Abstract
A novel method for the collection and transportation of dried-blood-plasma samples, SampleTanker (ST), was developed and compared to standard shipping protocols for frozen-plasma specimens containing human immunodeficiency virus type 1 (HIV-1) and/or hepatitis C virus (HCV). Matched frozen and dried 1-ml EDTA-containing plasma samples were collected and analyzed by several molecular-based virologic assays. After addition of 1.175 ml of reconstitution buffer, 1.035 ml of dried plasma was recovered. Mean intra-assay variances were 0.05, 0.05, and 0.06 log10 copies/ml for the Versant, Amplicor, and NucliSens QT HIV-1 load assays, respectively (P, not significant). However, mean HIV-1 viral load was consistently reduced in dried samples by 0.32 to 0.51 log10 copies/ml, depending on assay type (P < 0.05). Infectious HIV-1 was not recovered from dried ST plasma. There was no significant difference in HIV-1 viral load results obtained using ST after 8 weeks of storage at ambient temperature. Compared to frozen plasma, HIV-1 genotypic results were >99% concordant at the nucleotide and amino acid levels, as well as for resistance-associated mutations. We further demonstrated successful detection of multiple analytes, including HIV-1 viral load, HIV-1 antiretroviral resistance genotype, and HCV genotype, from a single ST unit. Dried plasma collected with ST yielded comparable results to frozen samples for multiple-analyte clinical testing. As such, ST could be a useful alternative for virologic tests and clinical trials worldwide by significantly diminishing transportation cost and the sample volume restrictions associated with dried-blood-spot technology.
Diagnostic and therapeutic monitoring assays for human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) infections are useful surrogate markers in the management and treatment of these infections (29). In addition, HIV-1 and HCV viral load, HCV genotype, and HIV-1 resistance testing are strongly correlated with response to antiviral therapy (27). Currently, blood tests which utilize HIV-1 and HCV viral nucleic acid require plasma separation and freezing at −70°C or greater within a few hours of collection. Most clinical samples and those associated with clinical trials require overnight shipment to reference laboratories under frozen conditions, which is expensive and cumbersome. In addition, the processing, storage, and transportation requirements for these assays limit their utility and accessibility in resource-poor environments.
The use of dried blood, serum, or plasma transport medium for viral diagnostics and therapeutic monitoring would offer several advantages over current sample collection and transportation requirements. The utility of using a dry blood spot (DBS) on filter paper has been a well-established whole-blood collection method for congenital and inherited metabolic neonatal screening programs and was demonstrated to be effective in the collection of serum containing thyroid hormone several decades ago (16). The utility of DBSs was reported early in the AIDS epidemic for HIV-1 antibody detection (12) and continues to be evaluated for antibodies to various subtypes and for p24 antigen testing (4, 17, 18, 21, 31). The DBS has also been used for detection of antibodies against other viral infections such as HCV, human T-cell leukemia virus type 1, and hepatitis B surface antigen (10), as well as HIV-1 proviral DNA in peripheral blood mononuclear cells (PBMCs) and HIV and HCV RNA viral load using PCR (6, 7, 13, 14, 28, 30). Dried-blood samples have been used for HIV-1 subtype analysis and CD4+ T-cell and plasma antiretroviral drug concentration quantification (8, 19, 24, 26, 33), and antiretroviral (ARV) genotypic resistance testing has been demonstrated (5, 22, 32, 35). Phenotypic ARV resistance testing or the feasibility to assay multiple viral nucleic acid analytes simultaneously from a single sample has not been described.
In this study, we describe a novel, non-paper-based matrix sample collection and transportation device called SampleTanker (ST), which was used to collect dried blood plasma for virologic testing. We evaluated the feasibility, precision, and short-term stability of using ST as a transportation method for shipping specimens at ambient temperature and for subsequent individual or simultaneous testing using HIV-1 viral load, HIV genotypic resistance testing, and HCV genotyping assays.
MATERIALS AND METHODS
Institutional review boards at the University of Texas Medical Branch, Galveston, TX, and Stanford University, Stanford, CA, approved the protocol. Whole-blood samples containing EDTA as an anticoagulant were collected from HIV-1-infected subjects, and separated blood plasma was stored at −80°C. Deidentified samples were pulled from freezer storage and shipped to Research Think Tank, Inc., Buford, GA, on dry ice for next-day arrival. Samples arrived frozen with dry ice still visible in the shipping container. Samples were pooled and mixed thoroughly to create a master pool, and nine 1-ml aliquots were taken for viral load determination using the Cobas Amplicor HIV-1 Monitor Standard assay (Roche Molecular, Branchburg, NJ). The master pool was determined to have an HIV-1 viral load of approximately 350,000 copies/ml. A 1:100 dilution was performed using the master pool and Base Matrix (normal human plasma; SeraCare Life Sciences, Milford, MA) to obtain a low-viral-load determinant point of approximately 3,500 copies/ml as determined by the Amplicor HIV-1 Monitor standard viral load assay.
Five 1-ml replicates each were created representing high (350,000 copies/ml) and low (3,500 copies/ml) viral load control reference standards and immediately frozen at −80°C. One milliliter of the experimental plasma at the high and low determinant points was directly applied to five replicate ST units to determine short-term stability under the temperature conditions room temperature (23°C, 42% humidity), 37°C (incubator, 42% humidity), and −80°C and the termination points day 2, day 7, day 14, day 24, and day 56. ST units were then placed on the front grill of a Baker class II A/B3 externally venting biological safety cabinet. All drying ST units were left with the cabinet running overnight with a relative humidity reading of the interior hood of 32 to 35%.
Once ST units reached the termination time point for the condition and time noted above, they were pulled and observed for color change of the desiccant in the transport indicator capsule. Color was logged, and each respective experimental ST unit was rehydrated and incubated with 1.175 ml of rehydration buffer for approximately 30 min at ambient room temperature before recovery. Reconstituted plasma was recovered using the ST sample recovery kit. The reconstituted plasma was then used for molecular testing.
All stability experiment samples were run using the Amplicor HIV-1 Monitor standard viral load assay. The control frozen plasma reference standards were only run with the day 2 ST experimental samples. These frozen plasma viral load values were used as the reference for comparisons to all subsequent ST stability experimental samples. The additional samples were all reconstituted, and viral load was determined on the day specified above.
HIV-1 and HIV-1-HCV-coinfected clinical specimens were selected for testing using precharacterized samples with known results for HIV-1 viral load and HIV and HCV genotyping tests. Specimens were unlinked to identifying information including date, patient identifier, patient name, and health center identifier. Replicate, deidentified, and coded 1-ml frozen-plasma samples, collected in standard EDTA-containing blood collection or plasma preparation tubes (Becton Dickinson, Franklin Lakes, NJ), and matched ST samples were prepared for shipping simultaneously. Frozen samples were shipped overnight on dry ice to the testing facility (Research Think Tank, Inc., Buford, GA) according to standard shipping requirements. ST samples were prepared using the ST sample recovery kit (Research Think Tank, Inc., Buford, GA). One thousand microliters of plasma was added directly to the ST matrix and completely air dried for a minimum of 5 h at ambient room temperature in a certified biological safety cabinet. STs were shipped at ambient temperature in air-tight, sealed, desiccant-containing ST transport and storage tubes, via U.S. mail or a commercial courier service using standard mailing envelopes according to the CDC guidelines for dried-blood shipment (9).
Additional patient samples were used to determine intra-assay variability and possible differences in HIV-1 viral load from paired frozen- and reconstituted-plasma replicates using the Versant HIV-1 RNA 3.0 assay (Siemens, Tarrytown, NY) and NucliSens HIV-1 QT assay (bioMerieux, Durham, NC), in addition to using Amplicor HIV-1 Monitor standard or ultrasensitive test, version 1.5 (Roche Diagnostics, Branchburg, NJ). HCV genotyping was performed using the TruGene HCV 5′NC genotyping kit (Siemens, Tarrytown, NY). HCV genotyping results were compared using the automated reporting of the HCV TruGene 5′NC assay and OpenGene system. HIV-1 genotypic resistance testing was performed using the TruGene HIV-1 resistance genotyping kit (Siemens, Tarrytown, NY). To assess short-term stability for HIV-1 genotypic resistance testing, three dried-plasma samples were compared to frozen samples and evaluated at day 1 and day 7 of storage. All plasma samples used for genotyping were extracted using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). All sequencing, data processing, and reporting were performed using the OpenGene DNA sequencing system (Siemens, Tarrytown, NY). HIV-1 genotypic analysis for dried specimens was accomplished using FASTA nucleotide and translated amino acid sequences in direct comparison to the corresponding sequence derived from frozen plasma. The FDA-approved TruGene sequence from frozen plasma served as the standard or reference sequence for comparison to the paired TruGene sequence obtained from ST samples. Stringent scoring of sequence and detected mixtures was accomplished using universal IUB base coding. As a secondary and confirmatory analysis, the polymorphic fingerprints or differences in nucleotide and amino acid sequences as compared to a fixed reference of HIV-1LAI were also compared.
Dried-plasma samples were cultured for infectious HIV using the following protocol. Briefly, 1 ml of EDTA plasma from an HIV-uninfected donor was spiked with approximately 1 × 105 tissue culture infectious dose (TCID)/ml of the HIV-1 NL4-3 strain. Infectious plasma samples were applied to STs in triplicate, allowed to dry, and eluted as described above. Reconstituted ST plasma was incubated with 1 million phytohemagglutinin-stimulated PBMCs in medium (RPMI 1640, 10% fetal calf serum) for 1 h. PBMCs were washed with phosphate-buffered saline twice, resuspended in 1 ml of complete culture medium, and transferred to 24-well plates. Cultures were held for a total of 14 days, with a medium exchange and additional PBMCs added on day 7. Day 14 culture supernatants were assayed for HIV p24 antigen using a commercially available kit (Zeptometrix, Buffalo, NY). Positive controls included cultures of NL4-3-spiked EDTA-containing plasma that was not applied to STs, including dilutions (105 to 101 TCID/ml) of the spiked plasma that were used to verify the infectious titer. Negative controls included cultures of non-virus-spiked HIV-1-negative plasma.
Descriptive statistics were used to calculate mean variances of log10-transformed viral load copy number/ml and percent change in log10/ml copy number, and the nucleotide differences comparing paired frozen and dried samples. Student's t test or Wilcoxon's signed-rank test was utilized to measure differences in log10 viral load copy number or changes in log10 percent comparing frozen to dried samples and dried-sample intra-assay variability as appropriate. Significance was defined as P < 0.05.
RESULTS
ST stability results.
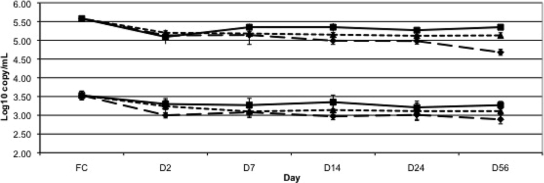
HIV-1 viral load results were determined from replicate samples stored on ST at 37°C, 23°C, and −80°C for 2, 7, 14, 24, and 56 days. After reconstitution, the ST specimens yielded a mean recovered plasma volume of 1.035 ± 0.041 ml. Compared to baseline frozen plasma, there were mean reductions in viral load of 0.38 to 0.45 and 0.23 to 0.53 log10 copies/ml for the high- and low-copy-number samples, respectively, at day 2 depending on the temperature of storage (all comparisons, P < 0.05). However, when day 2 ST was compared to day 56 ST, there was no significant difference in HIV log10 copy number out to 56 days of storage at 23°C or −80°C and out to 24 days at 37°C, but there was a further significant decline in viral load in samples held at 37°C for 56 days (P < 0.05) (Fig. 1). The log10 copy/ml percent loss between baseline frozen plasma and day 2 ST was 6.4 to 8.8% at the high copy number and 7.1 to 14.8% at the low copy number (P < 0.05 for all temperatures). When day 2 ST log10 copy/ml was compared to day 56 ST, the percent loss ranged from <1 to 9% at the high copy number (P < 0.05 for the 37°C samples only) and 2.6 to 10.7% at the low copy number, which were not significantly different at any temperature.
FIG. 1.
HIV-1 viral load results at high (350,000 copies/ml) and low (3,500 copies/ml) copy numbers expressed as log10 copies/ml according to time and temperature variation. Squares with solid lines, −80°C; triangles with dotted lines, 23°C; diamonds with hatched lines, 37°C. Error bars indicate standard deviation. FC, frozen control.
Multiple HIV-1 viral load assay results.
HIV-1 RNA from frozen and dried plasma samples was successfully obtained in all samples with known prior detectable viral load. Intra-assay variation was determined in 12 patient samples by analyzing multiple ST replicates from three different clinical samples (six replicates each) using the Versant assay, three replicates from six clinical samples (three replicates each) using the Amplicor HIV-1 Monitor assay, and five replicates from three clinical samples (five replicates each) using the NucliSens QT assay. Intra-assay overall mean variances were 0.05, 0.05, and 0.06 log10 copies/ml for the Versant, Monitor, and NucliSens QT assays, respectively (data not shown; P not significant).
Comparisons between 31 matched frozen- and dried-plasma samples demonstrated mean differences of 0.36, 0.51, and 0.32 log10 copies/ml for the Monitor (n = 9), Versant (n = 12), and NucliSens QT (n = 10) assays, respectively (Table 1) (P < 0.05 for individual assay analyses and for all three assay results combined). HIV-1 viral loads obtained from the ST matrix were consistently lower than those obtained from frozen plasma regardless of the HIV-1 viral load assay used and resulted in significant losses of 8%, 12.7%, and 8.3% in mean log10 copies/ml for the Monitor, Versant, and NucliSens assays, respectively (P < 0.05 for individual assay analyses and for all assays combined).
TABLE 1.
HIV-1 viral load comparison between frozen- and dried-plasma samples
| Assay | No. of samples | Mean log10 copies/ml in plasma
|
Difference in log10 copies/ml | |
|---|---|---|---|---|
| Frozen | Dried | |||
| Amplicor HIV-1 Monitor, v1.5 | 9 | 4.72 | 4.36 | 0.36 |
| Versant HIV 3.0 | 12a | 4.38 | 3.87 | 0.51 |
| NucliSens HIV-1 QT | 10 | 4.07 | 3.75 | 0.32 |
Three samples were <75 (1.87 log10) copies/ml for both frozen and dried plasma.
HIV-1 genotypic resistance testing.
Twenty archived frozen-plasma-derived HIV-1-positive samples with viral loads ranging from 3,100 to 258,000 copies/ml (3.49 to 5.41 log10 copies/ml) were tested using the TruGene HIV-1 genotyping assay. Mean accuracy and concordance of HIV-1 genotypic results between frozen and dried plasma were 99.7% at the nucleotide level (n = 924 nucleotides sequenced/sample) and 99.8% at the amino acid level (n = 308 amino acids). Reported resistance-associated mutations (RAMs) as defined by the TruGene assay demonstrated 99.9% concordance for genotyping assays (Table 2). There was no difference in the RAMs detected when day 1 and 7 dried-plasma samples were compared, and the overall mean sequence accuracy was ≥99.9% (data not shown). These results are comparable to results published in the TruGene product insert comparing duplicate frozen-plasma accuracy and reproducibility experiments with the U.S. FDA-approved kit.
TABLE 2.
HIV-1 genotypic resistance test results comparing frozen- and dried-plasma specimens
| Sample no. | Amt of frozen plasma RNA (log10 copies/ml) | % Similarity score for frozen vs. dried plasmaa
|
|||
|---|---|---|---|---|---|
| RAMs
|
Polymorphic fingerprint
|
||||
| Nucleotides (n = 165) | Amino acids (n = 55) | Nucleotides (n = 924) | Amino acids (n = 308) | ||
| 1 | 3.49 | 99.4 | 100 | 99.9 | 100 |
| 2 | 3.59 | 99.7 | 100 | 98.3 | 97.6 |
| 3 | 4.14 | 100 | 100 | 99.7 | 100 |
| 4 | 4.23 | 99.4 | 99.1 | 99.7 | 99.4 |
| 5 | 4.34 | 100 | 100 | 99.8 | 99.8 |
| 6 | 4.40 | 100 | 100 | 99.6 | 100 |
| 7 | 4.45 | 100 | 100 | 100 | 100 |
| 8 | 4.50 | 99.4 | 99.1 | 99.4 | 99.4 |
| 9 | 4.59 | 100 | 100 | 99.6 | 99.7 |
| 10 | 4.62 | 99.7 | 100 | 99.7 | 99.8 |
| 11 | 4.68 | 99.7 | 100 | 99.7 | 100 |
| 12 | 4.81 | 100 | 100 | 100 | 100 |
| 13 | 4.84 | 100 | 100 | 99.5 | 100 |
| 14 | 4.86 | 100 | 100 | 99.7 | 100 |
| 15 | 4.94 | 100 | 100 | 99.5 | 100 |
| 16 | 4.97 | 99.7 | 99.1 | 99.7 | 99.7 |
| 17 | 4.99 | 100 | 100 | 99.9 | 100 |
| 18 | 5.00 | 100 | 100 | 100 | 100 |
| 19 | 5.26 | 100 | 100 | 99.9 | 100 |
| 20 | 5.41 | 100 | 100 | 99.9 | 100 |
| Mean score | 99.9 | 99.9 | 99.7 | 99.8 | |
Specimens were air dried and stored at ambient temperature for 4 to 7 days prior to testing. TruGene HIV-1 genotyping kit-derived real-time first-pass sequences were used as the “gold standard” reference sequence for comparison to dried ST specimens. Stringency of scoring includes IUB base codes for mixture detection.
Multiple-analyte testing of HIV-1-HCV-coinfected samples.
Three HIV-1-HCV-coinfected clinical samples were used to explore the feasibility of detecting multiple molecular analytes from a single dried ST specimen. HIV-1 viral load, ARV genotype, and HCV genotype results were successfully determined for each unique sample (Table 3). Comparison of dried with frozen plasma indicated >99% genotypic concordance in the detection of RAMs by TruGene (Table 3). It was also noted that the signal/noise ratio and overall peak signal intensity of dried ST specimens using the TruGene assay were improved over those of frozen plasma (data not shown). The TruGene HCV 5′ NC genotyping assay demonstrated identical type determinations for dried- versus frozen-plasma samples (Table 3), whereas, subtyping results were also concordant for two of three samples. One frozen-plasma sample did not subtype, while the corresponding dried specimen did give a subtype result.
TABLE 3.
Multiple-analyte comparison of frozen- and dried-plasma specimens from HIV-1-HCV-coinfected samples by HIV-1 viral load, genotypic resistance testing, and HCV genotyping
| Sample no. | Amt of dried plasma RNA (log10 copies/ml) | % Similarity score for frozen plasma reference vs. dried plasmaa
|
HCV genotype from plasmab
|
||||
|---|---|---|---|---|---|---|---|
| RAMs
|
Polymorphic fingerprint
|
||||||
| Nucleotides (n = 165) | Amino acids (n = 55) | Nucleotides (n = 924) | Amino acids (n = 308) | Frozen | Dried | ||
| 1 | 2.57 | 99.2 | 100 | 99.4 | 99.8 | 1a | 1a |
| 2 | 4.01 | 99.1 | 99.1 | 99.4 | 99.4 | 2b | 2b |
| 3 | 4.53 | 100 | 100 | 99.9 | 100 | 3c | 3a |
| Mean score | 99.4 | 99.7 | 99.6 | 99.7 | |||
HIV-1 viral load (analyte 1) was determined by the Amplicor HIV-1 Monitor test, version 1.5 (standard method). Specimens were air dried and stored ambient for 7 days prior to testing. For genotypic resistance testing (analyte 2), TruGene HIV-1 genotyping kit-derived real-time first-pass sequences were used as the “gold standard” reference sequence for comparison to dried ST specimens. Stringency of scoring includes IUB base codes for mixture detection.
HCV genotype results represent analyte 3. Results are from the TruGene HCV 5′NC genotyping kit.
Subtype not resolved.
HIV-1 infectivity testing.
HIV culture results were positive in all fresh plasma replicates at 105 to 101 TCID/ml, whereas all HIV cultures from ST dried plasma were negative for HIV-1 p24 antigen (data not shown).
DISCUSSION
We have shown that quantitative and qualitative virologic results obtained from dried-plasma samples collected and shipped using ST are comparable to those obtained from normally processed samples. Viral load results among dried samples using ST with U.S. FDA-approved quantitative HIV-1 viral load assays were highly reproducible. Intra-assay results were within previously published acceptable limits for all assays (25). Although quantitation of HIV-1 viral load assays using ST dried plasma yielded somewhat lower values, consistent with findings using DBS (2), results were generally within the variation described for replicate samples previously described. Additional experiments with 750 and 500 μl of elution volume still yielded similar reduced HIV viral load results (data not shown). Thus, concentration of ST dried plasma did not correct for lower HIV viral loads seen with larger eluent volume. This indicates that there is likely some loss in HIV RNA as a result of the drying process. Evaluation is ongoing to determine optimal conditions for drying that could reduce or eliminate this discrepancy. However, the data on comparability and reproducibility suggest that ST has promise for clinical care and in clinical trial management of HIV- and HCV-infected individuals.
There was no significant difference in sensitivity on nucleic acid sequences, as demonstrated by the high correlation compared with frozen plasma. Genotype accuracy and reproducibility using ST were comparable to data published in the TruGene product inserts for frozen plasma samples (15, 20). Finally, HCV genotyping was concordant with matched frozen- and ST plasma specimens. As proof of concept, three clinical samples were tested for multiple analytes and demonstrated that HIV viral load, HIV genotypic resistance testing, and HCV genotyping results could all be obtained from a single dried 1-ml sample. Improved raw data were noted for the dried ST specimens over the neat plasma when using the HIV-1 or HCV genotyping assays. This suggests the possible removal of interfering substances after the drying and subsequent rehydration steps using the ST sample recovery kit.
As previously mentioned, multiple studies have been published evaluating filter paper as an efficient method of whole-blood or serum collection for a variety of analytes. Although an efficient method of collection, the assessment of multiple analytes from DBSs is limited due to sample volumes of 25 to 50 μl per spot. Thus, multiple DBSs or filter cards maybe necessary in order to perform multiple assays. In addition, there is concern about how filter card samples are processed and handled and their ability to withstand various temperature and humidity conditions. A recent study analyzing the stability of DBSs held at 37°C and 85% humidity for 3 months with a desiccant found ARV genotypic results to be highly correlated with frozen plasma (5). In addition, ARV genotypic analyses performed on DBSs stored for 5 to 6 years found that those stored at −20°C and −70°C, as opposed to those stored at room temperature, were comparable to frozen plasma (23). HIV viral load results were found to be highly concordant between DBSs and frozen plasma stored at ambient temperature or 37°C for 7 days (1). No long-term studies using DBSs for viral load measurement have been performed. We have shown that samples held at ambient room temperature or at 37°C for over 3 weeks are extremely stable, do not gain moisture (assessed by dessicant color change), and yield comparable, although slightly lower, viral load results to fresh frozen-plasma samples or immediately processed ST samples.
Most nucleic acid assays using DBSs require separation by cutting or punching out the spots and placing them into a buffer prior to nucleic acid extraction. There is significant risk of contamination between samples if the equipment used in cutting the DBSs is not sufficiently decontaminated between samples or subsequent pooling of multiple spots if required, although recent data analyzing HIV DNA (not plasma HIV-1 viral load) suggest this might not be as great a concern as previously thought (11). However, whole-blood DBSs may not be as sensitive as plasma DBSs, limiting their utility for HIV-1 viral load testing (3). ST removes these concerns as each sample utilizes its own sample collection kit, the recovered volume of reconstituted plasma is sufficient to assess multiple analytes, no cutting or pooling is required, and samples are transported within a sealed vial. Therefore, there is minimal risk of contamination compared to DBS samples prepared on filter paper. Furthermore, recent data indicate that successful amplification of nucleic acid for HIV-1 genotyping from DBS was approximately 82 to 93%, depending on storage conditions, ability to resolve mixtures, whether plasma RNA or cellular DNA was tested, and which HIV-1 gene was analyzed (5, 22, 32, 34). We demonstrated no such loss in sensitivity in detecting nucleic acid for HIV-1 genotyping from plasma dried on the ST matrix.
DBSs have been determined to be noninfectious and thus do not require specialized handling (9). Although ST contains significantly more plasma volume, and hence a greater possibility of infectious HIV, we did not find this to be the case using standard HIV culture techniques and high-infectious-titer reconstruction experiments. Whether other infectious agents such as HCV retain their infectivity in ST dried plasma requires further study. Thus, if blood samples applied to ST were also declared not infectious, they would offer the advantage of using standard mail or commercial shipment without the use of dry or wet ice, special biocontainment vessels, and the requirement for hazardous shipment designation. The average cost for commercial shipment of biohazardous material ranges from $50 to $100 for U.S. domestic shipping to several hundred dollars for international shipping. Transition to dried-blood specimens for virologic testing would result in millions of dollars saved in associated shipping costs. As more countries respond to the global AIDS crisis, the need for virologic assays will increase. Limited resources should not be used to pay for costly, specialized sample shipment. Transportation and storage systems like ST could help facilitate clinical trials and clinical practice to proceed in resource-limited areas by allowing samples to be dried, temporarily stored, and sent to regional or international reference laboratories with the assurance that results are valid and reproducible.
In summary, ST is a convenient, cost-effective alternative to the storage and shipping of frozen plasma for nucleic acid testing. ST could eliminate the need for frozen specimen shipments for HIV clinical trials worldwide. Further comparison studies are ongoing for virologic as well as biochemical analytes and show promise for ST use in a wider array of clinical testing.
Acknowledgments
The ST collection system is a product that is currently being developed by Research Think Tank, Inc., Buford, GA. The following coauthors are or were employees of Research Think Tank: R. M. Lloyd, Jr., D. A. Burns, J. T. Huong, R. L. Mathis, J. L. Wetshtein, W. O'Brien, and P. M. Feorino. D. R. McClernon has an equity position with respect to Research Think Tank, Inc. A. De La Rosa is a board member of Research Think Tank, Inc. None of the other coauthors have any conflict of interest regarding the development and evaluation of this product.
This work was supported in part by a grant from the Department of Veterans Affairs (Mark Holodniy).
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Alvarez-Munoz, M. T., S. Zaragoza-Rodriguez, O. Rojas-Montes, G. Palacios-Saucedo, G. Vazquez-Rosales, A. Gomez-Delgado, J. Torres, and O. Munoz. 2005. High correlation of human immunodeficiency virus type-1 viral load measured in dried-blood spot samples and in plasma under different storage conditions. Arch. Med. Res. 36382-386. [DOI] [PubMed] [Google Scholar]
- 2.Amellal, B., C. Katlama, and V. Calvez. 2007. Evaluation of the use of dried spots and of different storage conditions of plasma for HIV-1 RNA quantification. HIV Med. 8396-400. [DOI] [PubMed] [Google Scholar]
- 3.Ayele, W., R. Schuurman, T. Messele, W. Dorigo-Zetsma, Y. Mengistu, J. Goudsmit, W. A. Paxton, M. P. de Baar, and G. Pollakis. 2007. Use of dried spots of whole blood, plasma, and mother's milk collected on filter paper for measurement of human immunodeficiency virus type 1 burden. J. Clin. Microbiol. 45891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barin, F., J. C. Plantier, D. Brand, S. Brunet, A. Moreau, B. Liandier, D. Thierry, F. Cazein, F. Lot, C. Semaille, and J. C. Desenclos. 2006. Human immunodeficiency virus serotyping on dried serum spots as a screening tool for the surveillance of the AIDS epidemic. J. Med. Virol. 78(Suppl. 1)S13-S18. [DOI] [PubMed] [Google Scholar]
- 5.Bertagnolio, S., L. Soto-Ramirez, R. Pilon, R. Rodriguez, M. Viveros, L. Fuentes, P. R. Harrigan, T. Mo, D. Sutherland, and P. Sandstrom. 2007. HIV-1 drug resistance surveillance using dried whole blood spots. Antivir. Ther. 12107-113. [PubMed] [Google Scholar]
- 6.Cassol, S., M. J. Gill, R. Pilon, M. Cormier, R. F. Voigt, B. Willoughby, and J. Forbes. 1997. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J. Clin. Microbiol. 352795-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassol, S., T. Salas, M. Arella, P. Neumann, M. T. Schechter, and M. O'Shaughnessy. 1991. Use of dried blood spot specimens in the detection of human immunodeficiency virus type 1 by the polymerase chain reaction. J. Clin. Microbiol. 29667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassol, S., B. G. Weniger, P. G. Babu, M. O. Salminen, X. Zheng, M. T. Htoon, A. Delaney, M. O'Shaughnessy, and C. Y. Ou. 1996. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 121435-1441. [DOI] [PubMed] [Google Scholar]
- 9.CDC. 1995. Guidelines for the shipment of dried blood spot specimens. Centers for Disease Control and Prevention, Atlanta, GA.
- 10.Das, P. C., A. H. de Vries, R. L. McShine, and C. T. Sibinga. 1996. Dried sera for confirming blood-borne virus infections (HCV, HTLV-I, HIV & HBsAg). Transfus. Med. 6319-323. [DOI] [PubMed] [Google Scholar]
- 11.Driver, G. A., J. C. Patton, J. Moloi, W. S. Stevens, and G. G. Sherman. 2007. Low risk of contamination with automated and manual excision of dried blood spots for HIV DNA PCR testing in the routine laboratory. J. Virol. Methods 146397-400. [DOI] [PubMed] [Google Scholar]
- 12.Farzadegan, H., T. Quinn, and B. F. Polk. 1987. Detecting antibodies to human immunodeficiency virus in dried blood on filter papers. J. Infect. Dis. 1551073-1074. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, A., C. Lejczak, C. Lambert, J. Servais, N. Makombe, J. Rusine, T. Staub, R. Hemmer, F. Schneider, J. C. Schmit, and V. Arendt. 2004. Simple DNA extraction method for dried blood spots and comparison of two PCR assays for diagnosis of vertical human immunodeficiency virus type 1 transmission in Rwanda. J. Clin. Microbiol. 4216-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiscus, S. A., D. Brambilla, L. Grosso, J. Schock, and M. Cronin. 1998. Quantitation of human immunodeficiency virus type 1 RNA in plasma by using blood dried on filter paper. J. Clin. Microbiol. 36258-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, R. M., D. R. Kuritzkes, V. A. Johnson, J. W. Mellors, J. L. Sullivan, R. Swanstrom, R. T. D'Aquila, M. Van Gorder, M. Holodniy, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J. Clin. Microbiol. 411586-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irie, M., K. Enomoto, and H. Naruse. 1975. Measurement of thyroid-stimulating hormone in dried blood spot. Lancet ii1233-1234. [DOI] [PubMed] [Google Scholar]
- 17.Knuchel, M. C., B. Jullu, C. Shah, Z. Tomasik, M. P. Stoeckle, R. F. Speck, D. Nadal, H. Mshinda, J. Boni, M. Tanner, and J. Schupbach. 2007. Adaptation of the ultrasensitive HIV-1 p24 antigen assay to dried blood spot testing. J. Acquir. Immune Defic. Syndr. 44247-253. [DOI] [PubMed] [Google Scholar]
- 18.Knuchel, M. C., Z. Tomasik, R. F. Speck, R. Luthy, and J. Schupbach. 2006. Ultrasensitive quantitative HIV-1 p24 antigen assay adapted to dried plasma spots to improve treatment monitoring in low-resource settings. J. Clin. Virol. 3664-67. [DOI] [PubMed] [Google Scholar]
- 19.Koal, T., H. Burhenne, R. Romling, M. Svoboda, K. Resch, and V. Kaever. 2005. Quantification of antiretroviral drugs in dried blood spot samples by means of liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 192995-3001. [DOI] [PubMed] [Google Scholar]
- 20.Kuritzkes, D. R., R. M. Grant, P. Feorino, M. Griswold, M. Hoover, R. Young, S. Day, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Performance characteristics of the TRUGENE HIV-1 Genotyping Kit and the Opengene DNA Sequencing System. J. Clin. Microbiol. 411594-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, C. C., K. D. Seidel, R. W. Coombs, and L. M. Frenkel. 2005. Detection and quantification of human immunodeficiency virus type 1 p24 antigen in dried whole blood and plasma on filter paper stored under various conditions. J. Clin. Microbiol. 433901-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masciotra, S., C. Garrido, A. S. Youngpairoj, A. McNulty, N. Zahonero, A. Corral, W. Heneine, C. de Mendoza, and J. G. Garcia-Lerma. 2007. High concordance between HIV-1 drug resistance genotypes generated from plasma and dried blood spots in antiretroviral-experienced patients. AIDS 212503-2511. [DOI] [PubMed] [Google Scholar]
- 23.McNulty, A., C. Jennings, D. Bennett, J. Fitzgibbon, J. W. Bremer, M. Ussery, M. L. Kalish, W. Heneine, and J. G. Garcia-Lerma. 2007. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J. Clin. Microbiol. 45517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei, J. V., W. H. Hannon, T. L. Dobbs, C. J. Bell, C. Spruill, and M. Gwinn. 1998. Radioimmunoassay for monitoring zidovudine in dried blood spot specimens. Clin. Chem. 44281-286. [PubMed] [Google Scholar]
- 25.Murphy, D. G., L. Côté, M. Fauvel, P. René, and J. Vincelette. 2000. Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1.5, Organon Teknika NucliSens QT with Extractor, and Bayer Quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 384034-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mwaba, P., S. Cassol, R. Pilon, C. Chintu, M. Janes, A. Nunn, and A. Zumla. 2003. Use of dried whole blood spots to measure CD4+ lymphocyte counts in HIV-1-infected patients. Lancet 3621459-1460. [DOI] [PubMed] [Google Scholar]
- 27.National Institutes of Health. 2002. NIH consensus statement on management of hepatitis C: 2002. NIH Consens. State Sci. Statements 191-46. [PubMed] [Google Scholar]
- 28.Ou, C. Y., H. Yang, S. Balinandi, S. Sawadogo, V. Shanmugam, P. M. Tih, C. Adje-Toure, S. Tancho, L. K. Ya, M. Bulterys, R. Downing, and J. N. Nkengasong. 2007. Identification of HIV-1 infected infants and young children using real-time RT PCR and dried blood spots from Uganda and Cameroon. J. Virol. Methods 144109-114. [DOI] [PubMed] [Google Scholar]
- 29.Panel on Antiretroviral Guidelines for Adults and Adolescents. 29 October 2004, posting date. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. Department of Health and Human Services. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf.
- 30.Patton, J. C., E. Akkers, A. H. Coovadia, T. M. Meyers, W. S. Stevens, and G. G. Sherman. 2007. Evaluation of dried whole blood spots obtained by heel or finger stick as an alternative to venous blood for diagnosis of human immunodeficiency virus type 1 infection in vertically exposed infants in the routine diagnostic laboratory. Clin. Vaccine Immunol. 14201-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patton, J. C., G. G. Sherman, A. H. Coovadia, W. S. Stevens, and T. M. Meyers. 2006. Ultrasensitive human immunodeficiency virus type 1 p24 antigen assay modified for use on dried whole-blood spots as a reliable, affordable test for infant diagnosis. Clin. Vaccine Immunol. 13152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plantier, J. C., R. Dachraoui, V. Lemee, M. Gueudin, F. Borsa-Lebas, F. Caron, and F. Simon. 2005. HIV-1 resistance genotyping on dried serum spots. AIDS 19391-397. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro, H. M., F. F. Mandy, T. F. Rinke de Wit, and P. Sandstrom. 2004. Dried blood spot technology for CD4+ T-cell counting. Lancet 363164-165. (Letter.) [DOI] [PubMed] [Google Scholar]
- 34.Steegen, K., S. Luchters, E. Demecheleer, K. Dauwe, K. Mandaliya, W. Jaoko, J. Plum, M. Temmerman, and C. Verhofstede. 2007. Feasibility of detecting human immunodeficiency virus type 1 drug resistance in DNA extracted from whole blood or dried blood spots. J. Clin. Microbiol. 453342-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziemniak, C., A. George-Agwu, W. J. Moss, S. C. Ray, and D. Persaud. 2006. A sensitive genotyping assay for detection of drug resistance mutations in reverse transcriptase of HIV-1 subtypes B and C in samples stored as dried blood spots or frozen RNA extracts. J. Virol. Methods 136238-247. [DOI] [PubMed] [Google Scholar]