Abstract
Acanthamoeba keratitis (AK) is a rare but sight-threatening ocular infection. Outbreaks have been associated with contaminated water and contact lens wear. The epidemiology and pathology may be associated with unique genotypes. We determined the Rns genotype for 37 clinical isolates from 23 patients presenting at the University of Miami Bascom Palmer Eye Institute with confirmed AK infections in 2006 to 2008. The genus-specific ASA.S1 amplicon allowed for rapid genotyping of the nonaxenic cultures. Of the 37 isolates, 36 were of the T4 genotype. Within this group, 13 unique diagnostic fragment 3 sequences were identified, 3 of which were not in GenBank. The 37th isolate was a T5, the first in the United States and second worldwide to be found in AK. For five patients with isolates from the cornea and contact lens/case, identical sequences within each patient cluster were observed, confirming the link between contact lens contamination and AK infection. Genotyping is an important tool in the epidemiological study of AK. In this study, it allowed for the detection of new strains and provided an etiological link between source and infection. Additionally, it can allow for accurate categorizing of physiological differences, such as strain virulence, between isolates and clades.
The genus Acanthamoeba is comprised of a group of free-living amoebae that are responsible for causing Acanthamoeba keratitis (AK), a rare but sight-threatening corneal infection. In recent years, the number of AK cases has been on the increase, especially among wearers of contact lenses, who make up 85 to 90% of the AK cases (8, 24, 31, 35). Diagnosis of AK is problematic due to clinical features which are similar to those of herpetic, bacterial, and fungal infections. For example, the stromal ring infiltrate associated with AK is only observed ∼6% of the time in early cases and ∼16% of the time in late cases (2, 3, 4, 14). AK can be the primary infection or be present as a suprainfection in combination with other infectious organisms, like bacteria or fungi, thereby complicating diagnosis and treatment. The encystment capability of Acanthamoeba species also confounds treatment due to the recalcitrant nature of the cyst to most treatment options allowing reemergence of amoebae after treatment cessation.
Acanthamoeba genotyping is a useful tool for studying taxonomic and epidemiological relationships and thereby allowing correlations between the infectious isolates and disease phenotypes, such as virulence factors, drug susceptibility, and/or species-clinical outcome correlations, to be explored. The gene targeted most often in Acanthamoeba genotyping is the nuclear small-subunit rRNA gene (Rns), and utilizing a 5% sequence dissimilarity cutoff point, 15 or more genotype clades, designated T1, T2, T3, etc., have been identified (12, 13, 15, 17, 29). Isolates from six of the genotypic clades (T3, T4, T5, T6, T11 and T15) are confirmed to be causative agents of AK (10, 13, 19, 21, 28, 29, 34, 36). The most prevalent Acanthamoeba genotype in both clinical and environmental samples is the T4 genotype (6, 7). Within the genotype clades, multiple species designations can be observed. This is primarily due to the traditional classification method's reliance on changeable morphological characteristics, such as cyst morphology, creating inconsistent species identification (25, 32). Therefore, it was proposed that each genotypic clade be equated with a single species (29). For example, all isolates in the T4 clade could be reclassified as Acanthamoeba castellanii since the T4 genotype includes the type strain for that species.
In this study, 37 isolates from corneal scrapes, contact lenses, and lens cases of 23 patients presenting with AK at the Anne Bates Leach Eye Hospital, Bascom Palmer Eye Institute, University of Miami, from 2006 to 2008 were examined to assess the Rns genotypes responsible for the infections. Acanthamoeba species can be rapidly genotyped by targeting a highly variable region designated diagnostic fragment 3 (DF3) within the genus-specific Rns ASA.S1 amplicon (5, 26); therefore, this region was chosen for analysis. The genotypes identified in this study were also compared to strains identified in other studies in order to examine the prevalence of the DF3 sequence types within genotype clades (5, 37, 38).
MATERIALS AND METHODS
Cultures.
Thirty-seven Acanthamoeba isolates cultured from corneal scrapings, biopsies, contact lenses, or lens cases were recovered from 23 patients presenting with AK at the University of Miami Bascom Palmer Eye Institute between January 2006 and February 2008 (Table 1). Patients' ages ranged from 14 to 83 years. The risk factor for all patients involved the use of contact lenses. Diagnosis of AK was based on the detection of cysts or trophozoites in corneal sample smears and/or growth on nonnutrient agar plates overlaid with live Escherichia coli.
TABLE 1.
Isolates used for sequencing of the Rns ASA.S1 amplicon
| Culture designationa | Culture source | Rns genotype/ DF3 sequenceb | GenBank accession no. |
|---|---|---|---|
| BP:P1:RCS | Right corneal scrape | T4/13 | FJ422513 |
| BP:P2:CB | Corneal button | T4/6 | FJ422515 |
| BP:P2:CS | Corneal scrape | T4/6 | FJ422514 |
| BP:P3:RCS | Right corneal scrape | T4/6 | FJ422517 |
| BP:P3:RCS[2] | Right corneal scrape | T4/6 | FJ422523 |
| BP:P3:LCS | Left corneal scrape | T4/6 | FJ422522 |
| BP:P3:RCL | Right contact lens | T4/6 | FJ422516 |
| BP:P3:RLC | Right lens case | T4/6 | FJ422520 |
| BP:P3:LLC | Left lens case | T4/6 | FJ422521 |
| BP:P4:RCS | Right corneal scrape | T4/6 | FJ422518 |
| BP:P5:LLC | Left lens case | T4/6 | FJ422519 |
| BP:P6:LCS | Left corneal scrape | T4/14 | FJ422524 |
| BP:P7:LCL | Left contact lens | T4/14 | FJ422525 |
| BP:P7:RCL | Right contact lens | T4/15 | FJ422526 |
| BP:P8:LCS | Left corneal scrape | T4/12 | FJ422512 |
| BP:P9:LCS | Left corneal scrape | T4/17 | FJ422537 |
| BP:P9:RCL | Right contact lens | T4/17 | FJ422538 |
| BP:P9:LCL | Left contact lens | T4/17 | FJ422539 |
| BP:P10:RCL | Right contact lens | T4/2 | FJ422528 |
| BP:P10:RCB | Right corneal button | T4/2 | FJ422529 |
| BP:P10:RCS | Right corneal scrape | T4/2 | FJ422530 |
| BP:P11:RCS | Right corneal scrape | T4/2 | FJ422531 |
| BP:P12:LCS | Left corneal scrape | T4/2 | FJ422532 |
| BP:P13:CB | Corneal button | T4/20 | FJ422536 |
| BP:P14:LCS | Left corneal scrape | T4/18 | FJ422533 |
| BP:P14:LC | Lens case | T4/18 | FJ422534 |
| BP:P15:RCS | Right corneal scrape | T4/16 | FJ422527 |
| BP:P16:RCS | Right corneal scrape | T4/21 | FJ422541 |
| BP:P16:LC | Lens case | T4/21 | FJ422540 |
| BP:P16:LC[2] | Lens case | T4/21 | FJ422543 |
| BP:P17:LCS | Left corneal scrape | T4/21 | FJ422542 |
| BP:P18:LCS | Left corneal scrape | T4/21 | FJ422544 |
| BP:P19:RCS | Right corneal scrape | T4/21 | FJ422545 |
| BP:P20:LCS | Left corneal scrape | T4/11 | FJ422511 |
| BP:P21:LCS | Left corneal scrape | T4/11 | FJ422510 |
| BP:P22:LCS | Left corneal scrape | T4/19 | FJ422535 |
| BP:P23:LCS | Left corneal scrape | T5 | FJ422546 |
Explanation of culture designations: BP, Bascom Palmer; P, patient; RCS, right corneal scrape; CB, corneal button; LCS, left corneal scrape; LLC, left lens case; RLC, right lens case; LC, lens case; LCL, left contact lens; RCL, right contact lens. [2] indicates second isolates obtained from the same source.
The DF3 sequence nomenclature used in this study is the same as that reported by Booton et al. (5). The first part is the Rns genotype and the second part is a unique code assigned to the specific DF3 sequence.
Genotyping.
Acanthamoeba isolates were harvested from agar plates and rinsed in phosphate-buffered saline (pH 7.4), and DNA was extracted using the UNSET method (18). PCR amplification of the Rns amplicon ASA.S1 was generated using the genus-specific primer set JDP1 (5′-GGCCCAGATCGTTTACCGTGAA-3′) and JDP2 (5′-TCTCACAAGCTGCTAGGGGAGTCA-3′), which encodes the highly variable DF3 region (26). Two or more PCR products were pooled or independently sequenced using the amplification primers JDP1 and JDP2, in addition to the conserved primers 892 (5′-CCAAGAATTTCACCTCTGAC-3′) and 892C (5′-GTCAGAGGTGAAATTCTTGG-3′). Sequencing of the PCR products was performed by Genewiz, Inc. (South Plainfield, NJ). The DF3 sequence designation is based on nomenclature described by Booton et al. (5). The first part is the Rns genotype of the isolate. The second part is a unique code assigned to a specific DF3 sequence type. The Booton et al. study (5) identified 10 DF3 sequence types. The numbers used to define the DF3 sequence type in this study are a continuation of that system.
Phylogenetic analysis.
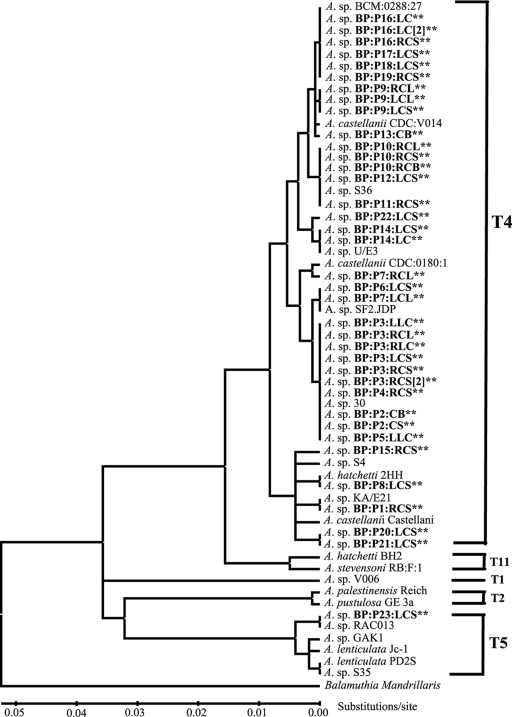
Alignments and phylogenetic reconstructions were performed using the phylogenetic computer program MEGA4 (Molecular Evolutionary Genetic Analysis software, version 4) (30). The evolutionary distances were computed using the Kimura 2 parameter distance algorithm (20) and are in the units of the number of base substitutions per site. All positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons. A total of 449 positions were used in the final data set. The bootstrap consensus tree is inferred from 1,000 replicates (11). Balamuthia mandrillaris, a close phylogenetic relative of Acanthamoeba, was used as the outgroup to root the trees. Phylogenetic reconstructed gene trees were generated using maximum-parsimony, neighbor-joining, UPGMA or minimum evolution methods in MEGA4 were compared. The neighbor-joining tree is displayed in Fig. 1.
FIG. 1.
Rns DF3 linearized neighbor-joining gene tree. Isolates from this study are shown in boldface text and with asterisks. The tree was constructed using 1,000 bootstrap replications. The T1, T2, T4, T5, and T11 designations shown on the tree correspond to strains previously determined to be of that particular genotype (26, 29).
Nucleotide sequence accession numbers.
The 37 sequences determined in this study were deposited in GenBank under accession numbers FJ422510 to FJ422546. The other Acanthamoeba sequences used in this study are available in GenBank under the following accession numbers: Acanthamoeba castellanii strain CDC:0180:1, U07405; Acanthamoeba hatchetti strain 2HH, AF26022; Acanthamoeba castellanii strain castellani, U07413; Acanthamoeba sp. strain KA/E21, EF140633; Acanthamoeba sp. strain U/E3, AY026747; Acanthamoeba sp. strain S36, EU146073; Acanthamoeba sp. strain S30, DQ087313; Acanthamoeba sp. strain SF2.JDP, EU338518; Acanthamoeba sp. strain S4, DQ087320; Acanthamoeba castellanii strain CDC:0184:V014, U07401; Acanthamoeba sp. strain BCM:0288:27, U07409; Acanthamoeba hatchetti strain BH2, AF019068; Acanthamoeba stevensoni strain RB:F:1, AF019069; Acanthamoeba sp. strain V006, U07400; Acanthamoeba palestinensis strain Reich, U07411; Acanthamoeba pustulosa strain GE 3a, AF019050; Acanthamoeba sp. strain RAC013, AB327060; Acanthamoeba sp. strain GAK1, AY944575; Acanthamoeba lenticulata strain Jc-1, U94739; Acanthamoeba lenticulata strain PD2S, U94741; Acanthamoeba sp. strain S35, EU146072; Balamuthia mandrillaris, AF477022.
RESULTS
DF3 sequences.
The variable DF3 regions of the Rns genes of 37 isolates from 23 patients identified 14 unique DF3 sequences (Fig. 2 and Table 1). Of the 14 sequence types obtained, 13 correspond to 36/37 (97%) of the isolates examined, and these were identical or similar to previously described isolates of the T4 genotype (Fig. 1), herein referred to as T4/2, T4/6, and T4/11 to T4/21. Three of the sequence types (T4/11, T4/17, and T4/19) represent new T4 sequences not found in GenBank. The remaining isolate possessed a DF3 sequence most similar to sequences of Acanthamoeba lenticulata isolates, which are classified as genotype T5.
FIG. 2.
Primary sequence alignment of a subset area of the highly variable and informative region of DF3 (stem 29-1, 18S rRNA) of Bascomb Palmer Eye Institute isolates. Sequences are aligned by similarity. Gaps are represented by dashes.
Rns T4 genotype isolates.
Table 1 summarizes the genotype/DF3 sequence type of all the isolates examined in this study. All 23 patients were contact lens wearers, and of these, 5 (patients BP:P3, BP:P9, BP:P10, BP:14, and BP:P16) had the Rns sequence type determined for the cultures grown from their contact lens paraphernalia and corneal scrapes. In all cases, identical DF3 sequences were observed in the corneal scrape specimens and the contact lens paraphernalia, which suggests that the contact lens paraphernalia can be a source of the infection (Table 1 and Fig. 1).
Patient BP:P7 was unusual in that the sequence types of the isolated Acanthamoeba strains were different between the right and left lens case. The Acanthamoeba strain isolated from the right lens case was genotype T4/15, whereas the genotype of the Acanthamoeba strain in the left lens case was T4/14. No corneal scrape specimen was available for patient BP:P7; therefore, it is unknown which, if either, caused the keratitis.
Identical sequence types were observed not only within different sources from a single patient, but also between different patients. Five of the sequence types, T4/2, T4/6, T4/21, T4/14, and T4/11, showed identical sequence types between different patients, suggesting infection by similar if not identical Acanthamoeba strains. Alignments with sequences from GenBank showed that the majority of the sequence types have been observed in multiple patients with keratitis worldwide.
Rns T5 genotype isolate.
Of the 37 Acanthamoeba cultures examined, 1 isolate was determined to have the rare T5 genotype. This isolate's DF3 sequence was identical to that of Acanthamoeba sp. RAC013, an isolate from drinking water in Osaka, Japan, and this isolate is the first case of a T5 Acanthamoeba isolate causing AK in the United States.
DISCUSSION
The genotyping data obtained in this study of amoebae isolated from AK patients further confirms T4 as the predominant genotype, a trend observed in previous studies (5, 37, 38). A comparison of genotypes from this study with those from other studies that investigated multiple AK isolates revealed that our study had the T4/6 and T4/2 genotypes in common with the study of Hong Kong isolates (5). Our study and the Zhang et al. (38) results for North China had the T4/2, T4/12, and T4/13 genotypes in common, and our study and the Yera et al. study (37) from France had only the T4/2 genotype in common. Although based on limited datasets, the T4/2 genotype appears to be the geographically predominate sequence type.
With the worldwide prevalence of the T4 genotype regardless of region, it is not surprising that 90% of Acanthamoeba isolates associated with AK are genotype T4. What is of particular interest is that the second most abundant environmental clade, T5, is dramatically underrepresented in AK cases (6, 7). This study is only the second study to describe a T5 isolate causing AK and the first in the United States. It is unlikely a lack of exposure that explains the low infection rate, as the T5 genotype has been detected in human mucosa without amoebic infection (9). Further complicating the issue are experimental animal and tissue culture models that have shown T5 isolates to be capable of a high degree of pathogenicity (33, 34). Additionally, studies comparing T4 and T5 resistance to multipurpose contact lens cleaning solutions, interestingly, show that the T5 genotype possesses a better resistance (16, 27). It is possible that the majority of T5 Acanthamoeba isolates may not be pathogenic to humans, but as the number of people that wear contact lenses continues to grow, the risk of encountering pathogenic T5 isolates may increase.
An interesting observation was the lack of the T3 genotype in this study. Several studies that have determined genotypes of Acanthamoeba strains from AK and contact lens/cases each identified the presence of the T3 genotype, which, based on environmental distribution, is less prevalent than T5 (5, 7, 37, 38). Also, like T5 isolates, T3 isolates can show more resistance to multipurpose contact lens cleaning solutions than do isolates of the T4 genotype (27). Understanding what makes T4 more virulent to humans is an important area of study. Multiple factors contribute to Acanthamoeba pathogenicity, such as extracellular protease production and amoeba cell surface adherence ability. In studies that examined pathogenicity predictive factors, the T3, T5, and T4 genotypes all displayed high pathogenicity (1, 22, 33, 34), although the T3 results were not always consistent between isolates. The T4 genotypes did show increased cell surface binding compared to that of T3 (1); however, it is essential to realize the small number of T3 and T5 genotypes examined in these studies compared to the number of T4. These observations do suggest that a different rationale must exist to explain the underrepresentation observed with T3 and T5 genotypes. It should be noted that these studies used in vitro cell culture models to compare the pathogenicities of isolates, which emphasizes the need for a good clinical animal model.
Obviously, there are certain properties within the T4 genotype that make it more virulent. Therefore, the need for accurate genotyping of Acanthamoeba strains from different environments along with an analysis of their virulence factors and, in clinical AK cases, an examination of outcome would greatly enhance and stimulate research. Also, the integration of a PCR-based assay in the detection of Acanthamoeba strains, in addition to genotypic information that can be obtained, offers a rapid diagnostic tool. Utilized alongside the conventional method of smear examination, an AK diagnosis can be ideally accomplished in less than a day and would be more cost-effective than fluorescence- or in vivo confocal microscopy-based methods. The use of a PCR-based assay offers all the hallmarks of a good diagnostic test: high sensitivity, high specificity, and high positive and negative predictive values (23, 36).
Acknowledgments
This work was supported by the University of Miami Wallace H. Coulter Center for Translational Research, Research to Prevent Blindness Unrestricted Grant to the University of Miami, Public Health Service grant EY014801 from the National Eye Institute, and a gift from Bausch & Lomb, Inc.
At the time this work was performed, M.E.F. held the Walter G. Ross Chair in Ophthalmic Research at the University of Miami.
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Alsam, S., K. S. Kim, M. Stins, A. O. Rivas, J. Sissons, and N. A. Khan. 2003. Acanthamoeba interactions with human brain microvascular endothelial cells. Microb. Pathog. 35234-235. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, A. S., J. K. Dart, L. A. Ficker, M. M. Matheson, and P. Wright. 1993. Acanthamoeba keratitis. The value of early diagnosis. Ophthalmology 1001238-1243. [DOI] [PubMed] [Google Scholar]
- 3.Bacon, A. S., D. G. Frazer, J. K. Dart, M. Matheson, L. A. Ficker, and P. Wright. 1993. A review of 72 consecutive cases of Acanthamoeba keratitis, 1984-1992. Eye 7719-725. [DOI] [PubMed] [Google Scholar]
- 4.Bernauer, W., G. I. Duguid, and J. K. Dart. 1996. Early clinical diagnosis of Acanthamoeba keratitis. A study of 70 eyes. Klin. Monatsbl. Augenheilkd. 208282-284. [DOI] [PubMed] [Google Scholar]
- 5.Booton, G. C., D. J. Kelly, Y. W. Chu, D. V. Seal, E. Houang, D. S. Lam, T. J. Byers, and P. A. Fuerst. 2002. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J. Clin. Microbiol. 401621-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booton, G. C., A. Rogerson, T. D. Bonilla, D. V. Seal, D. J. Kelly, T. K. Beattie, A. Tomlinson, F. Lares-Villa, P. A. Fuerst, and T. J. Byers. 2004. Molecular and physiological evaluation of subtropical environmental isolates of Acanthamoeba spp., causal agent of Acanthamoeba keratitis. J. Eukaryot. Microbiol. 51192-200. [DOI] [PubMed] [Google Scholar]
- 7.Booton, G. C., G. S. Visvesvara, T. J. Byers, D. J. Kelly, and P. A. Fuerst. 2005. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J. Clin. Microbiol. 431689-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2007. Acanthamoeba keratitis multiple states, 2005 to 2007. MMWR Morb. Mortal. Wkly. Rep. 56532-534. [PubMed] [Google Scholar]
- 9.De Jonckheere, J. F., and R. Michel. 1988. Species identification and virulence of Acanthamoeba strains from human nasal mucosa. Parasitol. Res. 74314-316. [DOI] [PubMed] [Google Scholar]
- 10.Di Cave, D., R. Monno, P. Bottalico, S. Guerriero, S. D'Amelio, C. D'Orazi, and F. Berrilli. 2008. Acanthamoeba T4 and T15 genotypes associated with keratitis infections in Italy. Eur. J. Clin. Microbiol. Infect. Dis. doi: 10.1007/s10096-008-0682-4. [DOI] [PubMed]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39783-791. [DOI] [PubMed] [Google Scholar]
- 12.Gast, R. J. 2001. Development of an Acanthamoeba-specific reverse dot-blot and the discovery of a new ribotype. J. Eukaryot. Microbiol. 48609-615. [DOI] [PubMed] [Google Scholar]
- 13.Gast, R. J., D. R. Ledee, P. A. Fuerst, and T. J. Byers. 1996. Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J. Eukaryot. Microbiol. 43498-504. [DOI] [PubMed] [Google Scholar]
- 14.Goodall, K., A. Brahma, and A. Ridgway. 1996. Acanthamoeba keratitis: masquerading as adenoviral keratitis. Eye 10643-644. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt, M. K., B. S. Robinson, P. T. Monis, and C. P. Saint. 2003. Identification of a new Acanthamoeba 18S rRNA gene sequence type, corresponding to the species Acanthamoeba jacobsi Sawyer, Nerad, and Visvesvara, 1992 (Lobosea: Acanthamoebidae). Acta Protozool. 42325-329. [Google Scholar]
- 16.Hiti, K., J. Walochnik, E. M. Haller-Schober, C. Faschinger, and H. Aspöck. 2002. Viability of Acanthamoeba after exposure to a multipurpose disinfecting contact lens solution and two hydrogen peroxide systems. Br. J. Ophthalmol. 86144-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn, M., T. R. Fritsche, T. Linner, R. K. Gautom, M. D. Harzenetter, and M. Wagner. 2002. Obligate bacterial endosymbionts of Acanthamoeba spp. related to the beta-Proteobacteria: proposal of “Candidatus Procabacter Acanthamoebae” gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 52599-605. [DOI] [PubMed] [Google Scholar]
- 18.Hugo, E. R., V. J. Stewart, R. J. Gast, and T. J. Byers. 1992. Purification of amoeba mtDNA using UNSET procedure, p. D-7.1. In A. T. Sold and J. J. Lee (ed.), Protocols in protozoology. Allen Press, Lawrence, KS.
- 19.Khan, N. A., E. L. Jarroll, and T. A. Paget. 2002. Molecular and physiological differentiation between pathogenic and nonpathogenic Acanthamoeba. Curr. Microbiol. 45197-202. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16111-120. [DOI] [PubMed] [Google Scholar]
- 21.Ledee, D. R., J. Hay, T. J. Byers, D. V. Seal, and C. M. Kirkness. 1996. Acanthamoeba griffini. Molecular characterization of a new corneal pathogen. Investig. Ophthalmol. Vis. Sci. 37544-550. [PubMed] [Google Scholar]
- 22.Lorenzo-Morales, J., A. Ortega-Rivas, E. Martínez, M. Khoubbane, P. Artigas, M. V. Periago, P. Foronda, N. Abreu-Acosta, B. Valladares, and S. Mas-Coma. 2006. Acanthamoeba isolates belonging to T1, T2, T3, T4 and T7 genotypes from environmental freshwater samples in the Nile Delta region, Egypt. Acta Trop. 10063-69. [DOI] [PubMed] [Google Scholar]
- 23.Pasricha, G., S. Sharma, P. Garg, and R. K. Aggarwal. 2003. Use of 18S rRNA gene-based PCR assay for diagnosis of Acanthamoeba keratitis in non-contact lens wearers in India. J. Clin. Microbiol. 413206-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel, A., and K. Hammersmith. 2008. Contact lens-related microbial keratitis: recent outbreaks. Curr. Opin. Ophthalmol. 19302-306. [DOI] [PubMed] [Google Scholar]
- 25.Sawyer, T. 1971. Acanthamoeba griffini, a new species of marine amoeba. J. Protozool. 18650-654. [Google Scholar]
- 26.Schroeder, J. M., G. C. Booton, J. Hay, I. A. Niszl, D. V. Seal, M. B. Markus, P. A. Fuerst, and T. J. Byers. 2001. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 391903-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoff, M., A. Rogerson, S. Schatz, and D. Seal. 2007. Variable responses of Acanthamoeba strains to three multipurpose lens cleaning solutions. Optom. Vis. Sci. 84202-207. [DOI] [PubMed] [Google Scholar]
- 28.Spanakos, G., K. Tzanetou, D. Miltsakakis, E. Patsoula, E. Malamou-Lada, and N. C. Vakalis. 2006. Genotyping of pathogenic Acanthamoebae isolated from clinical samples in Greece—report of a clinical isolate presenting T5 genotype. Parasitol. Int. 55147-149. [DOI] [PubMed] [Google Scholar]
- 29.Stothard, D. R., J. M. Schroeder-Diedrich, M. H. Awwad, R. J. Gast, D. R. Ledee, S. Rodriguez-Zaragoza, C. L. Dean, P. A. Fuerst, and T. J. Byers. 1998. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J. Eukaryot. Microbiol. 4545-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 31.Thebpatiphat, N., K. M. Hammersmith, F. N. Rocha, C. J. Rapuano, B. D. Ayres, P. R. Laibson, R. C. Eagle, Jr., and E. J. Cohen. 2007. Acanthamoeba keratitis: a parasite on the rise. Cornea 26701-706. [DOI] [PubMed] [Google Scholar]
- 32.Visvesvara, G. S. 1991. Classification of Acanthamoeba. Rev. Infect. Dis. 13(S5)S369-S372. [DOI] [PubMed] [Google Scholar]
- 33.Walochnik, J., A. Obwaller, and H. Aspöck. 2000. Correlations between morphological, molecular biological, and physiological characteristics in clinical and nonclinical isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 664408-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walochnik, J., E. Haller-Schober, H. Kölli, O. Picher, A. Obwaller, and H. Aspöck. 2000. Discrimination between clinically relevant and nonrelevant Acanthamoeba strains isolated from contact lens-wearing keratitis patients in Austria. J. Clin. Microbiol. 383932-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watt, K., and H. A. Swarbrick. 2005. Microbial keratitis in overnight orthokeratology: review of the first 50 cases. Eye Contact Lens 31201-208. [DOI] [PubMed] [Google Scholar]
- 36.Yera, H., O. Zamfir, T. Bourcier, T. Ancelle, L. Batellier, J. Dupouy-Camet, and C. Chaumeil. 2007. Comparison of PCR, microscopic examination and culture for the early diagnosis and characterization of Acanthamoeba isolates from ocular infections. Eur. J. Clin. Microbiol. Infect. Dis. 26221-224. [DOI] [PubMed] [Google Scholar]
- 37.Yera, H., O. Zamfir, T. Bourcier, E. Viscogliosi, C. Noël, J. Dupouy-Camet, and C. Chaumeil. 2008. The genotypic characterisation of Acanthamoeba isolates from human ocular samples. Br. J. Ophthalmol. 921139-1141. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., X. Sun, Z. Wang, R. Li, S. Luo, X. Jin, S. Deng, and W. Chen. 2004. Identification of 18S ribosomal DNA genotype of Acanthamoeba from patients with keratitis in North China. Investig. Ophthalmol. Vis. Sci. 451904-1907. [DOI] [PubMed] [Google Scholar]