Abstract
Fluoroquinolones are not indicated for use for the treatment of pneumonia in children; however, non-levofloxacin-susceptible Streptococcus pneumoniae (NLSSP) has emerged in South Africa among children receiving treatment for multidrug-resistant tuberculosis. This study aimed to genotypically characterize NLSSP isolates. Invasive isolates were collected through active national laboratory-based surveillance for invasive pneumococcal disease (IPD) from 2000 through 2006 (n = 19,404). Carriage studies were conducted at two hospitals for patients with tuberculosis in two provinces. Phenotypic characterization was performed by determination of MICs and serotyping. Fluoroquinolone resistance mutations were identified, and clonality was investigated by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing. Twelve non-levofloxacin-susceptible cases of IPD were identified, and all were in children <15 years of age. Ten isolates were serotype 19F and formed two clusters according to their PFGE profiles, antibiogram types, and fluoroquinolone resistance-conferring mutations. All nine carriage isolates from children in hospital A were NLSSP, serotype 19F, were indistinguishable by PFGE, and were related to invasive isolates in cluster 2. Of 26 child carriers in hospital B, 22 (85%) were colonized with NLSSP. The isolates were indistinguishable by PFGE, although they displayed two serotypes, serotypes 19F and 23F. The isolates were related to invasive isolates in cluster 1; however, higher levofloxacin MICs and different fluoroquinolone resistance mutations were suggestive of horizontal gene transfer. A serotype 23F carriage isolate displayed increased fitness compared with the fitness of an otherwise indistinguishable serotype 19F carriage isolate. These data suggest that a low-level non-levofloxacin-susceptible strain transformed into a highly resistant strain under antibiotic pressure and underwent capsular switching in order to have increased fitness.
The emergence of non-levofloxacin-susceptible Streptococcus pneumoniae (NLSSP) in hospitalized children receiving fluoroquinolones as part of treatment for multidrug-resistant (MDR) tuberculosis (TB) in South Africa has recently been reported (26). Prior to this report, only two cases of invasive NLSSP disease among children had been described, and one of these was from South Africa (11, 27). Even though fluoroquinolones are not currently indicated for use for the treatment of pneumonia in children (16), von Gottberg et al. (26) showed that the use of fluoroquinolones for the treatment of MDR TB in children may lead to the development and spread of NLSSP.
Fluoroquinolones inhibit bacterial DNA synthesis by targeting the DNA gyrase (GyrA-GyrB) and topoisomerase IV (ParC-ParE) proteins, which have essential functions in DNA replication. Fluoroquinolone resistance in pneumococci is mediated by stepwise mutations in the quinolone resistance-determining regions (QRDRs) of the respective genes and occurs more commonly in parC and gyrA (3, 6). Fluoroquinolone resistance has also been shown to occur by horizontal gene transfer (1, 21). Low-level resistance may be conferred by the increased level of expression of an efflux pump, PmrA.
In this study, the invasive and carriage NLSSP isolates described previously (26) were characterized at the molecular level to determine the fluoroquinolone resistance mutations as well as investigate the clonality and fitness of the isolates.
MATERIALS AND METHODS
Invasive disease surveillance.
Isolates were collected as part of a national laboratory-based surveillance system for invasive pneumococcal disease (IPD). Clinical isolates and the clinical and demographic data for the patients from whom they were recovered were sent to the National Institute for Communicable Diseases in Johannesburg, South Africa, from approximately 120 laboratories throughout South Africa. A case of IPD was defined as the isolation of S. pneumoniae from a normally sterile site (e.g., cerebrospinal fluid, blood, or joint fluid) from January 2000 through December 2006. In addition, cases included patients with specimens testing positive by latex agglutination and supported by positive results by either Gram stain microscopy or PCR. Only one isolate per case was included in the analysis. Repeat isolates from the same patient received within 21 days of receipt of the initial isolate were excluded. If an isolate was received from the same patient more than 21 days after the initial isolate was received, it was treated as an isolate responsible for a new episode of IPD. Non-levofloxacin susceptibility was defined as a levofloxacin MIC of ≥4 mg/liter. Children were defined as individuals <15 years of age.
Carriage study.
To determine the prevalence of NLSSP carriage, cross-sectional carriage studies were carried out at two hospitals for patients with TB where invasive NLSSP disease was detected: a hospital in Gauteng Province (hospital A) in August 2006 and a hospital in Western Cape Province (hospital B) in May 2007. Details of the study have been described previously (26). Both hospitals treat chronic TB and specialize in the treatment of MDR TB. Patients receive fluoroquinolones as part of treatment for MDR TB. Other reasons for the use of fluoroquinolones include syndromic therapy for sexually transmitted infections in adults.
Phenotypic characterization.
Isolates were identified as pneumococci by standardized methodologies (23). Isolates were screened for fluoroquinolone resistance with 5-μg ofloxacin disks (Mast Diagnostics, Merseyside, United Kingdom); and if they were found to be resistant, ofloxacin, levofloxacin, and moxifloxacin MICs were determined by agar dilution (4) or Etest (AB-Biodisk, Solna, Sweden). MDR was defined as nonsusceptibility to antibiotics in three or more antimicrobial classes. Pneumococci were serotyped by the Quellung method with specific antisera (Statens Serum Institut, Copenhagen, Denmark).
PFGE.
Isolates were characterized by pulsed-field gel electrophoresis (PFGE) by previously described methods (15, 17). Restriction profiles were analyzed with GelCompar software (Applied Maths, Kortrijk, Belgium), and dendrograms were generated by the unweighted pair group method with arithmetic averages. The banding patterns were analyzed with the Dice coefficient, and an optimization of 1.5% and a position tolerance of 1.5% were used for the band migration distance. The PFGE profiles were visually examined, and clusters were assigned on the basis of three band differences or less (25), congruent with a similarity index of approximately 80%. Therefore, a cluster was defined as three or more isolates sharing ≥80% similarity on the dendrogram.
MLST.
Multilocus sequence typing (MLST) was performed, as described previously (7), on a random selection of isolates representative of the various clusters generated by PFGE. Sequence types (STs) were assigned by submission of the allele sequences to the MLST website (http://spneumoniae.mlst.net). Allelic profiles were compared with those of the global pneumococcal clones (http://www.sph.emory.edu/PMEN). Isolates with two allele differences or less were defined as related.
QRDR sequencing.
DNA was prepared by boiling bacterial cultures; and the QRDR regions of parC, parE, gyrA, and gyrB were amplified and sequenced as described previously (18). Mutations were identified by comparison of the sequences with those of fluoroquinolone-susceptible pneumococcal strain R6.
Growth studies.
Growth studies were performed for one invasive isolate and one carriage isolate from hospital A and one invasive isolate and two carriage isolates (of serotypes 19F and 23F, respectively) from hospital B. Pneumococci were inoculated from glycerol stocks (optical density at 600 nm, 0.3) into tryptone soy broth (dilution, 1:100) and incubated at 37°C in 5% CO2. Growth was monitored by measurement of the optical density at 600 nm at intervals of 30 min, until the cultures reached the stationary phase of growth.
RESULTS
Invasive disease surveillance.
From January 2000 through December 2006, 21,521 cases of IPD were identified. Of these, viable isolates could be obtained from 19,404 (90%). Of 19,572 cases whose ages were known, 44% (n = 8,692) were children. Twenty-two of 19,404 (0.1%) isolates were nonsusceptible to ofloxacin, and 12 of these were NLSSP. All 12 NLSSP were isolated from children, 9 of whom were known to be receiving TB treatment. Of the cases with known outcomes (n = 11), 5 (45%) died, 1 with a clinical diagnosis of meningitis (26).
Carriage study.
At hospital A, 116 of 139 (83%) adult patients and all 19 pediatric inpatients were swabbed. Nine of 19 (47%) children and no adults were found to be pneumococcal carriers. All nine isolates were NLSSP. Forty-six of 47 (98%) children at hospital B were swabbed. Fifty-seven percent (26/46) carried a pneumococcus, and of these, 85% (22/26) carried NLSSP. Two children were found to be carriers of multiple pneumococci during antimicrobial susceptibility testing and/or serotyping.
Characterization of NLSSP strains.
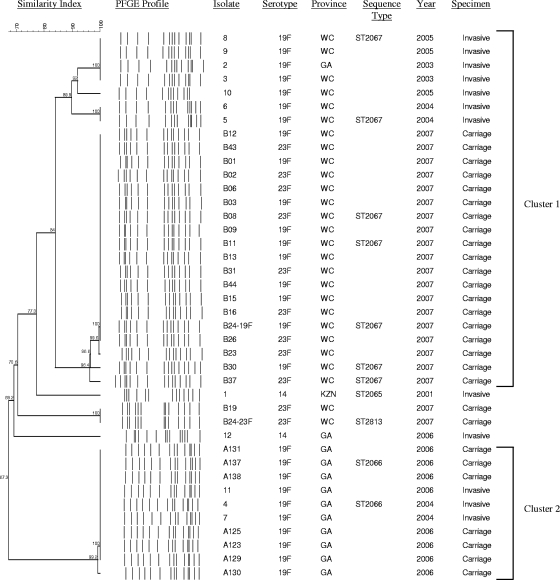
Of the 12 invasive isolates, 10 (83%) were serotype 19F and formed two clusters by PFGE (Fig. 1). The remaining two isolates were serotype 14 and were unrelated by PFGE. Seven invasive isolates formed cluster 1. These isolates were characterized by the mutations D78N in ParC, S81F in GyrA, and R447C in ParE (Table 1) and were nonsusceptible to trimethoprim-sulfamethoxazole and rifampin (rifampicin). Two randomly selected invasive isolates from this cluster were shown to be ST2067 (Fig. 1).
FIG. 1.
PFGE dendrogram indicating the clonality of the fluoroquinolone-resistant invasive and carriage pneumococcal isolates. An “A” in the isolate number indicates carriage isolates from hospital A; a “B” indicates carriage isolates from hospital B. GA, Gauteng Province; WC, Western Cape Province; KZN, KwaZulu-Natal Province.
TABLE 1.
Non-levofloxacin-susceptible pneumococci causing invasive disease in children <15 years in South Africa, 2000 to 2006
| Isolate | Serotype | MIC (mg/liter)
|
Resistance antibiograma | PFGE cluster | QRDR mutation(s)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Ofloxacin | Levofloxacin | Moxifloxacin | ParC | GyrA | ParE | ||||
| 1 | 14 | 32 | 16 | 8 | PEN, RIF, SXT | Unrelated | S79F, K137N | S81F | I460V |
| 2 | 19F | 32 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 3 | 19F | 32 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 4 | 19F | >32 | >32 | 8 | PEN, TET, ERY, CLI, RIF, SXT | 2 | S52G, S79I, N91D | S81Y, S114G | |
| 5 | 19F | 16 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 6 | 19F | 16 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 7 | 19F | >32 | >32 | 8 | PEN, TET, ERY, CLI, RIF, SXT | 2 | S52G, S79I, N91D | S81Y, S114G | |
| 8 | 19F | 32 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 9 | 19F | 32 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 10 | 19F | 32 | 4 | 2 | RIF, SXT | 1 | D78N | S81F | R447C |
| 11 | 19F | >32 | >32 | 8 | PEN, TET, ERY, CLI, RIF, SXT | 2 | S52G, S79I, N91D | S81Y, S114G | |
| 12 | 14 | 4 | 4 | 0.25 | PEN, TET, ERY, CLI, RIF, SXT | Unrelated | S52G, S79I, N91D, P140S | I460V | |
PEN, penicillin; TET, tetracycline; ERY, erythromycin; CLI, clindamycin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole.
Three invasive isolates formed cluster 2 and contained the mutations S52G, S79I, and N91D in ParC and mutations S81Y and S114G in GyrA (Table 1). The cluster 2 isolates were nonsusceptible to penicillin, tetracycline, erythromycin, clindamycin, rifampin, and trimethoprim-sulfamethoxazole. In addition, they had higher levofloxacin MICs than the cluster 1 isolates. MLST of one of these three invasive isolates identified the strain as ST2066 (Fig. 1), a double-locus variant of ST236, which is representative of the Taiwan19F-14 global clone.
All nine carriage isolates from hospital A were serotype 19F; had levofloxacin MICs >32 mg/liter; and were nonsusceptible to penicillin, erythromycin, clindamycin, rifampin, and trimethoprim-sulfamethoxazole. PFGE profiles were available for seven of the nine carriage isolates. They were indistinguishable from each other and the invasive isolates in cluster 2, including the invasive isolates from the same hospital (isolates 4 and 7) (Fig. 1). The fluoroquinolone resistance-determining mutations identified in the carriage isolates from this hospital were identical to those identified in the invasive isolates (Table 1).
At hospital B, 24 isolates were obtained from 22 patients carrying NLSSP, of which 12 each were serotype 19F and serotype 23F. These included two patients carrying both serotype 19F and serotype 23F (isolates B24 and B33, respectively). Isolate B33-23F was fluoroquinolone susceptible and was therefore excluded from further analysis. All 23 NLSSP carriage isolates were also nonsusceptible to rifampin and trimethoprim-sulfamethoxazole. PFGE profiles were available for 21 of 23 isolates, of which serotype 19F (n = 10) and serotype 23F (n = 9) isolates, including isolate B24-19F, were related by PFGE. They were classified as belonging to cluster 1 and were related to the invasive isolates from the same hospital (isolates 3, 5, 6, and 8 in cluster 1) (Fig. 1). However, carriage isolates had higher levofloxacin MICs (>32 mg/liter) and had mutations S52G, S79I, and N91D in ParC and mutations S81Y and S114G in GyrA. Five randomly selected carriage isolates were ST2067. Strain B24-23F, which contained the S81F mutation in GyrA and the D435N mutation in ParE, was ST2813 and was identical by PFGE to one other serotype 23F carriage isolate from hospital B (Fig. 1). ST2813 is a single-locus variant of ST242, which is representative of the Taiwan23F-15 global clone.
Growth studies.
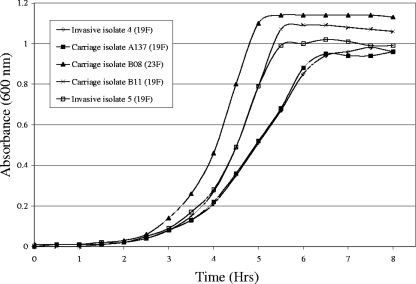
Growth curves are shown in Fig. 2. The invasive and carriage isolates from hospital A, which were identical by PFGE and MLST and which had identical fluoroquinolone resistance mutations, displayed almost identical growth curves. The invasive and carriage serotype 19F isolates from hospital B, which were related by PFGE and which were both ST2067, also displayed highly similar growth curves, despite their different QRDR mutations. However, the serotype 23F carriage isolate from hospital B reached the exponential and stationary phases of growth faster than the serotype 19F carriage isolate, which was otherwise identical to the serotype 23F isolate by PFGE, MLST, and QRDR mutations.
FIG. 2.
Growth curves at 37°C of a non-levofloxacin-susceptible invasive isolate (isolate 4) and a carriage isolate (isolate A137) from hospital A (PFGE cluster 2) and an invasive isolate (isolate 5) and carriage isolates (isolates B08 and B11) from hospital B (PFGE cluster 1). Serotypes are indicated in parentheses.
DISCUSSION
The emergence of NLSSP among children receiving treatment for MDR TB in South Africa has recently been described (26). The carriage rates of non-levofloxacin-susceptible pneumococci among the children were notably high and likely due to nosocomial spread. In this study, we genotypically characterized these invasive and carriage isolates to investigate their clonality as well as the molecular dynamics of these strains in this setting of children in close contact and receiving fluoroquinolone treatment for MDR TB.
The carriage isolates recovered from hospital A in 2006 belonged to the same clone as invasive isolates causing disease in that hospital recovered from 2004 to 2006. The isolates had identical antibiograms, resistance mutations, and PFGE patterns and were the same ST, indicating that this MDR clone had persisted and remained genetically stable over the 3-year period.
The carriage and invasive isolates from hospital B were related by PFGE and MLST. However, the carriage isolates, collected in May 2007, harbored different fluoroquinolone resistance-determining mutations and had higher levofloxacin MICs compared to the strains previously causing invasive disease in this hospital. The complete change in QRDR mutations between the invasive and the carriage strains suggests that horizontal gene transfer is likely responsible for the development of the highly resistant strain under the selective pressure of fluoroquinolone treatment. Pneumococci are naturally transformable (14), and therefore, the cocolonization of strains in the nasopharynx makes intra- and interspecies recombination possible (9). Interspecies and intraspecies horizontal gene transfer of fluoroquinolone resistance-conferring genes contributes to the spread of fluoroquinolone resistance in S. pneumoniae (1, 8, 24). Interspecies recombination has been shown to occur frequently in the parC gene of Streptococcus pyogenes (5); however, this is uncommon in the fluoroquinolone resistance-conferring genes in invasive pneumococci in the United States (19). The similar growth characteristics of the serotype 19F invasive and carriage isolates from hospital B suggest that the more recently identified QRDR mutations of the carriage isolates in the hospital B clone are not associated with increased fitness. The QRDR mutations of the carriage isolates are likely to have replaced the original mutations due to the increased levels of fluoroquinolone resistance that they confer under antibiotic pressure.
Furthermore, while the carriage isolates from hospital B displayed identical PFGE patterns, STs, and mutations, they expressed two different serotypes (serotypes 19F and 23F). Again, this indicates that genetic material was transferred between strains that were carried simultaneously, resulting in capsular switching (12). Capsular switching in a levofloxacin-resistant Spain23F-1 clone has been described previously (10). The growth studies indicated that the serotype 23F carriage isolate had increased fitness in comparison with the fitness of the serotype 19F isolate, and this may have been the driving force for the switch in capsule.
The invasive NLSSP isolates in South Africa formed two serotype 19F clusters, one of which was related to the Taiwan19F-14 global clone. In addition, two serotype 23F strains identified in carriers in hospital B were related to the international clone, Taiwan23F-15. Increases in fluoroquinolone resistance have been shown to be due to clonal spread (13, 19). In Hong Kong, fluoroquinolone-resistant isolates were related to the multidrug-resistant Spain23F-1 clone (10). This clone was also found to be predominant among levofloxacin-resistant isolates in the United States (20). Non-levofloxacin-susceptible pneumococci related to the Taiwan19F-14 global clone have been detected in the United States (22).
All the carriage and invasive NLSSP isolates were nonsusceptible to rifampin. Whereas fluoroquinolones inhibit bacterial DNA synthesis, rifampin binds to RNA polymerase and inhibits transcription (2). Resistance to rifampin is conferred by mutations in the RNA polymerase gene, rpoB. It is therefore likely that the rifampin resistance-conferring mutations developed independently from the fluoroquinolone resistance-conferring mutations in these organisms during treatment for MDR TB.
This study highlights the ability of pneumococci to undergo genetic recombination under antibiotic pressure. An environment in which children are in close contact, such as day care centers and hospitals, provides the ideal habitat for the cocolonization of multiple strains and species. In this study, all of the invasive and carriage isolates were vaccine serotypes 19F, 14, and 23F; and therefore, infection with these strains is preventable by vaccination. However, carriage of a non-vaccine-serotype strain and selective pressure due to the pneumococcal conjugate vaccine may result in fluoroquinolone-resistant strains whose serotypes are not covered by the vaccine and that have the potential to spread in these environments.
There are no data on the rates of carriage of resistant pneumococci in the South African community; however, we hypothesize that the low number of levofloxacin-resistant strains that were detected among more than 19,000 invasive pneumococcal isolates tested suggests that fluoroquinolone-resistant pneumococci are not able to ascend in frequency in South Africa, given the current low levels of fluoroquinolone use by children and adults outside of the hospital environment.
Acknowledgments
We thank all laboratory and clinical staff throughout South Africa for contributing to national surveillance and all the patients and guardians involved in the carriage study. We also thank Azola Fali, Olga Hattingh, Lenny Lengwati, Kedibone Mothibeli, Ruth Mpembe, Thomas Rafundisani, Happy Skosana, and Anthony Smith for technical expertise and assistance and Muzi Hlanzi and Ethel Maringa for data management. We also acknowledge the Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) for their efforts in collecting the South African surveillance and carriage isolates.
This study received funding from the National Institute for Communicable Diseases of the National Health Laboratory Service and was supported in part by funds from the United States Agency for International Development's Antimicrobial Resistance Initiative, transferred through cooperative agreement U60/CCU022088 from the Centers for Disease Control and Prevention (CDC), Atlanta, GA (2002 to 2005); and for 2006, it was supported in part by the DHHS CDC, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention (NCHHSTP), Global AIDS Program (GAP), through cooperative agreement U62/PSO022901.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC.
The work described here was performed at the Respiratory and Meningeal Pathogens Research Unit, National Institute for Communicable Diseases, Private Bag X4, Sandringham, 2131, Gauteng, South Africa.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Balsalobre, L., M. J. Ferrandiz, J. Linares, F. Tubau, and A. G. de la Campa. 2003. Viridans group streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 472072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calfee, D. P. 2005. Rifamycins, p. 374-388. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 6th ed. Elsevier Churchill Livingstone, Philadelphia, PA.
- 3.Chen, F. J., and H. J. Lo. 2003. Molecular mechanisms of fluoroquinolone resistance. J. Microbiol. Immunol. Infect. 361-9. [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing: 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Duesberg, C. B., S. Malhotra-Kumar, H. Goossens, L. McGee, K. P. Klugman, T. Welte, and M. W. Pletz. 2008. Interspecies recombination occurs frequently in quinolone resistance-determining regions of clinical isolates of Streptococcus pyogenes. Antimicrob. Agents Chemother. 524191-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eliopoulos, G. M. 2004. Quinolone resistance mechanisms in pneumococci. Clin. Infect. Dis. 38(Suppl. 4)S350-S356. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 1443049-3060. [DOI] [PubMed] [Google Scholar]
- 8.Ferrandiz, M. J., A. Fenoll, J. Linares, and A. G. de la Campa. 2000. Horizontal transfer of parC and gyrA in fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardes, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 692477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, P. L., T. L. Que, S. S. Chiu, R. W. Yung, T. K. Ng, D. N. Tsang, W. H. Seto, and Y. L. Lau. 2004. Fluoroquinolone and other antimicrobial resistance in invasive pneumococci, Hong Kong, 1995-2001. Emerg. Infect. Dis. 101250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, T. D., L. Avrain, G. de Bilderling, and Y. Glupczynski. 2006. Streptococcus pneumoniae clinical isolate highly resistant to fluoroquinolones in a child. Pediatr. Infect. Dis. J. 251195-1196. [DOI] [PubMed] [Google Scholar]
- 12.Jefferies, J. M., A. Smith, S. C. Clarke, C. Dowson, and T. J. Mitchell. 2004. Genetic analysis of diverse disease-causing pneumococci indicates high levels of diversity within serotypes and capsule switching. J. Clin. Microbiol. 425681-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klugman, K. P. 2003. The role of clonality in the global spread of fluoroquinolone-resistant bacteria. Clin. Infect. Dis. 36783-785. [DOI] [PubMed] [Google Scholar]
- 14.Lacks, S. A. 2004. Transformation, p. 89-115. In E. I. Tuomanen, T. J. Mitchell, D. A. Morrison, and B. G. Spratt (ed.), The pneumococcus. ASM Press, Washington, DC.
- 15.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 312724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandell, L. A., L. R. Peterson, R. Wise, D. Hooper, D. E. Low, U. B. Schaad, K. P. Klugman, and P. Courvalin. 2002. The battle against emerging antibiotic resistance: should fluoroquinolones be used to treat children? Clin. Infect. Dis. 35721-727. [DOI] [PubMed] [Google Scholar]
- 17.McEllistrem, M. C., J. E. Stout, and L. H. Harrison. 2000. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J. Clin. Microbiol. 38351-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan, X. S., J. Ambler, S. Mehtar, and L. M. Fisher. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 402321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pletz, M. W., L. McGee, B. Beall, C. G. Whitney, and K. P. Klugman. 2005. Interspecies recombination in type II topoisomerase genes is not a major cause of fluoroquinolone resistance in invasive Streptococcus pneumoniae isolates in the United States. Antimicrob. Agents Chemother. 49779-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pletz, M. W., L. McGee, J. Jorgensen, B. Beall, R. R. Facklam, C. G. Whitney, and K. P. Klugman. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob. Agents Chemother. 483491-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pletz, M. W., L. McGee, C. A. Van Beneden, S. Petit, M. Bardsley, M. Barlow, and K. P. Klugman. 2006. Fluoroquinolone resistance in invasive Streptococcus pyogenes isolates due to spontaneous mutation and horizontal gene transfer. Antimicrob. Agents Chemother. 50943-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter, S. S., K. P. Heilmann, S. E. Beekmann, N. J. Miller, C. L. Rice, and G. V. Doern. 2005. The molecular epidemiology of Streptococcus pneumoniae with quinolone resistance mutations. Clin. Infect. Dis. 40225-235. [DOI] [PubMed] [Google Scholar]
- 23.Ruoff, K. L., R. A. Whiley, and D. Beighton. 2003. Streptococcus, p. 405-421. In P. R. Murray, E. J. Baron, M. A. Pfaller, J. H. Jorgensen, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 24.Stanhope, M. J., S. L. Walsh, J. A. Becker, M. J. Italia, K. A. Ingraham, M. N. Gwynn, T. Mathie, J. A. Poupard, L. A. Miller, J. R. Brown, and H. Amrine-Madsen. 2005. Molecular evolution perspectives on intraspecific lateral DNA transfer of topoisomerase and gyrase loci in Streptococcus pneumoniae, with implications for fluoroquinolone resistance development and spread. Antimicrob. Agents Chemother. 494315-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Gottberg, A., K. P. Klugman, C. Cohen, N. Wolter, L. de Gouveia, M. du Plessis, R. Mpembe, V. Quan, A. Whitelaw, R. Hoffmann, N. Govender, S. Meiring, A. M. Smith, and S. Schrag. 2008. Emergence of levofloxacin-non-susceptible Streptococcus pneumoniae and treatment for multidrug-resistant tuberculosis in children in South Africa: a cohort observational surveillance study. Lancet 3711108-1113. [DOI] [PubMed] [Google Scholar]
- 27.von Gottberg, A., H. Ludewick, S. Bamber, C. Govind, A. W. Sturm, and K. P. Klugman. 2003. Emergence of fluoroquinolone-resistant Streptococcus pneumoniae in a South African child in a tuberculosis treatment facility. Pediatr. Infect. Dis. J. 221020-1021. [DOI] [PubMed] [Google Scholar]