCR targets because of their sequence conservation, which has allowed the use of universal primers that enable the amplification of targets from unknown species. A second advantage of using the ribosomal DNA (rDNA) genes as an amplification target is the copy number, which can be 10 to 100 times that of single-copy genes (29, 30). However, in A. fumigatus, it is unclear whether all strains have the same number of rDNA subunits. With other fungi, the rDNA copy number is known to vary (4, 15, 16, 20, 29), although these observations have been made with fungi that are not frequently recovered as human pathogens. Given what is known for other organisms about the variability of the rDNA copy number and the importance of A. fumigatus as a human pathogen, this study was performed in order to determine if rDNA copy number is constant or strain specific in A. fumigatus.
MATERIALS AND METHODS
Strains and media.
Strains used in this study are shown in Table 1 and were confirmed to be A. fumigatus by colony morphology and DNA sequencing of the internal transcribed spacer (ITS) and D1/D2 regions. Each strain was grown on Sabouraud's dextrose (Difco Laboratories, Detroit, MI) or potato dextrose broth or potato dextrose agar (PDA) (Fisher Scientific, Pittsburgh, PA) for all assays, unless otherwise indicated. Agar media were prepared from broth by solidification with 2% agar. RPMI 1640 without l-glutamine (Mediatech, Inc., Herndon, VA) was prepared by filter sterilizing and added to an autoclaved solution of 2% dextrose and 2% agar (BD Diagnostic Systems, Franklin Lakes, NJ).
TABLE 1.
Strains used in this studya
| Strainb | Contributor, strain alias, and/or reference |
|---|---|
| AF293 | R. Aramayo |
| WSA-172 | M. Rinaldi, no. 98-407 |
| WSA-270 | ATCC 64746 |
| WSA-271 | ATCC 14110 |
| WSA-419 | K. J. Kwon-Chung, no. B-5233, 40 |
| WSA-445 | T. Patterson, no. MTFP0009 |
| WSA-446 | M. Rinaldi, no. 99-1900 |
| WSA-621 | B. Lutz, no. 1 |
All strains were obtained from clinical sources.
WSA isolates are from the Wickes laboratory culture collection.
DNA isolation.
Individual strains were inoculated into 200 ml of Sabouraud's dextrose broth in a 500-ml flask from a 7-day-old suspension of ∼5 × 108 conidia harvested from a PDA plate. The hyphae were recovered after 24 h by filtering through an 18.5-cm, 0.45-mm-pore-size Whatman disk (Whatman, Florham Park, NJ) and washed with sterile saline. DNA isolation consisted of methods reported elsewhere (22, 41), with slight modifications. After the saline wash, approximately 200 mg of wet hyphae were briefly dried by blotting between Whatman paper (Whatman) and then placed into a sterile mortar and frozen for 10 min at −70°C. Fungal cell walls were mechanically broken by grinding with a pestle for 1 to 2 min after the addition of sterile sand and 2 ml of Masterpure yeast DNA purification kit lysis buffer (Masterpure yeast DNA purification kit; Epicentre Technologies, Madison, WI). The slurry was transferred to 2- by 1.5-ml Microfuge tubes and spun at low speed (500 × g) for 15 s to pellet the sand. Four hundred microliters of the supernatant were transferred to a 2.0-ml screw-cap Microfuge tube and incubated at 65°C for 2 h, after the addition of 6 μl of proteinase K (50 μg/ml) from the DNA purification kit. Samples were processed from this point as described previously (22). After the final wash, the dried pellets were resuspended in 200 μl ultra pure water (Invitrogen, Carlsbad, CA). DNA was assessed for quality and quantified by gel electrophoresis and a 260-nm/280-nm absorbance ratio.
Due to the possibility of contamination of Aspergillus DNA with polysaccharides in crude DNA preps, DNA was further purified prior to performance of qPCR assays. DNA was run in a 1.0% low-melting-point agarose (InCert; FMC BioProducts, Rockland, ME) gel to separate it from contaminating materials. Gel fragments containing DNA were recovered, placed into 1.5-ml Microfuge tubes, and then treated with Gelase (Epicentre) according to the manufacturer's instructions. Purified DNA was assessed and quantitated by spectrophotometer and agarose electrophoresis as described above. Yields were 100 μg to 500 μg.
Growth conditions to evaluate stress effect on rDNA copy number.
In order to measure the effect of colony age on rDNA copy number, DNA was prepared from A. fumigatus strain AF293 grown for 3 days, 5 days, 10 days, and 25 days on PDA plates at 30°C. AF293 was also tested for the effect of temperature on copy number by preparing DNA from cultures grown at 30°C and 45°C for 5 days on PDA plates. DNA was isolated and processed from each condition, as previously described (22).
The effect of antifungal exposure on copy number was measured by harvesting AF293 grown in the presence of itraconazole (Oakdell Pharmacy, San Antonio, TX) using a modification of the standard MIC assay. Conidia were harvested from a 5-day-old PDA plate grown at 30°C overnight and used to prepare inoculums containing 4.5 × 106 CFU/ml. Each inoculum (10 ml) was then grown overnight at 30°C in the presence of different itraconazole concentrations (0 μg/ml, 0.03 μg/ml, 0.06 μg/ml, 0.125 μg/ml, 0.25 μg/ml, 0.5 μg/ml, 1.0 μg/ml, and 2.0 μg/ml) under modified MIC conditions described by the National Committee for Clinical Laboratory Standards (33). DNA was then recovered as described above.
In order to determine what effect morphology had on copy number, AF293 DNA was isolated from pure conidia and hyphae. Conidial cultures were prepared from PDA plates grown for 11 days at 30°C and harvested by washing with 10 ml of sterile PBS-0.1% Tween 20. The suspension was pelleted by centrifugation at 4,800 × g for 10 min. The supernatant was discarded, and the conidial pellet was transferred to a 1.7-ml microcentrifuge tube and washed once with 500 μl of sterile water and once with 500 μl of 0.1 M MgCl2. Hyphae were prepared as described previously (22). Conidial and hyphal DNA were recovered as described above.
The effect of growth in vivo during animal model infection on copy number was determined by passing AF293 through animals as follows. Nonimmunosuppressed mice and guinea pigs were infected as described by Sheppard et al. (35). Lungs and kidneys were harvested 5 days postinfection. DNA was extracted from tissue according to the Standard Operating Procedures for Invasive Aspergillosis Animal Models (http://www.sacmm.org/sop.html) and recovered in 100 μl of QIAamp DNA minikit elution buffer (Qiagen, Valencia, CA). After quantitation, DNA was stored at −20°C until analyzed.
PCR and qPCR primer and probe design.
The PCR primer and probe sequences used to quantitate and amplify A. fumigatus target genes are shown in Table 2. Primers for qPCR were designed using Primer Express software version 2.0, which is application-based design software provided by ABI (Applied Biosystems, Inc., Foster City, CA), or were designed based on previously published reports. The primers and probe for the A. fumigatus FKS1 gene were designed according to Costa et al. (10). The primers and probe for the 18s rDNA sequence were also based on a previous study (6). The FKS1 gene was chosen because it is a known single-copy gene in A. fumigatus involved in β(1-3) glucan synthesis (3) and was used as an internal control. The pyrG gene, which encodes orotidine-5′-monophosphate decarboxylase, was also included as a second single-copy reference gene (11, 44) and used to confirm FKS1 copy number determination. ARG4, which encodes carbamoyl-phosphate synthase, was the third single-copy reference gene used in this study and was identified from the genome sequence.
TABLE 2.
PCR primer and probe sequences
| Primer or probeb | Sequence | Reference or source |
|---|---|---|
| 18S rDNA.F | 5′-GGCCCTTAAATAGCCCGGT-3′ | 10 |
| 18S rDNA.R | 5′-TGAGCCGATAGTCCCCCTAA-3′ | 10 |
| 18S rDNA probea | 6-FAM-AGCCAGCGGCCCGCAAATG-MGBNFQ | 10 |
| AFKS.F | 5′-GCCTGGTAGTGAAGCTGAGCGT-3′ | 6 |
| AFKS.R | 5′-CGGTGAATGTAGGCATGTTGTCC-3′ | 6 |
| AFKS probe | 6-FAM-TCACTCTCTACCCCCATGCCCGAGCC-MGBNFQ | 6 |
| AFKS probe | 6-VIC-TCACTCTCTACCCCCATGCCCGAGCC-MGBNFQ | 6 |
| ARG4.F | 5′-CAGCCCCGGGAAACTCA-3′ | This study |
| ARG4.R | 5′-TCCGCTCCCTTGACAGCTT-3′ | This study |
| ARG4 probe | 6-FAM-CCAGACCAATGTTCCTGAG-MGBNFQ | This study |
| pyrG.F | 5′-TGGCCCAGACCGCATCT-3′ | This study |
| pyrG.R | 5′-CAACAGTCCTCTCTCAGGACCAT-3′ | This study |
| pyrG probe | 6-VIC-CGCAAGACTTCCC-MGBNFQ | This study |
| arsC.F | 5′-GCCGCTGGGTTCCTTACTC-3′ | This study |
| arsC.R | 5′-CAGCGGAGCGAACCTCAATA-3′ | This study |
| arsC probe | 6-FAM-CCTCGCAGGTGATG-MGBNFQ | This study |
| Chr1arsC.F | 5′-GACCTCGACACCCTAAGAAGC-3′ | This study |
| Chr1arsC.R | 5′-TCAAATGATGAGAGGCCAGA-3′ | This study |
| Chr5arsC.F | 5′-TCCTCCATCTTCATTCCCTTA-3′ | This study |
| Chr5arsC.R | 5′-GAGCTGGAACCTCAGCGTAG-3′ | This study |
MGB probe dyes are incorporated into the primer sequences, i.e., 6-FAM-AGCCAGCGGCCCGCAAATG-MGBNFQ is an MGB probe labeled with FAM.
AFKS primers and probes were used for detection of the FKS1 gene. ARG4, pyrG, and arsC primers and probes were used for detection of the ARG4, pyrG, and arsC genes. Primers designated Chr1arsC or Chr5arsC are for routine PCR amplification of the two arsC alleles from chromosome 1 or 5.
In order to test our ability to discriminate multiple copy genes, a duplicated gene was selected for analysis. The arsC (arsenate reductase) gene is a duplicated gene found in some but not all strains of A. fumigatus and is present in the AF293 genome sequence as two copies (34). Since arsC is not present in all A. fumigatus strains, we reconfirmed that it was present in AF293 in two copies by use of a method independent of qPCR. Based on the sequences of arsC from the two chromosomal locations, allele-specific primers were designed that spanned an NcoI site within the coding sequence of each arsC allele (Table 2). The chromosome 1 arsC primers consisted of Ch1arsC.F and Ch1arsC.R. The chromosome 5 arsC primers consisted of Chr5arsC.F and Chr5arsC.R. Each allele was amplified using the following conditions: 94°C for 2 min; 32 cycles at 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s; and a final extension of 72°C for 2 min. The amplicons were then digested with 5 U of NcoI (New England Biolabs, Beverly, MA) at 37°C for 3 h and then separated on a 3% NuSieve GTG agarose gel (Cambrex Bio Science, Inc., Rockland, ME). Sizes were then compared to the sizes predicted from the genome sequence.
qPCR validation assays and calculations.
FKS1, pyrG, ARG4 (single-copy genes), arsC (two-copy gene), and 18S rDNA (multiple copies) gene copy number determinations were done by qPCR (TaqMan) assay according to the method of Townson et al. (39), with modifications. In order to determine the copy number of a variable gene (18S rDNA), a single-copy reference gene needed to be identified and confirmed to be present in one copy/genome. Since the FKS1 gene is highly conserved in fungi and has been shown in a number of reports to be present in single copy in A. fumigatus (3, 14, 34), we selected this gene to use as the single-copy reference probe in the quantitative reverse transcription-PCRs. Confirmation was performed by comparison to other A. fumigatus genes already known to be single copy. The single-copy genes pyrG and ARG4 were confirmed using relative quantification (ratios of one gene to another) to determine the number of copies present per genome. Quantification standards were run in conjunction with each set of samples after primers and probes for the FKS1, 18S rDNA, pyrG, ARG4, and arsC genes were optimized for template concentration and primer efficiencies (1).
qPCRs were performed in triplicate using an ABI Prism 7900 sequence detector system (ABI) to detect minor groove binder probe binding. FKS1 was quantitated using both VIC and 6-carboxyfluorescein (FAM) dyes and used as a reference for comparison to the 18S rDNA FAM probe from each strain. Six serial 1:2 dilutions (20.0, 10.0, 5.0, 2.5, 1.25, and 0.625 ng/μl) of genomic DNA from A. fumigatus AF293 were used to generate standard curves of CT (threshold cycle) value against the log DNA concentration on each PCR plate for the FKS1 and 18S rDNA genes. Each experiment was performed three separate times from one DNA preparation and run in duplicate. CT values were determined and then converted into template quantity. After the creation of standard curves, the copy number of each gene was determined by DNA quantification using TaqMan technology. PCR cycle numbers were plotted against the value of 5′ fluorescence signal, and then threshold values were plotted against the copy number of the template DNA that was used to generate standard curves (1).
Absolute quantification using the ABI Prism 7900 requires that the absolute quantities of the standards be determined by some independent means first. In this study, fungal DNA was used to prepare absolute standards. Concentration and DNA quality were measured by determining the A260 and by gel electrophoresis and converted to the number of copies by use of the molecular weight of the DNA. The equation CT = m (log quantity) + b from the equation for a line (y = mx + b) was constructed by plotting the standard curve of log quantity versus its corresponding CT value. If the curve demonstrated an r2 value of >0.980, the standard curve then was used to determine sensitivity, primer efficiencies, and dynamic range, as well as specificity and reproducibility of every assay (FKS1, 18S rDNA, pyrG, ARG4, and arsC). Amplification of serially diluted genomic DNA (standard curves) from A. fumigatus AF293 was repeated in triplicate, on different days, in order to test reproducibility, primer efficiencies, and DNA optimal dilutions for the rest of the genes (pyrG, ARG4, and arsC). DNA concentrations ranged from 20.0 to 0.625 ng/μl. Specificity for all the assays was assessed by using DNA extracted from Candida albicans SC5314, as well as mouse and guinea pig DNA (9, 28). Comparative copy numbers for confirmation experiments were determined using the relative quantification (ΔΔCT) 2−ΔΔCT method. The 18S rDNA copy numbers were determined by the absolute quantitation method, by which total copies were first calculated using the following equation: total 18S rDNA copies = 10([CT − b]/m). The number of 18S rDNA copies per genome was then determined by the following equation: 18S rDNA copies per genome = (total copies of 18S rDNA)/(total copies of FKS1). Copy number was calculated as the ratio of template quantity for 18S rDNA to the template quantity for FKS1.
Statistical methods.
In each experiment, we altered one factor at a time under controlled conditions. This approach minimized the sources of variability within an experiment and maximized statistical power for detecting effects of a single factor on differential copy numbers. Results after determination of 18S rDNA copy numbers were compared by the Wilcoxon rank sum test for morphology and temperature. The Wilcoxon signed-rank test was used to compare copy numbers from different tissues in the same animal, and the Kruskal-Wallis test was used to compare culture ages and antifungal susceptibilities. Statistical analysis was done at the University of Texas Health Science Center at the San Antonio Department of Epidemiology and Biostatistics. Two-tailed P values less than 0.05 were considered significant.
RESULTS
Copy number confirmation of FKS1.
A number of confirmatory assays were performed to verify that FKS1 was present as a single copy in AF293. First, absolute quantitation was performed using FKS1 probes labeled with two dyes, FAM and VIC. The slope of the VIC line was −3.9341 (from y = −39341x + 52.288), while the slope for the FAM line was −3.8971 (from y = −3.8971x + 51.593). The r2 values of the VIC and FAM lines were 0.9946 and 0.9982, respectively, demonstrating that comparable results could be obtained independently of dye type. The copy number of FKS1 was next determined in a subset of A. fumigatus strains (WSA-172, WSA-445, WSA-621, and WSA-419) by absolute quantitation using FKS1 labeled with FAM and VIC for each strain. FKS1 copy numbers determined by qPCR ranged from 0.93 to 1.10 copies and were rounded to 1 copy based on the close integer scoring method (18) so that it could be used as the single-copy reference gene when determining the copy numbers of other genes. We next compared the copy number of FKS1 to that of other known single-copy genes (ARG4 and pyrG) using absolute quantification. The corresponding calculations of copy numbers of the three genes in AF293 by comparison of the CT values confirmed that each gene was present in single copy. This outcome was also observed with other A. fumigatus isolates (Table 3) and confirmed that FKS1 was suitable as a single-copy control gene.
TABLE 3.
Confirmation of copy number of predicted single-copy genesa
| Strain | FKS1 avg CT | FKS1 copy no. | pyrG avg CT | pyrG copy no. | ARG4 avg CT | ARG4 copy no. |
|---|---|---|---|---|---|---|
| AF293 | 19.16 ± .012 | 1 | 19.33 ± .005 | 1.13 | 19.06 ± 0.008 | 1.07 |
| WSA-172 | 21.39 ± .012 | 1 | 21.34 ± .001 | 0.97 | 21.23 ± .007 | 1.11 |
| WSA-445 | 22.26 ± .049 | 1 | 22.55 ± .003 | 1.22 | 22.31 ± .004 | 0.97 |
| WSA-621 | 24.34 ± .014 | 1 | 23.52 ± .004 | 1.29 | 24.54 ± 0.007 | 1.10 |
| WSA-419 | 21.40 ± .050 | 1 | 21.82 ± .003 | 1.34 | 21.40 ± .005 | 1.00 |
CT values are means ± standard deviations (three samples, run in duplicate). The copy number of the test gene (pyrG or ARG4) was equal to 2−ΔΔCT.
Detection of a multicopy gene in A. fumigatus.
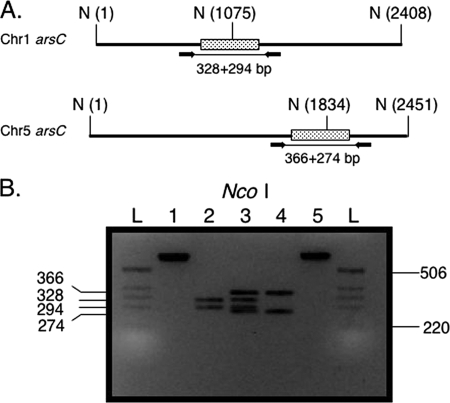
In order to accurately quantitate multicopy genes, it was necessary to demonstrate that FKS1 could be used to quantitate a multicopy gene of known copy number. Furthermore, we were interested in knowing how discriminatory our strategy would be with regard to copy number accuracy. To make this determination, we decided to use a multicopy gene that was present in low copy number and chose arsC as a target. Sequence analysis of arsC from the AF293 genome suggested that it was present in two copies, one copy on chromosome 1 and one copy on chromosome 5. Careful inspection of the two sequences by DNA alignment revealed that a combination of primer position and restriction digestion would confirm the presence of two copies, after gel electrophoresis, based on the predicted sizes of digestion products of the PCR (Fig. 1A). The data shown in Fig. 1B confirm that the predicted digestion patterns of the two arsC alleles matched what was observed with the gel after electrophoresis.
FIG. 1.
Confirmation of arsC copy number in AF293. (A) Priming sites for the two arsC alleles. The Chr1 arsC allele is located on chromosome 1, while the Chr5 arsC allele is located on chromosome 5. Primers are indicated by black arrows; PCR product is indicated by the line connecting the primers. The three NcoI sites (N) (one located within and two flanking the arsC genes), with locations given as base pairs, are indicated within parentheses. Stippled boxes are the arsC open reading frames. The predicted sizes of the fragments after NcoI digestion are indicated below each open reading frame. (B) NcoI digestion of arsC PCR products. Lane 1, uncut Chr1 arsC PCR product; lane 2, NcoI digest of Chr1 arsC; lane 3, mixture of both NcoI digestions; lane 4, NcoI digestion of Chr5 arsC PCR product; lane 5, uncut Chr5 arsC PCR product. Sizes are in base pairs. L, ladder. Ladder sizes are at the right of the gel; fragment sizes are at the left.
qPCR was next used to determine the copy number of arsC in AF293. The CT values were determined for the arsC sequences and compared to those of FKS1, which was used as the single-copy control. The output graph from the reaction shows an earlier CT for arsC than for FKS1 (Fig. 2), consistent with the greater copy number of arsC. Calculation of the copy number of arsC for AF293 and for other isolates demonstrated that arsC is present in two copies (Table 4), which confirmed that our strategy could determine the copy number of multicopy sequences.
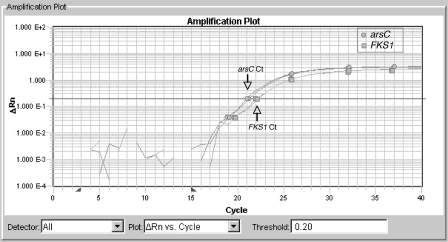
FIG. 2.
Amplification plot of AF293 arsC versus FKS1 TaqMan assays. TaqMan assays were performed using an arsC primer-probe combination and FKS1 primer-probe combination. The graph represents a sample plot from duplicate reactions run on aliquots of the same DNA sample. Amplification of the arsC gene is denoted by circles. Amplification of the FKS1 gene is denoted by squares. The CT value of the arsC line is approximately 21.1 (downward arrow), and the CT value of FKS1 is approximately 22.1 (upward arrow).
TABLE 4.
Determination of arsC copy number of all isolates by qRT-PCR in comparison to that of FKS1a
| Strain | FKS1 avg CT | FKS1 copy no. | arsC avg CT | arsC copy no.b |
|---|---|---|---|---|
| AF293 | 22.1 ± .107 | 1 | 21.1 ± .007 | 2 (2.00) |
| WSA-172 | 19.8 ± .010 | 1 | 18.7 ± .004 | 2 (2.14) |
| WSA-446 | 22.0 ± .052 | 1 | 21.1 ± .003 | 2 (1.89) |
| WSA-445 | 19.8 ± .014 | 1 | 18.7 ± .005 | 2 (2.16) |
| WSA-271 | 22.1 ± .025 | 1 | 21.0 ± .002 | 2 (2.07) |
| WSA-270 | 19.2 ± .042 | 1 | 18.2 ± .002 | 2 (2.00) |
| WSA-621 | 19.9 ± .060 | 1 | 18.8 ± .003 | 2 (2.01) |
| WSA-419 | 23.1 ± .014 | 1 | 21.9 ± .007 | 2 (2.42) |
CT values are means ± standard deviations (three samples, run in duplicate).
arsC copy numbers were determined using the formula 2−ΔΔCT. Results rounded to whole numbers are shown, with the unrounded results given in parentheses.
Determination of rDNA copy number.
Once FKS1 was established as a reliable single-copy control, this sequence was used to determine the copy numbers of the rDNA genes in A. fumigatus by quantitating the copy number of the 18S rDNA subunit. Since the copy number of the rDNA genes of AF293 has been determined from the genome sequence, this isolate was used in a pilot TaqMan assay in which FKS1 (single copy) was used to calculate the copy number of the 18S rDNA subunit. According to the genome sequence, AF293 has 35 copies of the rDNA genes per genome (34). Figure 3 shows an example of the plots of FKS1 versus 18S rDNA and clearly demonstrates that there are more copies of the 18S rDNA gene than the FKS1 gene. Calculation of the 18S rDNA copy numbers resulted in a value of 38 copies of 18S rDNA per genome in AF293, which is in fairly close agreement with the genomic copy number (38 versus 35 copies) for AF293 (34). The 18S rDNA copy numbers of the remaining isolates were then determined using FKS1 as the reference gene. The data show a range of 38 to 91 copies, with an average of 54 copies per genome (Table 5). These results show that for our set of isolates, 18S rDNA copy numbers are isolate specific and can vary substantially from strain to strain.
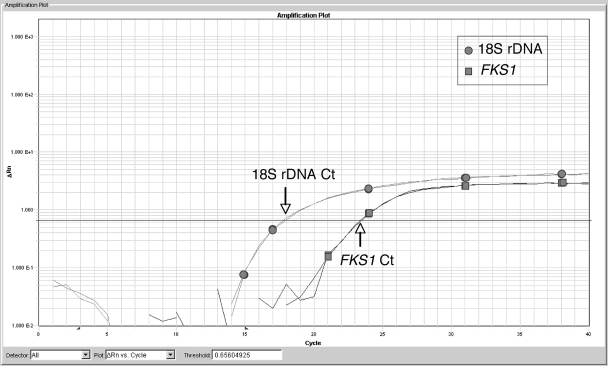
FIG. 3.
Amplification plot of 18S rDNA versus FKS1. An example of copy number determination of 18S rDNA using FKS1 as a single-copy control. The figure is an amplification plot of a TaqMan assay performed using the 18S rDNA primer-probe combination and FKS1 primer-probe combination. Template DNA was taken from the same DNA sample prepared from AF293 and run in duplicate. Note the earlier CT value of 18S rDNA (circles), which is approximately 18.0 (downward arrow) versus the FKS1 CT value (squares), which is approximately 23.4 (upward arrow). The lower CT value for 18S rDNA reflects the greater copy number of the target, since the fluorescence crosses the threshold at a much lower cycle number.
TABLE 5.
A. fumigatus 18S rDNA copy number determinations
| Strain | No. of 18S rDNA copiesa |
|---|---|
| AF293 | 38 ± 0.01 |
| WSA-172 | 46 ± 0.03 |
| WSA-446 | 47 ± 0.01 |
| WSA-445 | 49 ± 0.06 |
| WSA-271 | 49 ± 0.05 |
| WSA-270 | 53 ± 0.01 |
| WSA-621 | 70 ± 0.03 |
| WSA-419 | 91 ± 0.03 |
Values are mean ± standard deviations (three samples, run in duplicate).
Stability of rDNA copy number.
Since our results indicated that rDNA copy numbers could vary among strains of A. fumigatus, we investigated various environmental conditions to determine whether or not an effect on copy number could be observed. Factors that were investigated included morphology, growth temperature, culture age, antifungal exposure (itraconazole), and animal model organ site (lung versus kidney). The overall copy number mean was found to be 38.032 ± 0.13, which agrees with our initial copy number determination for AF293. However, Table 6 shows that some significant differences in copy numbers were observed among our growth conditions (morphology, growth temperature, and culture age). In spite of these differences, variation in copy numbers among all conditions tested was less than 1 copy and the copy numbers would all have been 38 copies if numbers were rounded.
TABLE 6.
A. fumigatus 18S rDNA copy number stability
| Condition | Subgroup | No. of 18S rDNA copiesa | P value |
|---|---|---|---|
| Morphology | Conidia | 38.02 ± 0.011 | 0.03 |
| Hyphae | 38.11 ± 0.01 | 0.03 | |
| Temperature | 30°C | 37.84 ± 0.044 | 0.03 |
| 45°C | 38.03 ± 0.015 | 0.03 | |
| Culture age | 3 days | 38.041 ± 0.024 | 0.004 |
| 5 days | 37.906 ± 0.059 | 0.004 | |
| 10 days | 38.321 ± 0.019 | 0.004 | |
| 25 days | 38.061 ± 0.017 | 0.004 | |
| Itraconazole concn | 0.00 μg/ml | 37.984 ± 0.049 | 0.17 |
| 0.03 μg/ml | 38.024 ± 0.015 | 0.17 | |
| 0.06 μg/ml | 38.039 ± 0.022 | 0.17 | |
| 0.125 μg/ml | 38.033 ± 0.012 | 0.17 | |
| 0.25 μg/ml | 38.001 ± 0.055 | 0.17 | |
| 0.5 μg/ml | 38.046 ± 0.009 | 0.17 | |
| 1.0 μg/ml | 38.039 ± 0.019 | 0.17 | |
| 2.0 μg/ml | 38.034 ± 0.038 | 0.17 | |
| Mouse | Lung | 38.056 ± 0.038 | 0.13 |
| Kidney | 38.136 ± 0.008 | 0.13 | |
| Guinea pig | Lung | 38.231 ± 0.008 | 0.13 |
| Kidney | 37.688 ± 0.059 | 0.13 |
Copy values are means ± standard deviations.
DISCUSSION
Timely diagnosis of IA is challenging due to the lack of specific clinical manifestations of infection. Unfortunately, symptoms can be nonspecific and include fever, cough, dyspnea, chest pain, and apnea. Therefore, diagnosis can be dependent on the combination of a strong index of suspicion and radiologic findings, serologic assays, or when possible, culture and/or histologic findings (24). For any of these methods, a quantitative estimate of fungal burden is difficult at best and can be expensive or time-consuming. In fact, even under controlled experimental conditions of animal modeling, colony counts can be misleading, as some studies have noted decreasing counts that are contradicted by other measurements with the same animal (36, 37). The ubiquitous nature of A. fumigatus in the environment and associated possibility of inhaling fungal elements that may or may not grow in vivo, but could be detected as CFU after lavage, further complicate making an accurate assessment.
Through advances in instrumentation and reagent chemistry, PCR continues to find new applications in clinically relevant areas. In spite of not being widely employed as a routine clinical diagnostic tool for detecting IA, PCR is proving increasingly useful as an investigational tool for studying aspergillosis both in vitro and in vivo and may ultimately find its way into the clinical laboratory as a routine diagnostic tool for IA. For in vivo applications of animal infections, qPCR is often used to make a determination of the number of CFU, which are frequently expressed as conidial equivalents in order to indicate one nucleus per conidium. While the number of CFU is fairly accurate for fungi that grow in a yeast morphology, the number of CFU obtained by plate counts can be difficult to interpret for filamentous organisms due to the inability to distinguish a single hypha that forms one colony from the same fragmented hypha that yields multiple colonies. In fact, using CFU for measuring A. fumigatus fungal loads has been shown to yield equivocal results (6, 36). Therefore, alternative methods that do not require obtaining viable colony counts but provide some indication of fungal burden are potentially useful for quantifying the fungal load of a given specimen. qRT-PCR is exceptionally well suited for this requirement. In fact, when all protocols are standardized, from infection model through tissue preparation, reproducible results can be obtained, even among interlaboratory studies (35).
The observations in this study add an important caveat for standardized procedures to now include working with the same A. fumigatus strain when qPCR quantitation using the rDNA genes is required. Our results have shown that using an 18S rDNA target requires prior knowledge of copy number of the strain of interest. With our small sample size, we found copy numbers to vary by as much as ∼2.5×. Neither the upper limit nor the lower limit of 18S rDNA copy number is known, but it is almost certain to vary by a larger amount than the amount that we observed for our set of isolates. Consequently, 18S rDNA copy number cannot be assumed based on another value previously determined from an unrelated strain. This observation presently does not have direct clinical implications, since qPCR is not routinely used to diagnose IA and fungal burden is rarely part of any diagnosis, since for at-risk patients a positive assay regardless of amount is always cause for concern. However, accurate quantitation of A. fumigatus CFU has numerous applications, many of which have clinically relevant consequences. These include data generated from more than one strain or testing unknown strains in experiments measuring tissue burdens, in vivo drug susceptibility testing, environmental quantitation, tracking CFU during disease progression, or comparison of different methods for measuring fungal load (2, 17, 32, 36, 42). Similarly, direct quantitative comparisons of the same or different strains that utilize qPCR versus some other method, such as CFU counts or galactomannan detection, can be erroneous in the absence of an accurate rDNA copy number. Finally, model systems that may use the same assay but different strains and report results in CFU, such as animal survival studies, typically use absolute numbers and therefore need to be calculated accurately if qPCR is part of the methodology. However, in spite of the variation between strains, our results suggest that within-strain variation, at least in the case of AF293, is negligible. Therefore, in studies that utilize the same strain and involve quantitation, qPCR using the rDNA genes should yield consistent results. We could not identify any condition that was able to cause the 18S subunit number to vary by more than 1 copy within AF293, in spite of investigating a number of stress conditions. However, we did identify some significant differences in our analyses. We suspect these differences may have been due to experimental error since qPCR accuracy requires precise technique. On the other hand, we know nothing about the mechanism by which copy number variation occurs and what, if any, phenotypic consequences are associated with changes in copy number within a strain. The fact that different strains of A. fumigatus have different rDNA copy numbers is evidence that variation occurs. Since our qPCR assay can detect only whole copies (a fraction of a copy would not yield a PCR product), the data could have arguably been rounded to the nearest whole copy. In this case, all copy numbers would round to 38, which matches the control AF293 number. However, since we cannot rule out copy number heterogeneity within a population, we chose not to round the data. Future studies of copy number should focus on whether changes are rapid, such as by an unequal recombination event that leads to large gains or losses of rDNA repeats, or gradual, which could result in small changes of a unit or two over longer periods of time. Understanding the mechanism may reveal whether or not the changes are responses to selection or are random, without clear phenotypic consequences.
In spite of the observed copy number variation within A. fumigatus, application of these results to other species of Aspergillus probably should not be done without empirical analysis. Aspergillus taxonomy can be complicated by the existence of sections, which may not be discriminated at the clinical level but can be discriminated at the molecular level. For example, in the Aspergillus section Fumigati, A. fumigatus may not be discriminated from other members, such as A. lentulus or A. brevipes. However, these species can be identified by sequencing select loci (i.e., β-tubulin). Therefore, rDNA variation could possibly indicate a separate subspecies. In our study, we confirmed that our strains were all A. fumigatus using β-tubulin sequencing (data not shown), but since so little is known at the molecular level about these subgenera, confirmational sequencing of additional loci may be required when trying to quantitate unknown isolates.
Although we targeted the 18S rDNA subunit in this study, determination of copy number should hold for targets that lie within the 28S subunit or between the two subunits (ITS1, ITS2, and 5.8S) as well, since the large and small ribosomal subunits, though multicopy and tandemly arrayed, are colinear and transcribed as a single transcript along with the intervening ITS region (21, 43). Therefore, based on what is known in model fungi, the copy numbers of the 18S and 28S genes, as well as the intervening sequences, should be the same in the same strain of A. fumigatus. The advantage of primer design in the more variable ITS1, ITS2, or even the D1/D2 region of the 28S subunit is that species specificity can be possible, subspecies issues as described above notwithstanding. If, on the other hand, the increased sensitivity of targeting the multicopy rDNA genes is not needed, a suitable single-copy gene (i.e., FKS1, ARG4, or pyrG) can be used with fairly high confidence that its copy number will be invariant among unrelated strains and equal to 1. Finally for presence or absence outcomes, copy number variation is probably not a concern; however, given that the ribosomal genes are usually targeted due to their increased sensitivity, if investigators are quantitating cell numbers using these genes, the strain-specific variability of rDNA copy number may be an important factor that affects the sensitivity of PCR assays for quantifying Aspergillus fumigatus.
Acknowledgments
This study was supported with federal funds from the National Institute of Allergy and Infectious Diseases under contract no. N01-AI-30041 to T.F.P. B.L.W. is supported by grant no. PR054228 from the U.S. Army Medical Research and Material Command, Office of Congressionally Directed Medical Research Programs.
We thank William R. Kirkpatrick, Laura Najvar, and Rose Bocanegra for technical assistance. We also thank Fei Du for help with statistical analysis.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Applied Biosystems. 2001. User bulletin #2. Relative quantitation of gene expression. Applied Biosystems, Foster City, CA.
- 2.Baskova, L., C. Landlinger, S. Preuner, and T. Lion. 2007. The Pan-AC assay: a single-reaction real-time PCR test for quantitative detection of a broad range of Aspergillus and Candida species. J. Med. Microbiol. 561167-1173. [DOI] [PubMed] [Google Scholar]
- 3.Beauvais, A., J. M. Bruneau, P. C. Mol, M. J. Buitrago, R. Legrand, and J. P. Latge. 2001. Glucan synthase complex of Aspergillus fumigatus. J. Bacteriol. 1832273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkhiri, A., and G. R. Klassen. 1996. Diverged 5s rRNA sequences adjacent to 5s rRNA genes in the rDNA of Pythium pachycaule. Curr. Genet. 29287-292. [PubMed] [Google Scholar]
- 5.Boudewijns, M., P. E. Verweij, and W. J. Melchers. 2006. Molecular diagnosis of invasive aspergillosis: the long and winding road. Future Microbiol. 1283-293. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, J. C., G. K. Abruzzo, J. W. Anderson, A. M. Flattery, C. J. Gill, V. B. Pikounis, D. M. Schmatz, P. A. Liberator, and C. M. Douglas. 2001. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob. Agents Chemother. 453474-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brookman, J. L., and D. W. Denning. 2000. Molecular genetics in Aspergillus fumigatus. Curr. Opin. Microbiol. 3468-474. [DOI] [PubMed] [Google Scholar]
- 8.Challier, S., S. Boyer, E. Abachin, and P. Berche. 2004. Development of a serum-based TaqMan real-time PCR assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 42844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamilos, G., and D. P. Kontoyiannis. 2006. Defining the diagnosis of invasive aspergillosis. Med. Mycol. 44(Suppl.)163-172. [DOI] [PubMed] [Google Scholar]
- 10.Costa, C., D. Vidaud, M. Olivi, E. Bart-Delabesse, M. Vidaud, and S. Bretagne. 2001. Development of two real-time quantitative TaqMan PCR assays to detect circulating Aspergillus fumigatus DNA in serum. J. Microbiol. Methods 44263-269. [DOI] [PubMed] [Google Scholar]
- 11.D'Enfert, C., M. Diaquin, A. Delit, N. Wuscher, J. P. Debeaupuis, M. Huerre, and J. P. Latge. 1996. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect. Immun. 644401-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26781-803, 804-805. [DOI] [PubMed] [Google Scholar]
- 13.Denning, D. W. 1996. Therapeutic outcome in invasive aspergillosis. Clin. Infect. Dis. 23608-615. [DOI] [PubMed] [Google Scholar]
- 14.Firon, A., A. Beauvais, J. P. Latge, E. Couve, M. C. Grosjean-Cournoyer, and C. D'Enfert. 2002. Characterization of essential genes by parasexual genetics in the human fungal pathogen Aspergillus fumigatus: impact of genomic rearrangements associated with electroporation of DNA. Genetics 1611077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, P., C. Gaillardin, M. A. Persuy, J. Klootwijk, and H. van Heerikhuizen. 1986. Heterogeneity in the ribosomal family of the yeast Yarrowia lipolytica: genomic organization and segregation studies. Gene 42273-282. [DOI] [PubMed] [Google Scholar]
- 16.Garber, R. C., B. G. Turgeon, E. U. Selker, and O. C. Yoder. 1988. Organization of ribosomal RNA genes in the fungus Cochliobolus heterostrophus. Curr. Genet. 14573-582. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Lopez, A., M. T. Martin-Gomez, P. Martin-Davila, P. Lopez-Onrubia, J. Gavalda, J. Fortun, A. Pahissa, J. L. Rodriguez-Tudela, and M. Cuenca-Estrella. 2006. Detection of fungal DNA by real-time polymerase chain reaction: evaluation of 2 methodologies in experimental pulmonary aspergillosis. Diagn. Microbiol. Infect. Dis. 56387-393. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, E., H. Kulkarni, H. Bolivar, A. Mangano, R. Sanchez, G. Catano, R. J. Nibbs, B. I. Freedman, M. P. Quinones, M. J. Bamshad, K. K. Murthy, B. H. Rovin, W. Bradley, R. A. Clark, S. A. Anderson, R. J. O'Connell, B. K. Agan, S. S. Ahuja, R. Bologna, L. Sen, M. J. Dolan, and S. K. Ahuja. 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 3071434-1440. [DOI] [PubMed] [Google Scholar]
- 19.Hohl, T. M., and M. Feldmesser. 2007. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot. Cell 61953-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howlett, B. J., B. D. Rolls, and A. J. Cozijnsen. 1997. Organisation of ribosomal DNA in the ascomycete Leptosphaeria maculans. Microbiol. Res. 152261-267. [DOI] [PubMed] [Google Scholar]
- 21.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 4087-109. [DOI] [PubMed] [Google Scholar]
- 22.Jin, J., Y. K. Lee, and B. L. Wickes. 2004. Simple chemical extraction method for DNA isolation from Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 424293-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan, Z. U., S. Ahmad, and A. M. Theyyathel. 2008. Detection of Aspergillus fumigatus-specific DNA, (1-3)-beta-d-glucan and galactomannan in serum and bronchoalveolar lavage specimens of experimentally infected rats. Mycoses 51129-135. [DOI] [PubMed] [Google Scholar]
- 24.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21161-172. [DOI] [PubMed] [Google Scholar]
- 25.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the light cycler system. J. Clin. Microbiol. 38586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeffler, J., K. Kloepfer, H. Hebart, L. Najvar, J. R. Graybill, W. R. Kirkpatrick, T. F. Patterson, K. Dietz, R. Bialek, and H. Einsele. 2002. Polymerase chain reaction detection of Aspergillus DNA in experimental models of invasive aspergillosis. J. Infect. Dis. 1851203-1206. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Ribot, J. L., R. K. McAtee, W. R. Kirkpatrick, S. Perea, and T. F. Patterson. 2000. Comparison of DNA-based typing methods to assess genetic diversity and relatedness among Candida albicans clinical isolates. Rev. Iberoam. Micol. 1749-54. [PubMed] [Google Scholar]
- 29.Maleszka, R., and G. D. Clark-Walker. 1990. Magnification of the rDNA cluster in Kluyveromyces lactis. Mol. Gen. Genet. 223342-344. [DOI] [PubMed] [Google Scholar]
- 30.Maleszka, R., and G. D. Clark-Walker. 1993. Yeasts have a four-fold variation in ribosomal DNA copy number. Yeast 953-58. [DOI] [PubMed] [Google Scholar]
- 31.Marr, K. A., R. A. Carter, M. Boeckh, P. Martin, and L. Corey. 2002. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood 1004358-4366. [DOI] [PubMed] [Google Scholar]
- 32.McDevitt, J. J., P. S. Lees, W. G. Merz, and K. J. Schwab. 2004. Development of a method to detect and quantify Aspergillus fumigatus conidia by quantitative PCR for environmental air samples. Mycopathologia 158325-335. [DOI] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi. Approved standard M38-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 34.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 4381151-1156. [DOI] [PubMed] [Google Scholar]
- 35.Sheppard, D. C., J. R. Graybill, L. K. Najvar, L. Y. Chiang, T. Doedt, W. R. Kirkpatrick, R. Bocanegra, A. C. Vallor, T. F. Patterson, and S. G. Filler. 2006. Standardization of an experimental murine model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 503501-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheppard, D. C., K. A. Marr, D. N. Fredricks, L. Y. Chiang, T. Doedt, and S. G. Filler. 2006. Comparison of three methodologies for the determination of pulmonary fungal burden in experimental murine aspergillosis. Clin. Microbiol. Infect. 12376-380. [DOI] [PubMed] [Google Scholar]
- 37.Singh, G., J. Imai, K. V. Clemons, and D. A. Stevens. 2005. Efficacy of caspofungin against central nervous system Aspergillus fumigatus infection in mice determined by TaqMan PCR and CFU methods. Antimicrob. Agents Chemother. 491369-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiess, B., D. Buchheidt, C. Baust, H. Skladny, W. Seifarth, U. Zeilfelder, C. Leib-Mosch, H. Morz, and R. Hehlmann. 2003. Development of a LightCycler PCR assay for detection and quantification of Aspergillus fumigatus DNA in clinical samples from neutropenic patients. J. Clin. Microbiol. 411811-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townson, J. R., L. F. Barcellos, and R. J. Nibbs. 2002. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur. J. Immunol. 323016-3026. [DOI] [PubMed] [Google Scholar]
- 40.Tsai, H. F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 1803031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Burik, J. A., R. W. Schreckhise, T. C. White, R. A. Bowden, and D. Myerson. 1998. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 36299-303. [PubMed] [Google Scholar]
- 42.van Vianen, W., S. de Marie, M. T. ten Kate, R. A. Mathot, and I. A. Bakker-Woudenberg. 2006. Caspofungin: antifungal activity in vitro, pharmacokinetics, and effects on fungal load and animal survival in neutropenic rats with invasive pulmonary aspergillosis. J. Antimicrob. Chemother. 57732-740. [DOI] [PubMed] [Google Scholar]
- 43.Warner, J. R. 1989. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev. 53256-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33378-385. [DOI] [PubMed] [Google Scholar]