Abstract
The diversity of spirochetes in primary endodontic infections of teeth with chronic apical periodontitis or acute apical abscesses was investigated using 16S rRNA gene clone library analysis. The prevalences of three common cultivable oral Treponema species were also determined using species-specific nested PCR. All detected spirochetes belonged to the genus Treponema. Overall, 28 different taxa were identified from the 431 clones sequenced: 9 cultivable and validly named species, 1 cultivable as-yet-uncharacterized strain, and 18 as-yet-uncultivated phylotypes, 17 of which were novel. The large majority of clones (94%) were from cultivable named species. The numbers of Treponema species/phylotypes per selected positive sample ranged from 2 to 12. Species-specific nested PCR detected T. denticola, T. socranskii, and T. maltophilum in 59 (66%), 33 (37%), and 26 (29%) of the 90 cases of primary endodontic infections, respectively. Clone library analysis revealed diverse Treponema species/phylotypes as part of the microbiota associated with asymptomatic and symptomatic (abscess) endodontic infections. Although several as-yet-uncultivated Treponema phylotypes were disclosed, including novel taxa, cultivable named species were more abundant and frequently detected.
In his milestone study published in 1894, Willoughby Dayton Miller first suggested that spirochetes could play a role in the pathogenesis of apical periodontitis, particularly in cases of acute apical abscesses (21). However, despite several studies revealing their occurrence in endodontic infections by microscopy (49, 50), it was not until approximately 100 years after Miller's study that spirochetes were consistently detected and identified by culture-independent molecular techniques in association with diverse forms of apical periodontitis and pathogenetic involvement was supported (30, 43, 46).
Phylogenetic analyses of 16S rRNA gene clone libraries suggest that all oral spirochetes belong to the genus Treponema (7), and so far, 10 oral species have been cultivated and validly named. They include four asaccharolytic species (Treponema denticola, T. medium, T. putidum, and “T. vincentii”) and six saccharolytic species (T. socranskii, T. pectinovorum, T. maltophilum, T. amylovorum, T. lecithinolyticum, and T. parvum) (36). All these species have been recently targeted and disclosed in primary endodontic infections by studies using molecular methods (4, 11, 12, 15, 27, 38-40, 42-44, 51). The most predominant treponemes in these infections are usually T. denticola and T. socranskii (4, 27, 40, 43), while T. parvum, T. maltophilum, and T. lecithinolyticum have been moderately prevalent (4, 15, 25, 38, 40). Among several other taxa, a recent study targeted all 10 cultivable and 4 as-yet-uncharacterized oral treponemes (26). Of the nine treponemes detected, T. denticola and T. socranskii were the most prevalent species, which is consistent with other studies (4, 27, 40, 43). The other most frequently found Treponema species/phylotypes included Treponema sp. oral taxon VI:G:G47 (16%), T. putidum (16%), Treponema sp. oral taxon II:10:D12 (14%), and T. parvum (12%) (26).
To the best of our knowledge, virtually all studies of the occurrence of spirochetes in endodontic infections have evaluated the occurrence of target species. An exception was a study using denaturing gradient gel electrophoresis with group-specific primers, which after sequencing of some denaturing gradient gel electrophoresis bands, revealed the occurrence of two novel as-yet-uncultivated phylotypes (41). Thus, the diversity of spirochetes in endodontic infections remains to be fully appreciated. Because there are nearly 60 oral Treponema species, approximately 80% of which remain uncultivated (7), a comprehensive analysis of the diversity of spirochetes associated with oral diseases, including apical periodontitis, is necessary and requires the utilization of culture-independent molecular approaches. The present study was undertaken to decipher the diversity of oral spirochetes in infections of endodontic origin using 16S rRNA gene clone library analysis. Additionally, the prevalences of three common cultivable oral Treponema species were assessed.
MATERIALS AND METHODS
Case description, sample taking, and DNA extraction.
Samples were taken from patients who had been referred for root canal treatment or emergency treatment to the Department of Endodontics, Estácio de Sá University, Rio de Janeiro, RJ, Brazil. Abscess samples were also taken from patients who were seeking emergency treatment in three hospitals in Rio de Janeiro. Only single-rooted teeth from adult patients (ages ranging from 18 to 74 years), all of them having carious lesions, necrotic pulps, and radiographic evidence of apical periodontitis, were included in this study. Selected teeth showed an absence of periodontal pockets deeper than 4 mm. In total, 90 samples of infections of endodontic origin were obtained. The teeth were grouped as follows: (i) 32 asymptomatic cases diagnosed as chronic apical periodontitis; (ii) 10 cases diagnosed as acute apical periodontitis, which showed symptoms such as tenderness to percussion and/or palpation, induced or spontaneous pain exacerbated by mastication, absence of pus in the canals, and no swelling; and (iii) 48 cases diagnosed as acute apical abscesses, which showed pain and localized or diffuse swellings, along with fever, lymphadenopathy, and/or malaise. No apparent communication from the abscess to the oral cavity or the skin surface was observed.
In cases of chronic or acute apical periodontitis, samples were obtained from the root canals using sterile paper points. Abscesses were sampled by aspiration of purulent exudate from the swollen mucosa using a sterile syringe. Sampling procedures were as described previously (45). DNA was extracted using the Qiaamp DNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Approval for the study protocol was obtained from the Ethics Committee of the Estácio de Sá University.
Amplification of the 16S rRNA gene.
Whole-genomic DNA extracts from clinical samples were used as templates in a 16S rRNA gene-based nested-PCR method for detection of spirochetes in endodontic samples. In the first PCR, a practically full-length 16S rRNA gene fragment was amplified using a universal 16S rRNA gene primer pair (Table 1). Aliquots of 5 μl of the DNA extracts from clinical samples were used as targets in the first PCR. Next, 2 μl of the resulting PCR product from each sample was used in the second round of amplification, which was specific for detection of spirochetes as a group or for T. denticola, T. socranskii, and T. maltophilum. A nested-PCR protocol was used to improve the performance of the method in samples with low DNA concentrations and to provide increased specificity (20, 37). Primer sequences are shown in Table 1.
TABLE 1.
PCR primers used for identification of Treponema species in primary endodontic infections
| Target | Primer sequences (5′-3′)a | Positions (bp) | Amplicon length (bp) | Reference |
|---|---|---|---|---|
| Spirochete specific | CAC ATT GGG ACT GAG ATA C | 312-1138 | 827 | 23 |
| TAC CTG TTA GTA ACY GGC AGT AG | ||||
| T. denticola | TAA TAC CGA ATG TGC TCA TTT ACA T | 193-508 | 316 | 3 |
| TCA AAG AAG CAT TCC CTC TTC TTC TTA | ||||
| T. maltophilum | AGA GTT TGA TYM TGG CTC AGb | 8-443 | 436 | 54 |
| CCT ATT GTG CTT ATT CAT CAG GC | ||||
| T. socranskii | GAT CAC TGT ATA CGG AAG GTA GAC A | 148-435 | 288 | 54 |
| TAC ACT TAT TCC TCG GAC AG | ||||
| Universal 16S rRNA gene | AGA GTT TGA TYM TGG CTC AG | 8-1541 | 1,534 | 9, 24 |
| GAA GGA GGT GWT CCA RCC GCA |
Y = T or C; M = A or C; W = A or T; R = G or A.
Forward universal primer.
All PCR amplifications were performed in 50 μl of reaction mixture containing a 1 μM concentration of each primer, 5 μl of 10× PCR buffer, 2 mM MgCl2, 2 U of Tth DNA polymerase, and 0.2 mM of each deoxyribonucleoside triphosphate (all reagents were from Biotools, Madrid, Spain). Positive controls used DNA extracted from T. denticola B1 (Forsyth Dental Institute, Boston, MA), T. socranskii S1 (Forsyth Dental Institute), and T. maltophilum ATCC 51939. One negative control consisting of sterile milliQ water instead of the sample was included for every five samples in all batches of samples analyzed.
Preparations were amplified in a DNA thermocycler (Mastercycler personal; Eppendorff, Hamburg, Germany). The PCR temperature profile for the first reaction using universal primers included an initial denaturation step at 95°C for 1 min; 26 cycles at 94°C for 45 s, 55°C for 45 s, and 72°C for 1.5 min; and a final step at 72°C for 15 min. PCR cycling conditions for the second round of amplification using primers specific for spirochetes included an initial denaturation step at 95°C for 4 min; 30 cycles at 94°C for 1 min, 52°C for 1 min, and 72°C for 2 min; and final extension at 72°C for 15 min. The amplicons were separated by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide, and viewed under UV transillumination. Temperature profiles for T. denticola, T. socranskii, and T. maltophilum were as described previously (27, 38, 40). PCR amplicons were identified in agarose electrophoretic gel and visualized for size using UV transillumination. Positive reactions were assigned based on the presence of clearly visible bands of the expected molecular size (Table 1). Representative amplicons were sequenced to check for specificity.
The prevalences of the three treponemes were recorded as the percentage of cases examined. The chi-square test with Yates correction was used to analyze the association between these species and abscesses or the overall occurrence of symptoms (joint cases of acute apical periodontitis and acute apical abscesses). Significance was set at 5% (P < 0.05).
Clone library analysis.
Nine samples positive for the spirochete-specific primers and exhibiting the strongest bands in the agarose gel were selected for further 16S rRNA gene clone library analysis (32). PCR products were purified using an UltraClean PCR Clean-up DNA purification kit (Mo Bio Laboratories, Inc., CA). Purified amplicon was ligated into the plasmid vector pCR2.1 and then transformed into One Shot INVαF′ competent cells using the Original TA Cloning Kit (Invitrogen, San Diego, CA). Plasmid DNAs were prepared from recombinants by using the Illustra TempliPhi DNA Amplification Kit (GE Healthcare, United Kingdom) and used as templates for sequencing. Attempts were made to sequence all clones (white colonies). Clones not showing the inserts were excluded. Sequencing was conducted using the spirochete-specific primers, a Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), and an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). All sequences were checked by the Chimera Check program of the Ribosomal Database Project II (6) to eliminate chimeras, which could result from the amplification of an accidental mixture of bacterial genes; no chimeric sequences were detected. Nucleotide sequences were analyzed by a BLASTN search (1) for the nearest matches. Database sequences with the highest similarities and scorebits to our sequences were chosen for their identification. The criterion to define a novel phylotype was set at sequences that differed from the closest GenBank entry by more than 2% (17). The sequences were aligned with the CLUSTAL W program (48) and corrected by manual inspection. A neighbor-joining phylogenetic tree was constructed from the alignment using the Molecular Evolutionary Genetics Analysis package (MEGA version 2.1) (18). Distances were calculated with the Jukes-Cantor algorithm. The robustness of the phylogeny was tested by bootstrap analysis with 500 iterations.
Nucleotide sequence accession numbers.
Sequences for the novel phylotypes were deposited in the GenBank database under accession numbers AB465691 to AB465707.
RESULTS
Spirochetal diversity by clone library analysis.
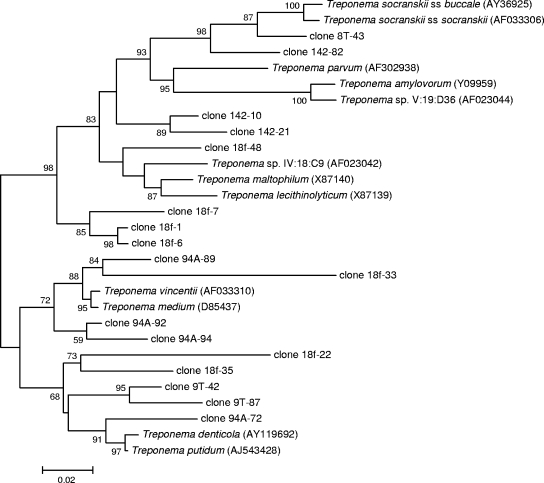
Of the nine samples selected for clone library analysis of spirochetal diversity, three were from root canals associated with chronic apical periodontitis and the other six were from pus aspirates from acute apical abscesses. The total number of clones sequenced from the nine samples was 431 (mean, 48; ranging from 10 to 78) (Table 2). No nontreponemal clones were found, indicating good specificity for the primers and protocol used. Overall, 28 different Treponema taxa were identified from the 431 clones sequenced: 9 cultivable and validly named species (not including the T. socranskii subspecies division), one as-yet-uncultivated phylotype (Treponema sp. oral taxon IV:18:C9), one cultivated but not-yet-validly named strain (Treponema sp. oral taxon V:19:D36), and 17 novel as-yet-uncultivated Treponema phylotypes, i.e., taxa that had never been detected (Fig. 1). Of the novel as-yet-uncultivated taxa, 3 were revealed in chronic cases and the other 14 were from abscesses.
TABLE 2.
Treponema taxa found in samples from primary endodontic infections by clone library analysis
| Taxon | No. of clones (%) in clinical samplesa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr
|
Abs
|
||||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 4 | 5 | 6 | |
| T. amylovorum | 2 (3) | 1 (1) | |||||||
| T. denticola | 5 (15) | 18 (53) | 26 (33) | ||||||
| T. lecithinolyticum | 3 (9) | ||||||||
| T. maltophilum | 52 (68) | 12 (35) | 3 (9) | 36 (80) | 57 (77) | 8 (10) | 1 (2) | 3 (8) | |
| T. medium | 6 (60) | 39 (50) | 1 (2) | 19 (50) | |||||
| T. parvum | 5 (7) | ||||||||
| T. putidum | 1 (1) | 1 (1) | |||||||
| T. socranskii subsp. buccale | 11 (14) | 4 (12) | 9 (20) | 2 (3) | 40 (95) | 1 (3) | |||
| T. socranskii subsp. socranskii | 6 (8) | 14 (41) | 2 (6) | 8 (11) | |||||
| “T. vincentii” | 2 (6) | 6 (16) | |||||||
| Treponema sp. oral taxon IV:18:C9 | 1 (10) | 1 (3) | |||||||
| Treponema sp. oral taxon V:19:D36 | 3 (30) | 1 (1) | |||||||
| Treponema clone 8T-43b | 1 (3) | ||||||||
| Treponema clone 9T-42b | 3 (9) | ||||||||
| Treponema clone 9T-87b | 1 (3) | ||||||||
| Treponema clone 142-10b | 1 (1) | ||||||||
| Treponema clone 142-21b | 2 (3) | ||||||||
| Treponema clone 142-82b | 1 (1) | ||||||||
| Treponema clone 18f-1b | 2 (5) | ||||||||
| Treponema clone 18f-6b | 1 (3) | ||||||||
| Treponema clone 18f-7b | 1 (3) | ||||||||
| Treponema clone 18f-22b | 1 (3) | ||||||||
| Treponema clone 18f-33b | 1 (3) | ||||||||
| Treponema clone 18f-35b | 1 (3) | ||||||||
| Treponema clone 18f-48b | 1 (3) | ||||||||
| Treponema clone 94A-72b | 1 (1) | ||||||||
| Treponema clone 94A-89b | 1 (1) | ||||||||
| Treponema clone 94A-92b | 1 (1) | ||||||||
| Treponema clone 94A-94b | 1 (1) | ||||||||
| Total no. of Treponema taxac | 4 | 5 | 6 | 2 | 3 | 8 | 8 | 3 | 12 |
| Total no. of Treponema clones | 76 | 34 | 34 | 45 | 10 | 74 | 78 | 42 | 38 |
chr, chronic apical periodontitis; abs, acute apical abscess.
Novel phylotype identified in this study.
For calculation of the total number of taxa in each sample, T. socranskii subspecies occurring together were considered one taxon.
FIG. 1.
Phylogenetic tree of Treponema species and phylotypes identified in samples from primary endodontic infections. The sequences were aligned with CLUSTAL W, and distances were calculated with the Jukes-Cantor algorithm. Bootstrap values (based on 500 replicates) are represented at each node when >50%. The scale bar represents a 2% difference in nucleotide sequences.
The huge majority of clones (404, or 94%) were from cultivable named species. Four clones were from Treponema sp. oral taxon V:19:D36, 2 were from Treponema sp. oral taxon IV:18:C9, and the other 21 clones were from novel phylotypes. Therefore, as-yet-uncultivated and/or uncharacterized phylotypes made up only a minor fraction of the Treponema diversity in endodontic infections, as they represented only 6% of the total number of clones sequenced.
Overall, the mean number of Treponema species/phylotypes per selected case was six. Chronic cases harbored a mean of 5 taxa (range, 4 to 6), while abscesses presented a mean of 6 taxa (range, 2 to 12) (Table 2). However, these figures are only illustrative, because the examined cases were selected based on the strengths of amplicons.
T. maltophilum was found in eight samples (three chronic cases and five acute abscesses), followed by T. socranskii subspecies buccale (six samples, two chronic and four acute), T. socranskii subspecies socranskii (four samples, three chronic and one acute), T. medium (four acute samples), and T. denticola (three samples, two chronic and one acute). T. amylovorum, T. putidum, “T. vincentii,” Treponema sp. oral taxon IV:18:C9, and Treponema sp. oral taxon V:19:D36 were found in two cases each. T. parvum, T. lecithinolyticum, and each of the novel phylotypes were found in only one sample each.
The most dominant species/phylotypes in the three root canals associated with chronic apical periodontitis were T. maltophilum (68% of the clones sequenced from case 1), T. socranskii subspecies socranskii (41% from case 2), and T. denticola (53% from case 3) (Table 2). Dominant Treponema taxa in the six abscess samples were T. medium (50% of the clones sequenced in one sample, 50% of another sample, and 60% of a third sample), T. maltophilum (77% and 80%), and T. socranskii subsp. buccale (95%) (Table 2).
Prevalences of three named Treponema species.
In general, species-specific nested PCR detected T. denticola, T. socranskii, and T. maltophilum in 59, 33, and 26 of the 90 cases of primary endodontic infections, respectively. No association with symptoms or abscesses was found for any of the test species (P > 0.05). The findings of the species-specific nested PCR are detailed in Table 3.
TABLE 3.
Frequency of detection of three Treponema species in primary endodontic infections by 16S rRNA gene-based nested-PCR analysis
| Species | Frequency ina:
|
|||
|---|---|---|---|---|
| Chronic apical periodontitis | Acute apical periodontitis | Acute apical abscess | Total | |
| T. denticola | 822/32 (69) | 8/10 (80) | 29/48 (60) | 59/90 (66) |
| T. maltophilum | 9/32 (28) | 5/10 (50) | 12/48 (25) | 26/90 (29) |
| T. socranskii | 12/32 (37.5) | 4/10 (40) | 17/48 (35) | 33/90 (37) |
Number of positive cases/number of samples examined (percentage).
DISCUSSION
Treponema species are examples of culture-difficult bacteria that have been identified in endodontic infections only by culture-independent molecular methods (4, 15, 27, 46). Most of the previous molecular studies focused on the detection of selected Treponema species or phylotypes (4, 11, 13, 15, 25-27, 38, 40, 43, 46). Of the several studies that have performed broad-range analyses for overall bacteria in primary endodontic infections (14, 22, 29, 31-33, 52, 53), only two reported the occurrence of spirochetes, and even those were a few species in a few cases (31, 32). These studies using universal bacterial primers may underestimate spirochetal populations, especially if they are in low numbers in the environment (5, 16). Indeed, one study revealed that no Treponema species were found in infected root canals associated with chronic apical periodontitis at levels of >105 (26).
To the best of our knowledge, this is the first attempt to examine the diversity of spirochetes in infections of endodontic origin using group-specific primers followed by cloning and 16S rRNA gene sequencing. Nine cases that presented strong amplicons after PCR amplification were chosen for this analysis. Up to 12 Treponema species/phylotypes were found per positive sample. Because of the obvious biases introduced by the criteria for sample selection, clone library analysis did not serve the purpose of revealing the mean number of treponemes per canal or the prevalence of the detected Treponema species, even though this information is provided here for illustrative purposes. Spirochetal diversity in endodontic infections was the main focus of this study.
Clone library analysis corroborated previous findings from species-specific PCR (4, 11, 15, 25, 27, 38, 40) or checkerboard studies (26, 35, 46, 47) in that virtually all cultivable Treponema species can be found in infections of endodontic origin. Of the 10 cultivable species, only T. pectinovorum was not detected here. As-yet-uncultivated and/or -uncharacterized Treponema phylotypes detected in a previous study (26) were not found in the present one. However, it was noteworthy that 18 as-yet-uncultivated phylotypes were detected, 17 of which were novel in the sense that they had never been found in other sites. Therefore, 64% of the Treponema species found in endodontic infections in the present study have yet to be cultivated and phenotypically characterized. These findings also indicate that spirochetal diversity in the oral cavity is still broader than previously reported (7). As for abundance, these phylotypes comprised only a minor proportion of the total number of clones sequenced (6%). This means that they are not dominant components of the spirochetal endodontic communities. This finding, however, does not imply a lack of relevance, as even low-dominance species may exert important ecological functions.
The cultivable Treponema species found to dominate the spirochetal community in most samples included T. maltophilum, T. socranskii, T. denticola, and T. medium. Of these, only the last has not been found at high prevalence in previous studies (4, 38). It is salient to point out that the fact that these species corresponded to most of the clones sequenced in this study actually means that they dominated the spirochetal community, but their proportion in relation to the whole bacterial community was not established. Hence, any inferences with regard to pathogenicity must be drawn with caution, even though these species have been suggested to play roles in other oral diseases, including marginal periodontitis (2, 8, 10, 34, 54).
Of the three species targeted in the species-specific nested-PCR assay, T. denticola was the most prevalent, which is in consonance with our previous findings (27, 40). However, data from clone library analysis revealed that the species was not the most prevalent. The differences may have been due to the fact that species-specific PCR assays arguably display higher sensitivity than group-specific or broad-range primers (19, 28). Nonetheless, because the cases for clone library analysis were selected on the basis of strong amplicon signal, the possibility exists that most cases positive for T. denticola were left out of the analysis. Indeed, of the six cases subjected to clone library analysis that were negative for T. denticola, only one was positive in species-specific nested PCR. Biases resulting from sample selection for analysis might also explain the opposite situation, i.e., why T. maltophilum was encountered more frequently in clone libraries than in species-specific nested PCR.
None of the three target species was positively associated with symptoms and/or abscessed cases. Although the three species were detected in several of the acute cases, they were also very frequent in cases of chronic apical periodontitis. However, the results presented in this study were only qualitative (presence/absence data). Further studies should quantify oral treponemes present in both asymptomatic and symptomatic endodontic infections so as to determine whether these bacteria are found in larger numbers in any particular clinical situation.
In conclusion, 16S rRNA gene group-specific PCR followed by cloning and sequencing revealed that diverse Treponema species/phylotypes can take part in the microbiota associated with asymptomatic and symptomatic (abscess) endodontic infections. Although several as-yet-uncultivated Treponema phylotypes were disclosed, including novel taxa, cultivable named species were more frequently detected and abundant. Species-specific nested PCR demonstrated that T. denticola, T. socranskii, and T. maltophilum, in decreasing order of prevalence, are frequently found in different forms of apical periodontitis.
Acknowledgments
This study was supported by grants from the RIKEN BioResource Center and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), a Brazilian governmental institution.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, Y., T. Jinno, H. Igarashi, Y. Ohyama, and T. Ogawa. 2002. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J. Clin. Microbiol. 403334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashimoto, A., C. Chen, I. Bakker, and J. Slots. 1996. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 11266-273. [DOI] [PubMed] [Google Scholar]
- 4.Baumgartner, J. C., S. U. Khemaleelakul, and T. Xia. 2003. Identification of spirochetes (treponemes) in endodontic infections. J. Endod. 29794-797. [DOI] [PubMed] [Google Scholar]
- 5.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Gobel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 621889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15196-202. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, A. M., D. Dymock, and H. F. Jenkinson. 2003. From tooth to hoof: treponemes in tissue-destructive diseases. J. Appl. Microbiol. 94767-780. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Bötter. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 177843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellen, R. P., and V. B. Galimanas. 2005. Spirochetes at the forefront of periodontal infections. Periodontology 2000 3813-32. [DOI] [PubMed] [Google Scholar]
- 11.Foschi, F., F. Cavrini, L. Montebugnoli, P. Stashenko, V. Sambri, and C. Prati. 2005. Detection of bacteria in endodontic samples by polymerase chain reaction assays and association with defined clinical signs in Italian patients. Oral Microbiol. Immunol. 20289-295. [DOI] [PubMed] [Google Scholar]
- 12.Gomes, B. P., R. C. Jacinto, E. T. Pinheiro, E. L. Sousa, A. A. Zaia, C. C. Ferraz, and F. J. Souza-Filho. 2006. Molecular analysis of Filifactor alocis, Tannerella forsythia, and Treponema denticola associated with primary endodontic infections and failed endodontic treatment. J. Endod. 32937-940. [DOI] [PubMed] [Google Scholar]
- 13.Gomes, B. P., F. Montagner, R. C. Jacinto, A. A. Zaia, C. C. Ferraz, and F. J. Souza-Filho. 2007. Polymerase chain reaction of Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia in primary endodontic infections. J. Endod. 331049-1052. [DOI] [PubMed] [Google Scholar]
- 14.Jacinto, R. C., B. P. Gomes, M. Desai, D. Rajendram, and H. N. Shah. 2007. Bacterial examination of endodontic infections by clonal analysis in concert with denaturing high-performance liquid chromatography. Oral Microbiol. Immunol. 22403-410. [DOI] [PubMed] [Google Scholar]
- 15.Jung, I. Y., B. Choi, K. Y. Kum, Y. J. Yoo, T. C. Yoon, S. J. Lee, and C. Y. Lee. 2001. Identification of oral spirochetes at the species level and their association with other bacteria in endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 92329-334. [DOI] [PubMed] [Google Scholar]
- 16.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 9614547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 433944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Maiwald, M. 2004. Broad-range PCR for detection and identification of bacteria, p. 379-390. In D. H. Persing, F. C. Tenover, J. Versalovic, Y.-W. Tang, D. Relman, and T. J. White (ed.), Molecular microbiology. Diagnostic principles and practice. ASM Press, Washington, DC.
- 20.McPherson, M. J., and S. G. Moller. 2000. PCR. BIOS Scientific Publishers Ltd., Oxford, United Kingdom.
- 21.Miller, W. D. 1894. An introduction to the study of the bacterio-pathology of the dental pulp. Dent. Cosmos 36505-528. [Google Scholar]
- 22.Munson, M. A., T. Pitt-Ford, B. Chong, A. Weightman, and W. G. Wade. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J. Dent. Res. 81761-766. [DOI] [PubMed] [Google Scholar]
- 23.Parrish, K. D., and E. P. Greenberg. 1995. A rapid method for extraction and purification of DNA from dental plaque. Appl. Environ. Microbiol. 614120-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rôças, I. N., and J. F. Siqueira, Jr. 2005. Occurrence of two newly named oral treponemes—Treponema parvum and Treponema putidum—in primary endodontic infections. Oral Microbiol. Immunol. 20372-375. [DOI] [PubMed] [Google Scholar]
- 26.Rôças, I. N., and J. F. Siqueira, Jr. 2008. Root canal microbiota of teeth with chronic apical periodontitis. J. Clin. Microbiol. 463599-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rôças, I. N., J. F. Siqueira, Jr., A. F. Andrade, and M. Uzeda. 2003. Oral treponemes in primary root canal infections as detected by nested PCR. Int. Endod. J. 3620-26. [DOI] [PubMed] [Google Scholar]
- 28.Rôças, I. N., and J. F. Siqueira, Jr. 2006. Characterization of Dialister species in infected root canals. J. Endod. 321057-1061. [DOI] [PubMed] [Google Scholar]
- 29.Rolph, H. J., A. Lennon, M. P. Riggio, W. P. Saunders, D. MacKenzie, L. Coldero, and J. Bagg. 2001. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 393282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rupf, S., S. Kannengiesser, K. Merte, W. Pfister, B. Sigusch, and K. Eschrich. 2000. Comparison of profiles of key periodontal pathogens in periodontium and endodontium. Endod. Dent. Traumatol. 16269-275. [DOI] [PubMed] [Google Scholar]
- 31.Saito, D., R. de Toledo Leonardo, J. L. M. Rodrigues, S. M. Tsai, J. F. Hofling, and R. B. Gonçalves. 2006. Identification of bacteria in endodontic infections by sequence analysis of 16S rDNA clone libraries. J. Med. Microbiol. 55101-107. [DOI] [PubMed] [Google Scholar]
- 32.Sakamoto, M., I. N. Rôças, J. F. Siqueira, Jr., and Y. Benno. 2006. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol. Immunol. 21112-122. [DOI] [PubMed] [Google Scholar]
- 33.Sakamoto, M., J. F. Siqueira, Jr., I. N. Rôças, and Y. Benno. 2007. Bacterial reduction and persistence after endodontic treatment procedures. Oral Microbiol. Immunol. 2219-23. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto, M., M. Umeda, and Y. Benno. 2005. Molecular analysis of human oral microbiota. J. Periodontal Res. 40277-285. [DOI] [PubMed] [Google Scholar]
- 35.Sassone, L. M., R. A. Fidel, M. Faveri, R. Guerra, L. Figueiredo, S. R. Fidel, and M. Feres. 2008. A microbiological profile of symptomatic teeth with primary endodontic infections. J. Endod. 34541-545. [DOI] [PubMed] [Google Scholar]
- 36.Siqueira, J. F., Jr. 2008. Microbiology of apical periodontitis, p. 135-196. In D. Ørstavik and T. Pitt Ford (ed.), Essential endodontology, 2nd ed. Blackwell Munksgaard Ltd., Oxford, United Kingdom.
- 37.Siqueira, J. F., Jr., and I. N. Rôças. 2005. Exploiting molecular methods to explore endodontic infections. Part 1: current molecular technologies for microbiological diagnosis. J. Endod. 31411-423. [DOI] [PubMed] [Google Scholar]
- 38.Siqueira, J. F., Jr., and I. N. Rôças. 2003. PCR-based identification of Treponema maltophilum, T amylovorum, T medium, and T lecithinolyticum in primary root canal infections. Arch. Oral Biol. 48495-502. [DOI] [PubMed] [Google Scholar]
- 39.Siqueira, J. F., Jr., and I. N. Rôças. 2003. Treponema socranskii in primary endodontic infections as detected by nested PCR. J. Endod. 29244-247. [DOI] [PubMed] [Google Scholar]
- 40.Siqueira, J. F., Jr., and I. N. Rôças. 2004. Treponema species associated with abscesses of endodontic origin. Oral Microbiol. Immunol. 19336-339. [DOI] [PubMed] [Google Scholar]
- 41.Siqueira, J. F., Jr., I. N. Rôças, C. D. Cunha, and A. S. Rosado. 2005. Novel bacterial phylotypes in endodontic infections. J. Dent. Res. 84565-569. [DOI] [PubMed] [Google Scholar]
- 42.Siqueira, J. F., Jr., I. N. Rôças, A. Favieri, J. C. Oliveira, and K. R. Santos. 2001. Polymerase chain reaction detection of Treponema denticola in endodontic infections within root canals. Int. Endod. J. 34280-284. [DOI] [PubMed] [Google Scholar]
- 43.Siqueira, J. F., Jr., I. N. Rôças, A. Favieri, and K. R. Santos. 2000. Detection of Treponema denticola in endodontic infections by 16S rRNA gene-directed polymerase chain reaction. Oral Microbiol. Immunol. 15335-337. [DOI] [PubMed] [Google Scholar]
- 44.Siqueira, J. F., Jr., I. N. Rôças, J. C. Oliveira, and K. R. Santos. 2001. Detection of putative oral pathogens in acute periradicular abscesses by 16S rDNA-directed polymerase chain reaction. J. Endod. 27164-167. [DOI] [PubMed] [Google Scholar]
- 45.Siqueira, J. F., Jr., I. N. Rôças, and A. S. Rosado. 2004. Investigation of bacterial communities associated with asymptomatic and symptomatic endodontic infections by denaturing gradient gel electrophoresis fingerprinting approach. Oral Microbiol. Immunol. 19363-370. [DOI] [PubMed] [Google Scholar]
- 46.Siqueira, J. F., Jr., I. N. Rôças, R. Souto, M. de Uzeda, and A. P. Colombo. 2000. Checkerboard DNA-DNA hybridization analysis of endodontic infections. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 89744-748. [DOI] [PubMed] [Google Scholar]
- 47.Siqueira, J. F., Jr., I. N. Rôças, R. Souto, M. Uzeda, and A. P. Colombo. 2001. Microbiological evaluation of acute periradicular abscesses by DNA-DNA hybridization. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 92451-457. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trope, M., L. Tronstad, E. S. Rosenberg, and M. Listgarten. 1988. Darkfield microscopy as a diagnostic aid in differentiating exudates from endodontic and periodontal abscesses. J. Endod. 1435-38. [DOI] [PubMed] [Google Scholar]
- 50.van Winkelhoff, A. J., A. W. Carlee, and J. de Graaff. 1985. Bacteroides endodontalis and other black-pigmented Bacteroides species in odontogenic abscesses. Infect. Immun. 49494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vianna, M. E., H. P. Horz, B. P. Gomes, and G. Conrads. 2005. Microarrays complement culture methods for identification of bacteria in endodontic infections. Oral Microbiol. Immunol. 20253-258. [DOI] [PubMed] [Google Scholar]
- 52.Vickerman, M. M., K. A. Brossard, D. B. Funk, A. M. Jesionowski, and S. R. Gill. 2007. Phylogenetic analysis of bacterial and archaeal species in symptomatic and asymptomatic endodontic infections. J. Med. Microbiol. 56110-118. [DOI] [PubMed] [Google Scholar]
- 53.Wade, W. G., D. A. Spratt, D. Dymock, and A. J. Weightman. 1997. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin. Infect. Dis. 25(Suppl. 2)S235-S236. [DOI] [PubMed] [Google Scholar]
- 54.Willis, S. G., K. S. Smith, V. L. Dunn, L. A. Gapter, K. H. Riviere, and G. R. Riviere. 1999. Identification of seven Treponema species in health- and disease-associated dental plaque by nested PCR. J. Clin. Microbiol. 37867-869. [DOI] [PMC free article] [PubMed] [Google Scholar]