Abstract
We assessed the feasibility of using dried serum spots (DSS) for the serological and molecular diagnosis of hepatitis A virus (HAV) infection. Sixty-eight sera spotted onto filter papers (Whatman International Ltd., United Kingdom) were used for detection of total anti-HAV antibodies, and 64 sera were used for detection of immunoglobulin M antibody to HAV. DSS were stored at 4°C, room temperature, and 37°C for 1, 2, and 4 weeks. Sensitivity and specificity of the serological assays were 100% regardless of temperature and storage duration. To assess the stability of HAV RNA, we performed qualitative and quantitative reverse transcription-PCRs (RT-PCRs) with human plasma spiked with serial dilutions of cultured HAV spotted on Flinders Technology Associates filter paper cards (Whatman International Ltd.). Filter papers were stored at room temperature and processed for RT-PCR assays. No reduction of viral load was observed after 5, 15, and 30 days of storage. The ∼10-fold reduction of sensitivity from DSS was attributable to a smaller sample input in DSS samples. This method was further evaluated using 35 frozen sera. HAV RNA amplification showed 100% specificity and 92.3% sensitivity, and sequence analysis from DSS and sera provided identical results. HAV RNA can be accurately recovered from DSS for molecular epidemiology purposes, and we confirm the reliability of blotted samples in the serological diagnosis of HAV infection. The DSS method facilitates storage and shipment of samples from routine laboratories to reference centers for further investigations and large epidemiological studies.
Hepatitis A virus (HAV) is a major cause of viral hepatitis worldwide. Infection is usually asymptomatic or anicteric in children younger than 6 years of age but can lead to fulminant hepatitis, particularly in adults older than 50 years of age. With the improvement in hygiene conditions in developed countries, there has been a striking reduction of HAV endemicity over the past few decades (13, 25). This decrease in natural immunization allows potentially massive outbreaks involving adults with a consequent number of severe clinical forms and an important socioeconomic impact.
The diagnosis of acute hepatitis A is based on the detection of the immunoglobulin M (IgM) antibody to HAV (HAVM). Determination of the total anti-HAV antibodies (HAVT) or anti-HAV IgG allows the determination of the HAV immune status. Investigation of outbreaks relies on epidemiological and serological studies, but only molecular investigation is able to link apparently sporadic cases or apparently distinct outbreaks (6, 24). This methodology requires costly devices, such as thermal cyclers and sequencers, and expertise in nucleotide sequence analysis that are available in only few reference centers. Many studies have demonstrated the good performances of blotted blood, serum, or plasma samples in serological and molecular diagnosis of different viral infections, including infection with cytomegalovirus (23), human immunodeficiency virus (HIV) (3, 18, 19), hepatitis C virus (9, 16), measles virus (11, 15, 17), or rubella virus (14). Dried blood spots are notably cost-effective as a blood sample collection device in epidemiologic field studies in developing countries. The use of dried serum spots (DSS), though not simplifying sample collection, may facilitate sample storage and shipment from routine laboratories to reference centers.
To date, three studies have reported the reliability of HAV serology with blotted blood or serum (10, 12, 21), but the impacts of time and temperature on serological results were not investigated. To our knowledge, the use of DSS for the molecular diagnosis of HAV infection has not been described. In the present study, we assessed both the feasibility of serological and molecular detection of HAV on DSS.
MATERIALS AND METHODS
Serological study. (i) Serum samples.
Sera collected between 2001 and 2007 were used for the study. Sera from the hospital staff had been tested for HAVT by using the ETI-AB-HAVK Plus kit (DiaSorin, Sallugia, Italy) for occupational medicine purposes. After the exclusion of sera with signal-to-cutoff (CO) ratios (i.e., optical density [OD]/CO) within a gray zone of ±20% around the CO, 68 sera were selected: 38 with an HAVT titer of <20 mIU/ml (considered negative) and 30 with an HAVT titer of >20 mIU/ml (considered positive). Of the 30 positive sera, 21 were obtained from patients with prior HAV infection and 9 were from vaccinated patients. Sixty-four samples had been tested for HAVM with the ETI-HA-IGMK Plus kit (DiaSorin) in the context of elevated aminotransferase levels: 32 were positive and 32 were negative. Samples were kept frozen at −20°C until further use.
(ii) DSS preparation.
Twenty-five microliters of serum or control (positive control, negative control, or calibrators) was spotted onto filter paper (S&S 903; Whatman International Ltd., United Kingdom) in one preprinted circle with a 1.1-cm diameter. Filter papers were air dried for 1 h at room temperature (20 to 25°C) and then packed individually in small, hermetic plastic bags. DSS were stored for 5, 15, or 30 days at +4°C and +37°C, with temperature monitored, and at room temperature (20 to 25°C).
(iii) Elution of HAVM and HAVT from DSS.
A circle with a 1.1-cm diameter was punched from each filter paper. It was then incubated at room temperature in 250 μl of the sample diluent provided in the DiaSorin kits for 1 h on a rotating device and then overnight without agitation. The original sera were thus diluted 10-fold after elution.
(iv) HAVT detection on DSS.
DSS eluates were analyzed for HAVT with the ETI-AB-HAVK Plus kit (DiaSorin) with a modified protocol. In the manufacturer's protocol, 50 μl of serum is added to 50 μl of diluent and incubated with 50 μl of neutralizing solution (HAV antigen). For DSS, 50 μl of the eluate was incubated with 15 μl of neutralizing solution without additional diluent to compensate for the dilution inherent to the elution step. The input of serum in enzyme-linked immunosorbent assay (ELISA) plates remained 4.3 times smaller for DSS. The following steps of the ELISA were performed without modifications. Spotted controls and calibrators were used for technical validation. As for serum samples, the CO value was the mean OD of the spotted 20-mIU/ml calibrator, tested in triplicate. Samples with absorbance values within ±20% of the CO value were considered to be in the gray zone. Within- and between-run variations were determined with the World Health Organization second international standard for anti-HAV human Ig purchased from the National Institute for Biological Standards and Control (Hertfordshire, United Kingdom). A 20-mIU/ml dilution of this standard was tested in quadruplicate in each assay.
(v) Detection of HAVM on DSS.
HAVM were detected in DSS eluates by using the ETI-HA-IGMK Plus kit (DiaSorin) with a modified protocol. In the manufacturer's protocol, 10 μl of serum is added to 990 μl of diluent, and 100 μl of this dilution is deposited into the microwells. For DSS, 10 μl of the eluate was added to 90 μl of diluent and deposited into microwells. Finally, the inputs of serum in ELISA plates were the same for sera and DSS. The following steps of the ELISA were performed without modifications. The CO value was determined by adding 0.100 to the mean OD value obtained with the spotted negative calibrator included in triplicate in each experiment. Samples with absorbance values within ±20% of the CO value were considered to be in the gray zone.
Molecular study. (i) Virus and cell cultures.
The HAV cell culture-adapted HM 175 was kindly provided by S. Perelle (from Agence Française de Sécurité Alimentaire). The virus was grown for 5 days in FrhK4 cells in minimal essential medium containing 10% fetal bovine serum in a 5% CO2 incubator at 37°C. The viral titer of the culture supernatant was 8 log copies/ml, as assessed by quantitative reverse transcription-PCR (RT-PCR) (see below).
(ii) Serum samples for molecular diagnosis.
Twenty-six HAV RNA-positive and 9 HAV RNA-negative sera collected between 2005 and 2007 were used for the study. These sera were collected from patients with elevated aminotransferase levels and positive HAVM detection results and originated from all regions of France. HAV RNA detection and sequencing of the VP1/2A junction region were prospectively performed at the time of reception at the National Reference Centre for HAV in Villejuif, France, as part of the French Observatoire of HAV strains. Samples were then frozen at −80°C until further use.
(iii) Dried spot preparation.
Seventy-five microliters of serum or cell culture supernatant was blotted onto Flinders Technology Associates filter paper cards (FTA cards; Whatman International Ltd.) in three separate spots of 25 μl each. Then, each filter paper was left to dry for 1 h at room temperature and then placed individually in small, hermetic plastic bags and stored at room temperature (20 to 25°C) for 1, 5, 15, or 30 days.
(iv) Elution of HAV RNA.
The three spots of each sample were cut out by a hole punch with a 1.1-cm diameter and incubated in 400 μl of Tris-EDTA buffer (10 mM Tris-Cl, pH 7.5, 1 mM EDTA) for 1 h at room temperature on a shaking platform prior to testing. The original sample was thus diluted 1:5.
(v) HAV RNA extraction.
Viral RNA was extracted from 140 μl of serum or eluate by using the QIAmp viral RNA kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. However, elution of DSS RNA from the QIAmp spin column was done with 50 μl of RNase/DNase-free water instead of 60 μl. HAV RNA extracts obtained from DSS samples were 4.5 times less concentrated than those from nonspotted samples.
(vi) HAV RNA detection.
Two distinct regions of the HAV genome were amplified in parallel. A 512-bp fragment encompassing the VP1/2A junction was amplified using previously described primers (5), and a 355-bp fragment encompassing the 5′ noncoding region (5′ NCR) was amplified with forward primer 5NCPB1 (GAT-ACC-TCA-CCG-TTT-G) and reverse primer 5NCPB2 (TAA-GAG-GTT-TCA-CCC-GTA-GCC). RT-PCR was performed with 10 μl of extracted RNA using the One-Step RT-PCR kit (Qiagen) according to the manufacturer's instructions.
(vii) HAV RNA quantification.
HAV RNA was quantified from 5 μl of extracted RNA by a real-time RT-PCR. Quantitative one-step RT-PCR was carried out in a LightCycler 1.0 instrument (Roche Diagnostics, Meylan, France), using the LightCycler HAV quantification kit (Roche Diagnostics) according to the manufacturer's recommendations. The lowest quantification limit of the assay is 2 log copies/ml for serum and 2.6 log copies/ml for DSS after correcting with the factor calculated above (i.e., 4.5).
(viii) Strain genotyping.
Nucleotide sequencing of the VP1/2A fragment amplified from DSS was carried out bidirectionally with the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) on an Applied Biosystems 3130 automatic sequencer, according to the manufacturer's protocol. Sequences from DSS were compared to the sequences prospectively obtained from serum at our National Reference Center for HAV. Sequences from serum and DSS were aligned with Clustal X software. Phylogenetic trees were constructed with MEGA software by the neighbor-joining method from a Kimura two-parameter distance matrix, and bootstrap values were determined from 1,000 bootstrap resamplings of the original data. The HAV genotype was determined by including reference sequences in the phylogenetic analysis. GenBank references were X75215 for genotype IA, M14707 for genotype IB, strain AY644670 for genotype IIB, AY644676 for genotype IIA, AJ299464 for genotype IIIA, and D00924 for genotype V.
(ix) Sensitivity of the PCR assays.
Serial 10-fold dilutions of the viral culture supernatant were prepared in HAV RNA-negative human plasma. In this experiment, 75 μl (three spots of 25 μl) of each dilution were spotted in duplicate onto two separate filter papers and kept at room temperature for 24 h. Spotted and nonspotted dilutions were extracted and amplified in the same experiment with the two qualitative RT-PCR assays described above.
To further assess a possible loss of HAV RNA induced by the blotting process, the viral load of each spotted and nonspotted dilution was determined by quantitative RT-PCR.
(x) Intra-assay reproducibility of the DSS method.
A 4-log copies/ml dilution of the HAV culture supernatant was spotted 10 times onto filter papers as described above and stored for 24 h at room temperature. Elution, extraction, and quantification of the 10 replicates were performed in the same experiment.
(xi) Effect of storage duration on HAV RNA stability.
Three dilutions of the HAV culture supernatant with HAV titers of 4, 5, and 6 log copies/ml were blotted in duplicate. Elution, detection, and quantification of viral loads were carried out as described above, after 5, 15, and 30 days of storage at room temperature.
Statistical analysis.
The sensitivity and specificity of the DSS assays were estimated with 95% confidence intervals (95% CI), using the results obtained with the nonspotted sera as a reference. Means were compared using the Student t test.
For immunoassays, positive and negative delta values were calculated by dividing the mean OD/CO ratios of positive and negative samples, respectively, by the standard deviation of each population (8). Delta values of >2 imply a good separation of the negative and positive populations, with a smaller potential of false-positive or -negative test results.
HAV RNA viral loads were expressed in log10 copies/ml. Quantification agreement between results from spotted and nonspotted samples was assessed by the Spearman correlation coefficient and the Bland and Altman model.
Nucleotide sequence accession numbers.
GenBank accession numbers for the VP1/2A nucleotide sequences reported here are FJ687492 to FJ687515.
RESULTS
Serological study. (i) Detection of HAVM on DSS.
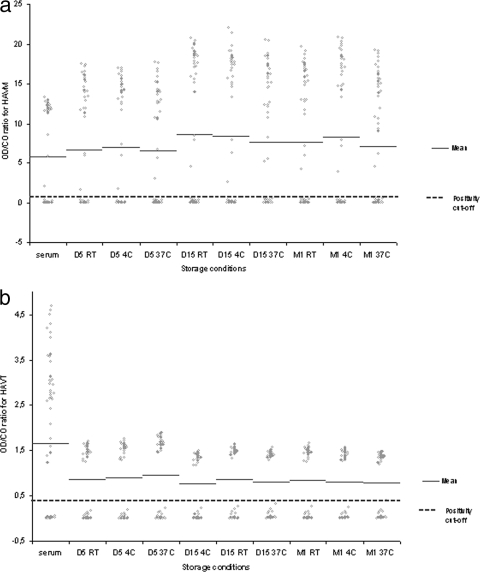
In the HAVM assay, there was no significant difference between the mean OD/CO ratios for DSS and matched sera, as shown in Fig. 1a. In blotting experiments, the values of positive and negative samples were clearly separated from the CO in all nine different conditions of temperature and storage duration, as reflected by positive and negative delta values of >2, except for the negative delta value after 1 month of storage at 37°C found at 1.85. Positive delta values ranged from 3.74 to 7.84, and negative delta values ranged from 1.85 to 3.39. For comparison, negative and positive delta values obtained from serum were 2.80 and 4.94, respectively. The sensitivity and specificity of the HAVM assay with DSS were 100% at all storage conditions.
FIG. 1.
OD/CO ratios of each serum or DSS sample are plotted, according to storage condition. The mean value of OD/CO ratios is shown for each condition. (a) The protocol used for detection of HAVM from DSS did not modify the input of serum in ELISA plates for this capture immunoassay; thus, mean OD/CO ratios of DSS and matched sera are not significantly different, whatever the storage condition. (b) The modified protocol used for HAVT detection from DSS samples implied the addition of a lower volume of neutralizing solution in this competitive ELISA (negative samples have OD values greater than the CO). This adaptation resulted in the OD/CO ratios for negative DSS being significantly lower than those for negative sera. However, OD values for positive and negative DSS samples are clearly separated from the CO, as further assessed by calculation of the delta values. Abbreviations: D, day; M, month; RT, room temperature.
(ii) Detection of HAVT on DSS.
In the HAVT assay, there was a significant difference between mean OD/CO ratios for DSS and matched sera, as shown in Fig. 1b. In this competitive assay, the mean OD/CO ratios of negative samples were higher in the serum assay than in all DSS assays (2.96 ± 0.96 versus 1.47 ± 0.13, respectively; P < 0.0001). This difference is attributable to the lower volume of neutralizing solution used in the adapted assay. Despite this difference, delta values for DSS were all >2. Negative delta values ranged from 2.40 to 2.77, and positive delta values from 4.41 to 8.62. For comparison, negative and positive delta values obtained from serum were 2.43 and 4.69, respectively. The sensitivity and specificity of the HAVT assay from DSS were 100% at all storage conditions.
Within-run variations of the modified HAVT assay, assessed with the 20-mIU/ml National Institute for Biological Standards and Control standard tested in quadruplicate after 5, 15, and 30 days of storage at 4°C, room temperature, and 37°C, in a total of nine experiments were low, with coefficients of variation ranging from 1.98 to 8.69%. The between-run coefficient of variation of the nine assays was 4.71%.
Molecular study. (i) Sensitivity of RT-PCR.
Serial 10-fold dilutions of the HAV culture supernatant with the 8-log copies/ml titer were submitted to RT-PCR for VP1/2A and 5′ NCR amplification. The sensitivities of the in-house RT-PCRs were 1 and 2 log copies/ml for VP1/2A and 5′ NCR amplifications, respectively; their sensitivities with matched spotted dilutions stored for 24 h at room temperature were 2 and 3 log copies/ml, respectively.
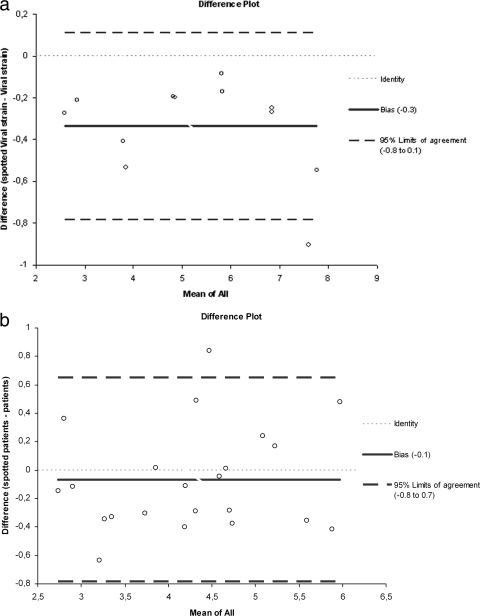
To search for a potential loss of RNA induced by the blotting step, we precisely quantified each dilution and each blotted dilution by quantitative RT-PCR and applied the 4.5 correction factor that accounts for the dilution of blotted specimen. Quantification results correlated very well (r = 0.99, 95% CI = 0.97 to 1.00). There was a nonsignificant bias toward lower viral loads obtained after blotting, as shown by the Bland and Altman model, indicating a mean difference of 0.34 log copies/ml (bias [i.e., mean difference between viral load results from spotted and nonspotted samples] 95% CI = −0.48 to −0.19) (Fig. 2a).
FIG. 2.
Serial 10-fold dilutions of the viral culture supernatant prepared in HAV RNA-negative human plasma were spotted and kept at room temperature for 24 h. Difference plots are shown. To assess a possible loss of HAV RNA induced by the blotting process, the viral loads of spotted and nonspotted dilutions were determined by quantitative RT-PCR. (a) After applying the 4.5 correction factor that accounts for the dilution of a blotted specimen, a nonsignificant bias toward lower viral loads obtained after blotting was shown by the Bland and Altman model, indicating a mean difference of 0.34 log copies/ml (bias 95% CI = −0.48 to −0.19). (b) HAV viral loads were determined in 26 HAV RNA-positive sera and in matched DSS samples kept at room temperature for 24 h. Four DSS samples could not be quantified after blotting: two matched sera had viral loads of <100 copies/ml, and two had 2.11 and 2.22 log copies/ml. Among the 22 quantifiable DSS, no significant bias toward lower viral loads obtained after blotting, as shown by the Bland and Altman model, indicating a mean difference of 0.1 log copies/ml (bias 95% CI = −0.48 to −0.19).
The 1-log loss in sensitivity is thus attributable mostly to a smaller sample input in RT-PCRs performed with DSS.
(ii) Intra-assay reproducibility of the DSS method for RNA detection.
A 4-log copies/ml dilution of the HAV culture supernatant was spotted 10 times. The mean viral load of 10 replicates was 3.84 (±0.11) copies/ml, providing an intra-assay coefficient of variation of 2.9%.
(iii) Effect of storage duration on HAV RNA detection.
Qualitative detection of duplicates of the HAV strains with titers of 4, 5, and 6 log copies/ml remained positive by RT-PCR amplification of the VP1/2A region and the 5′ NCR after 5, 15, and 30 days of storage at room temperature. No reduction of viral load was observed. Coefficients of variation between the three storage durations were 2.60%, 2.72%, and 0.69% for dilutions at 4, 5, and 6 log copies/ml, respectively, no higher than the intra-assay coefficient of variation.
(iv) Detection and quantification of HAV RNA from blotted serum.
Twenty-six HAV RNA-positive samples and 9 HAV RNA-negative samples were tested by quantitative RT-PCR at the time of DSS preparation. After 24 h of storage at room temperature, qualitative RT-PCRs were performed on all 35 spotted sera, and quantification was performed for the 26 positive ones.
All nine HAV RNA-negative sera were RT-PCR negative after spotting. Among the 26 HAV RNA-positive sera, 24 and 23 were successfully detected after spotting by amplification of the VP1/2A region and the 5′ NCR, respectively. Thus, the sensitivities of the RT-PCRs with DSS were 92.3% and 88.5% for VP1/2A and 5′ NCR amplifications, respectively.
Viral loads were quantifiable in 24/26 of the HAV RNA-positive sera and ranged from 2.11 to 6.08 log copies/ml (mean ± standard deviation, 4.12 ± 1.09 log copies/ml). Two samples yielding values of <100 copies/ml could not be amplified by any of the qualitative RT-PCRs after blotting. A sample with a viral load of 2.62 log copies/ml could be amplified only by the VP1/2A RT-PCR after blotting.
After blotting, the viral load was quantifiable in 22/26 sera. The two samples with viral loads of <100 copies/ml and two samples yielding 2.11 and 2.22 log copies/ml could not be quantified after blotting. Quantification results correlated very well (r = 0.99, 95% CI = 0.97 to 1.00). There was no significant bias toward lower viral loads obtained after blotting as shown by the Bland and Altman model, indicating a mean difference of 0.1 log copies/ml (bias 95% CI = −0.48 to −0.19) (Fig. 2b).
(v) Phylogenetic analysis of HAV RNA sequences amplified from blotted serum.
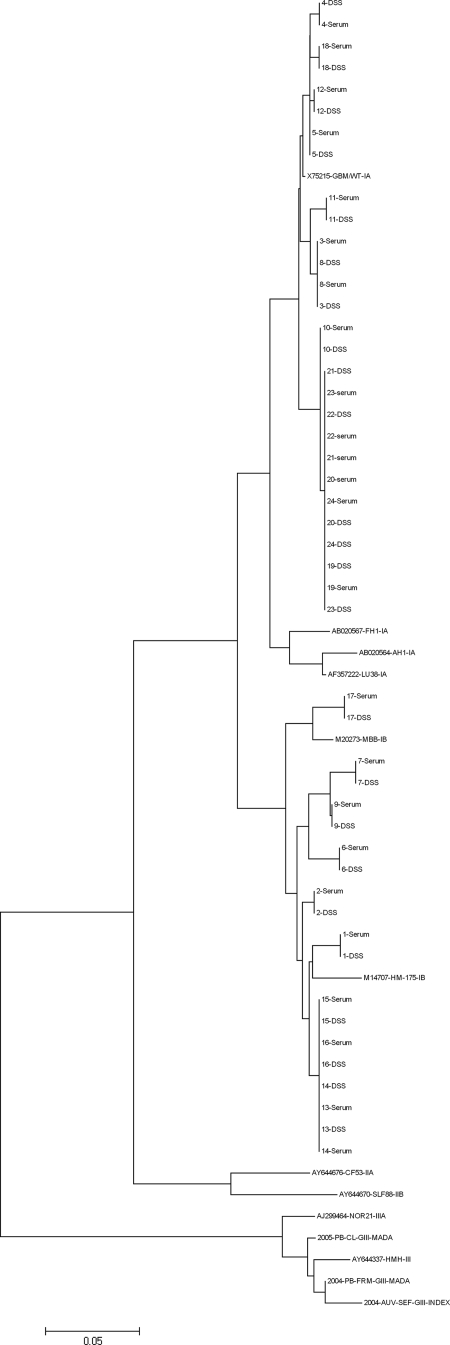
Filter papers were stored at room temperature for 24 h prior to amplification and sequencing reaction. Sequence analysis was performed on the 24 blotted samples that could be amplified by VP1/2A RT-PCR. Nucleotide sequences were compared to the sequences previously obtained at the National Reference Center for HAV. A 100% identity was found over the studied fragment, as illustrated by the phylogenetic tree including blotted and nonblotted sequences (Fig. 3).
FIG. 3.
VP1/2A sequences were obtained from 24 DSS. Phylogenetic analysis of sequences obtained from sera (named 1 to 24) and DSS show the same clustering for serum and matched DSS sequences. Sequences from serum and matched DSS presented a 100% identity over the studied fragment.
DISCUSSION
In the present study, we showed that HAV RNA can be accurately recovered from DSS for molecular epidemiology purposes. As well, we confirmed the reliability of blotted samples in the serological diagnosis of HAV infection. We provide a method to facilitate sample storage and transport from routine laboratories to reference centers for further investigations and large epidemiological studies.
Using dried human plasma spiked with cultured HAV, we showed that HAV RNA was stable at room temperature for up to one month without a loss of viral load. RNA loss induced by DSS methods has been reported for HIV RNA detection (2). The authors of that study attributed the median loss of 0.64 log10 copies/ml of HIV RNA to the susceptibility of HIV to desiccation. Two reasons may account for our better results in HAV RNA detection. First, HAV is a nonenveloped RNA virus which is not sensitive to desiccation. Second, we used chemically treated filter papers (FTA cards) specifically designed for collection and room temperature storage of biological samples for further RNA/DNA analysis. Nucleic acids are protected from degradation, and the chemical treatment is able to inactivate viruses, making the samples no longer infectious and allowing storage and transport without specific biohazard precautions (4, 19, 22).
In our study, the qualitative RT-PCR assays with DSS were about 1 log10 less sensitive than assays with serum. Such loss of sensitivity has been previously reported, for example, for detection of hepatitis C virus and dengue virus RNAs, and occasionally attributed to the elution method (1, 20). Several protocols to improve the final recovery of viral RNA have been described previously (1, 19, 20, 22). We found that elution with Tris-EDTA buffer generated the best results in terms of yield and that a smaller HAV RNA input in RT-PCRs with DSS is solely responsible for this loss of sensitivity. Actually, HAV RNA extracts from DSS were 4.5 times less concentrated than those from serum due to the smaller sample volume used (an input of 75 μl instead of 140 μl) and to the volume of elution (400 μl). When we corrected for this smaller input, we found no differences in HAV RNA viral loads either in virus-spiked human plasma or on patient sera.
As shown by other authors (1, 20), this loss of sensitivity may result in false-negative results for samples with extremely low virus titers for HAV. However, a detection CO of 100 copies/ml is usually sufficient for HAV RNA amplification. Costa-Mattioli et al. reported that the viral loads in clinical samples on the first day of clinical diagnosis ranged from 3.3 to 6.9 log copies/ml, with a mean value of 5.8 log copies/ml (7). The viral loads that we observed were about 1 log lower, ranging from 2.11 to 6.08 log copies/ml (mean, 4.12 log copies/ml). In our study, viral quantification was performed retrospectively for samples kept at −80°C but which were thawed at least twice, and this may have contributed to lower viral titers. We would have had a better sensitivity of HAV RNA amplification from DSS if we had used fresh sera. Nevertheless, VP1/2A amplification was obtained in 24/26 sera, and sequence analysis provided no discrepant results, demonstrating the reliability of the method for molecular epidemiology.
Three studies on the use of blotted samples for serological diagnosis of HAV infection have been published (10, 12, 21), but the present one provides new information regarding the influence of storage time and temperature on HAV antibody detection. We used filter papers different from those used for the molecular study, since FTA cards, by denaturing proteins, are not proper for immunoassays. Thus, these filter papers should be considered infectious, and we are further working on viral inactivation by beta-propiolactone before spotting.
We achieved 100% specificity and sensitivity for detection of HAVT from DSS stored up to one month, even at room temperature or at 37°C, despite a fourfold dilution of sample input due to the elution process. Several reasons may account for the sensitivity of our assay being better than the sensitivities of 89.6% and 91.3% reported in two previous studies (10, 12). Both studies used dried blood spots instead of serum spots and elution volumes larger than those used in our assay, thereby reducing the sample input. Then, in both cases, the competitive assays for HAVT were performed without blotting the calibrator used to determine the CO. And finally, both studies tested a larger number of samples, without excluding sera with values around the CO; thus, the probability of obtaining results discrepant between spotted and nonspotted samples was higher than in our study. By contrast, the 100% specificity and sensitivity of the HAVM assay with DSS are in line with published performances, also performed with DSS and with minimal adaptation of the immunoassay (21). For HAVT and HAVM assays and all storage conditions, we obtained positive and negative delta values of >2 in most cases. This clear separation of negative and positive populations from the CO reduces the probability of false-positive or -negative test results.
In conclusion, molecular and serological assays for HAV with DSS are reliable. The DSS method is valuable for use with small volumes and facilitates and secures the storage and transport of samples to reference centers.
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Abe, K., and N. Konomi. 1998. Hepatitis C virus RNA in dried serum spotted onto filter paper is stable at room temperature. J. Clin. Microbiol. 363070-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amellal, B., C. Katlama, and V. Calvez. 2007. Evaluation of the use of dried spots and of different storage conditions of plasma for HIV-1 RNA quantification. HIV Med. 8396-400. [DOI] [PubMed] [Google Scholar]
- 3.Barin, F., L. Meyer, R. Lancar, C. Deveau, M. Gharib, A. Laporte, J. C. Desenclos, and D. Costagliola. 2005. Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J. Clin. Microbiol. 434441-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharya, B., R. A. Weiss, C. Davis, H. Holmes, D. Hockley, and A. Fassati. 2004. Detection and quantitation of human immunodeficiency virus type-1 particles by confocal microscopy. J. Virol. Methods 12013-21. [DOI] [PubMed] [Google Scholar]
- 5.Bower, W. A., O. V. Nainan, X. Han, and H. S. Margolis. 2000. Duration of viremia in hepatitis A virus infection. J. Infect. Dis. 18212-17. [DOI] [PubMed] [Google Scholar]
- 6.Chironna, M., A. Grottola, C. Lanave, E. Villa, S. Barbuti, and M. Quarto. 2003. Genetic analysis of HAV strains recovered from patients with acute hepatitis from southern Italy. J. Med. Virol. 70343-349. [DOI] [PubMed] [Google Scholar]
- 7.Costa-Mattioli, M., S. Monpoeho, E. Nicand, M. H. Aleman, S. Billaudel, and V. Ferre. 2002. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J. Viral Hepat. 9101-106. [DOI] [PubMed] [Google Scholar]
- 8.Crofts, N., W. Maskill, and I. D. Gust. 1988. Evaluation of enzyme-linked immunosorbent assays: a method of data analysis. J. Virol. Methods 2251-59. [DOI] [PubMed] [Google Scholar]
- 9.Croom, H. A., K. M. Richards, S. J. Best, B. H. Francis, E. I. Johnson, E. M. Dax, and K. M. Wilson. 2006. Commercial enzyme immunoassay adapted for the detection of antibodies to hepatitis C virus in dried blood spots. J. Clin. Virol. 3668-71. [DOI] [PubMed] [Google Scholar]
- 10.de Almeida, L. M., R. S. Azevedo, A. A. Guimaraes, S. Coutinho Eda, C. J. Struchiner, and E. Massad. 1999. Detection of antibodies against hepatitis A virus in eluates of blood spotted on filter-paper: a pilot study in Rio de Janeiro, Brazil. Trans. R. Soc. Trop. Med. Hyg. 93401-404. [DOI] [PubMed] [Google Scholar]
- 11.De Swart, R. L., Y. Nur, A. Abdallah, H. Kruining, H. S. El Mubarak, S. A. Ibrahim, B. Van Den Hoogen, J. Groen, and A. D. Osterhaus. 2001. Combination of reverse transcriptase PCR analysis and immunoglobulin M detection on filter paper blood samples allows diagnostic and epidemiological studies of measles. J. Clin. Microbiol. 39270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil, A., A. Gonzalez, R. Dal-Re, V. Dominguez, P. Astasio, and L. Aguilar. 1997. Detection of antibodies against hepatitis A in blood spots dried on filter paper. Is this a reliable method for epidemiological studies? Epidemiol. Infect. 118189-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joussemet, M., P. Bourin, O. Lebot, G. Fabre, and R. Deloince. 1992. Evolution of hepatitis A antibodies prevalence in young French military recruits. Eur. J. Epidemiol. 8289-291. [DOI] [PubMed] [Google Scholar]
- 14.Karapanagiotidis, T., M. Riddell, and H. Kelly. 2005. Detection of rubella immunoglobulin M from dried venous blood spots using a commercial enzyme immunoassay. Diagn. Microbiol. Infect. Dis. 53107-111. [DOI] [PubMed] [Google Scholar]
- 15.Katz, R. S., M. Premenko-Lanier, M. B. McChesney, P. A. Rota, and W. J. Bellini. 2002. Detection of measles virus RNA in whole blood stored on filter paper. J. Med. Virol. 67596-602. [DOI] [PubMed] [Google Scholar]
- 16.McCarron, B., R. Fox, K. Wilson, S. Cameron, J. McMenamin, G. McGregor, A. Pithie, and D. Goldberg. 1999. Hepatitis C antibody detection in dried blood spots. J. Viral Hepat. 6453-456. [DOI] [PubMed] [Google Scholar]
- 17.Mercader, S., D. Featherstone, and W. J. Bellini. 2006. Comparison of available methods to elute serum from dried blood spot samples for measles serology. J. Virol. Methods 137140-149. [DOI] [PubMed] [Google Scholar]
- 18.O'Shea, S., J. Mullen, K. Corbett, I. Chrystie, M. L. Newell, and J. E. Banatvala. 1999. Use of dried whole blood spots for quantification of HIV-1 RNA. AIDS 13630-631. [DOI] [PubMed] [Google Scholar]
- 19.Picard-Meyer, E., J. Barrat, and F. Cliquet. 2007. Use of filter paper (FTA) technology for sampling, recovery and molecular characterisation of rabies viruses. J. Virol. Methods 140174-182. [DOI] [PubMed] [Google Scholar]
- 20.Prado, I., D. Rosario, L. Bernardo, M. Alvarez, R. Rodriguez, S. Vazquez, and M. G. Guzman. 2005. PCR detection of dengue virus using dried whole blood spotted on filter paper. J. Virol. Methods 12575-81. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez Lay, L. A., M. A. Ribas Antunez, B. Diaz Mendiondo, A. Roque Quintero, U. Aragon Rodriguez, R. Martinez Casanueva, and C. Rodriguez Valdes. 1998. Filter paper sampling for the detection of anti-hepatitis A IgM antibodies. Rev. Cubana Med. Trop. 5042-47. (In Spanish.) [PubMed] [Google Scholar]
- 22.Rogers, C. D., and L. A. Burgoyne. 2000. Reverse transcription of an RNA genome from databasing paper (FTA). Biotechnol. Appl. Biochem. 31219-224. [DOI] [PubMed] [Google Scholar]
- 23.Soetens, O., C. Vauloup-Fellous, I. Foulon, P. Dubreuil, B. De Saeger, L. Grangeot-Keros, and A. Naessens. 2008. Evaluation of different cytomegalovirus (CMV) DNA PCR protocols for analysis of dried blood spots from consecutive cases of neonates with congenital CMV infections. J. Clin. Microbiol. 46943-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stene-Johansen, K., P. A. Jenum, T. Hoel, H. Blystad, H. Sunde, and K. Skaug. 2002. An outbreak of hepatitis A among homosexuals linked to a family outbreak. Epidemiol. Infect. 129113-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stroffolini, T., R. D'Amelio, P. M. Matricardi, P. Chionne, A. Napoli, M. Rapicetta, S. Crateri, and P. Pasquini. 1993. The changing epidemiology of hepatitis A in Italy. Ital. J. Gastroenterol. 25372-374. [PubMed] [Google Scholar]