Abstract
The present study evaluates the performance of the staphylococcal interspersed repeat unit (SIRU) method applied to a diverse collection of 104 Staphylococcus aureus isolates previously characterized by pulsed-field gel electrophoresis (PFGE), spa typing, multilocus sequence typing (MLST), and staphylococcal cassette chromosome mec typing for methicillin-resistant S. aureus. The SIRU method distributed the 104 strains into 81 SIRU profiles that could be clustered into 12 groups and 29 singletons. The discriminatory power of the method at the profile level, translated by Simpson's index of diversity (SID), was similar to that of PFGE subtyping (SID = 99.23% versus 99.85%) and slightly higher than that of spa typing (SID = 97.61%). At the group level, the SIRU SID (93.24%) was lower than that of PFGE typing (95.41%) but higher than that of MLST (SID = 91.77%). The adjusted Rand (AR) coefficient showed that SIRU typing at the group level had the highest congruence with MLST (AR = 0.5736) and with clonal complex (CC) (AR = 0.4963) but the lowest congruence with PFGE subtype (AR = 0.0242). The Wallace coefficient indicated that in the present collection, two strains with the same SIRU profile have a 100% probability of belonging to the same CC, a 90% probability of sharing the same spa type, and an 83% probability of being classified in the same sequence type. The high discriminatory power of the SIRU method, along with its apparent concordance with MLST results, makes it potentially valuable for S. aureus short-term epidemiological investigations and population dynamics as well.
Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA), continues to be a major cause of health care-associated and, more recently, community-associated infections (40, 62). It is critical to have access to an accurate typing method to design cost-effective intervention and prevention strategies (45, 48, 61). A large number of molecular typing methods have been developed to assess strain relatedness for outbreak control, surveillance programs, and population structure and evolution studies (58, 61). The three most used typing methods for S. aureus have advantages and disadvantages, as follows. (i) Pulsed-field gel electrophoresis (PFGE), which is the “gold standard” typing method, has high discriminatory power and accuracy, but it is time-consuming and expensive, and the interlaboratory exchange of results is challenging. (ii) Sequence-based multilocus sequence typing (MLST) is easy to perform, and the results, given as an allelic profile, are portable and easy to exchange due to a public database available on the Internet (http://www.mlst.net), but it is expensive and not useful for local outbreak investigations. MLST is frequently combined with staphylococcal cassette chromosome mec (SCCmec) typing in order to define clonal types of MRSA (24). (iii) spa typing, a single-locus sequence typing method, is being used more frequently for S. aureus typing, and the development of a public database on the Internet (http://spaserver.ridom.de), as with MLST, ensured an international typing nomenclature and thus a great facility in exchanging typing data. By calculation of Simpson's index of diversity (SID), it was shown that spa typing is nearly as discriminatory as PFGE (1, 25), although it takes into account a single variable region of the protein A gene.
In choosing a new method, it is worth taking into consideration that PCR-based methods are commonly used in typing laboratories because of their accuracy, ease of use, low cost, and speed in retrieving results (in a few hours).
Many bacterial genomes carry loci of repetitive DNA, which may contain variable repeated units among strains (43, 60). Systems based on a multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) have been used extensively for typing of clinical isolates of several bacterial species and were shown to perform well compared to other genotyping methods (43, 61). S. aureus harbors a diverse population of DNA repeats, which allowed the design of various MLVA schemes (28, 30, 39, 53, 60). Hardy et al. (36, 38) developed a MLVA scheme for S. aureus where seven novel multiple tandem repeats with a high degree of similarity in the flanking regions were identified based on the alignment of seven S. aureus sequenced genomes (strains N315, MW2, Mu50, MSSA476, MRSA252, NCTC8325, and COL). Six of these seven loci were located on intergenic regions scattered around the S. aureus genome; the remaining locus corresponds to the protein A gene, spa. The method, designated staphylococcal interspersed repeat unit (SIRU) typing, relies on PCR amplification of the seven loci of repetitive DNA, using primers specific for the flanking regions of each locus, and on the determination of the size of each amplicon, which reflects the number of repeated units present on the targeted SIRU. To each of the seven loci is attributed the respective number of DNA repeats, generating a combination of seven numbers that characterizes each strain and corresponds to the allelic profile. This allelic profile makes the SIRU method amenable to interlaboratory comparisons and database management, comparable to MLST. So far, the SIRU method has been applied to S. aureus isolates from nosocomial outbreaks in the United Kingdom and Germany, mainly MRSA isolates, and therefore to highly related strains (29, 35-37). Very recently, a single study evaluated a MLVA scheme including the SIRU typing loci and the sspA gene, using a European collection of contemporary S. aureus isolates (39).
The aim of the present study was to evaluate the SIRU method with a more diverse collection of S. aureus isolates, including MRSA and methicillin-susceptible S. aureus (MSSA) isolates, from different continents, isolated throughout several decades and previously characterized by well-established typing methods (PFGE, spa typing, MLST, and SCCmec for MRSA).
MATERIALS AND METHODS
Bacterial isolates.
A collection of 104 strains (78 MRSA and 26 MSSA strains), previously characterized by PFGE (16), spa typing (1, 56), MLST (3), and SCCmec typing (for MRSA strains) (46), was selected from the Laboratory of Molecular Genetics collection at Instituto de Tecnologia Química e Biológica, Oeiras, Portugal (Table 1). The selected collection included hospital- and community-related strains isolated during a period of over 50 years (from 1943 to 2006) from 17 countries distributed over four continents. Efforts were made to select strains with various degrees of genetic relatedness. Therefore, the collection included (i) strains belonging to the five main clonal complexes (CCs) of S. aureus—CC5 (n = 23), CC8 (n = 32), CC22 (n = 4), CC30 (n = 7), and CC45 (n = 9); (ii) isolates belonging to minor CCs—CC1 (n = 7), CC509 (n = 3), CC50, CC59, CC97, CC101, CC228, CC398, and CC1021 (one isolate of each); (iii) two isolates belonging to CC80, ST80, and SCCmec type IV, identified as community-acquired MRSA; (iv) six single CC isolates, referred to as singletons (S1, sequence type 157 [ST157]; S2, ST447; S3, ST668; S4, ST707; S5, ST580; S6, ST445); and (v) four strains belonging to nondefined CC groups (CC assignments assessed by eBURST v3) and designated ND1 to ND4.
TABLE 1.
Characteristics of the 104 S. aureus isolates and SIRU typing results
| Strain | Isolation date (yr) | Country | No. of SIRU repeatsa
|
SIRU profileb | SIRU groupc | PFGE typed | spa type | ST | CCe | SCCmec typef | Reference(s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIRU01 | SIRU13 | SIRU15 | SIRU16 | SIRU21 | SIRU05 | SIRU07 | |||||||||||
| IPOP38 | 2001 | Portugal | 1 | 1 | 1 | 3 | 7 | 5g | 3 | 1 | G1 | T2 | t127 | 1 | 1 | MSSA | 5 |
| CV73 | 1997 | Cape Verde | 1 | 1 | 2 | 3 | 7 | 5g | 3 | 2 | G1 | T1 | t127 | 1 | 1 | MSSA | 11 |
| HU332 | 2003 | Hungary | 1 | 1 | 2 | 3 | 7 | 5g | 3 | 2 | G1 | F11 | t127 | 1 | 1 | II | 17 |
| HGSA240 | 2003 | Portugal | 1 | 1 | 2 | 3 | 7 | 2 | 3 | 3 | G1 | T7 | t127 | 81 | 1 | MSSA | 13 |
| HSA49 | 1993 | Portugal | 2 | 2 | 1 | 3 | 10 | 3 | 1 | 4 | G2 | P1 | t002 | 5 | 5 | IV | 7 |
| 122MEXU | 1998 | Mexico | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | M2 | t002 | 5 | 5 | II | 63 |
| 78MEXC | 1997 | Mexico | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | A2 | t002 | 5 | 5 | II | 63 |
| 79MEXC | 1997 | Mexico | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | N3 | t002 | 5 | 5 | II | 63 |
| ARG229 | 1995 | Argentina | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | A4 | t002 | 100 | 5 | II | 7, 9, 18 |
| BK2464 | 1990 | United States | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | A1 | t002 | 5 | 5 | II | 52 |
| BM18 | 1989 | United States | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | B2 | t002 | 5 | 5 | IV | 22, 51 |
| N315 | 1982 | Japan | 2 | 3 | 1 | 3 | 10 | 3 | 1 | 5 | G2 | Q2 | t002 | 5 | 5 | II | 41 |
| 47MEXU | 1997 | Mexico | 2 | 3 | 1 | 3 | 8 | 3 | 1 | 6 | G2 | A3 | t895 | 5 | 5 | II | 63 |
| COB3 | 1996 | Colombia | 2 | 3 | 1 | 3 | 7 | 3 | 1 | 7 | G2 | B3 | t045 | 5 | 5 | IV | 31 |
| HU245 | 2001 | Hungary | 2 | 3 | 1 | 3 | 10 | 5 | 1 | 8 | G2 | A1 | t002 | 5 | 5 | II | 17 |
| HU317 | 2003 | Hungary | 2 | 3 | 1 | 3 | 6 | 3 | 1 | 9 | G2 | T5 | t062 | 5 | 5 | II | 17 |
| HU363 | 2004 | Hungary | 2 | 3 | 1 | 3 | 6 | 3 | 1 | 9 | G2 | T6 | t062 | 5 | 5 | II | 17 |
| JP1 | 1997 | Japan | 2 | 3 | 1 | 3 | 10 | 1 | 1 | 10 | G2 | M1 | t002 | 5 | 5 | II | 8 |
| JP26 | 1997 | Japan | 2 | 3 | 1 | 2 | 10 | 2 | 1 | 11 | G2 | ZF | t002 | 5 | 5 | II | 7, 8 |
| PL72 | 1991 | Poland | 2 | 3 | 1 | 3 | 10 | 7 | 1 | 12 | G2 | P2 | t053 | 5 | 5 | IV | 42 |
| POL3 | 1992 | Poland | 2 | 3 | 1 | 3 | 10 | 8 | 1 | 13 | G2 | B4 | t053 | 5 | 5 | IV | 42 |
| ARG33 | 1996 | Argentina | 2 | 3 | 1 | 3 | 10 | 99 | 1 | 14 | G2 | Q1 | t001 | 85 | 5 | IIIA | 7, 9, 18 |
| ARG64 | 1996 | Argentina | 2 | 3 | 1 | 3 | 10 | 99 | 1 | 14 | G2 | S1 | t001 | 85 | 5 | I | 7, 9, 18 |
| HAR41 | 1998 | Germany | 2 | 3 | 1 | 3 | 10 | 2 | 1 | 15 | G2 | I2 | t001 | 228 | 228 | I | 47 |
| HU2 | 1996 | Hungary | 3 | 0 | 1 | 3 | 12 | 2 | 2 | 16 | G3 | D2 | t989 | 239 | 8 | III | 23 |
| HUSA304 | 1993 | United States | 3 | 0 | 1 | 3 | 12 | 2 | 2 | 16 | G3 | D1 | t1053 | 239 | 8 | III | 23 |
| HU270 | 2002 | Hungary | 3 | 0 | 1 | 2 | 6 | 2 | 2 | 17 | G3 | E4 | t030 | 239 | 8 | III | 17 |
| HU294 | 2003 | Hungary | 3 | 0 | 1 | 3 | 13 | 2 | 2 | 18 | G3 | D5 | t538 | 239 | 8 | IIIA | 17 |
| TUR1 | 1996 | Turkey | 3 | 0 | 1 | 3 | 6 | 2 | 2 | 19 | G3 | E7 | t030 | 239 | 8 | III | 7 |
| TUR27 | 1996 | Turkey | 3 | 0 | 1 | 3 | 6 | 2 | 2 | 19 | G3 | E8 | t030 | 239 | 8 | III | 7 |
| PL46 | 1995 | Poland | 4 | 0 | 1 | 2 | 6 | 2 | 2 | 20 | G3 | E6 | t030 | 157 | S1 | III | 42 |
| CPS22 | 1985 | Portugal | 4 | 0 | 1 | 3 | 6 | 1 | 2 | 21 | G4 | E1 | t421 | 239 | 8 | IIIvar | 21 |
| CPS68 | 1985 | Portugal | 4 | 0 | 1 | 3 | 6 | 1 | 2 | 21 | G4 | E3 | t421 | 239 | 8 | IIIvar | 21 |
| HSA10 | 1992 | Portugal | 4 | 0 | 1 | 3 | 6 | 1 | 2 | 21 | G4 | E2 | t421 | 239 | 8 | IIIvar | 7 |
| GRE108 | 1998 | Greece | 4 | 0 | 1 | 3 | 7 | 2 | 2 | 22 | G4 | C5 | t461 | 239 | 8 | III | 4, 7 |
| HGSA142 | 2003 | Portugal | 4 | 0 | 1 | 3 | 7 | 0 | 2 | 23 | G4 | D3 | t037 | 239 | 8 | IIIA | 13 |
| HGSA339 | 2003 | Portugal | 4 | 0 | 1 | 3 | 7 | 0 | 2 | 23 | G4 | C3 | t037 | 239 | 8 | IIIA | 13 |
| HGSA57 | 1995 | Portugal | 4 | 0 | 1 | 3 | 7 | 0 | 2 | 23 | G4 | C6 | t037 | 239 | 8 | IIIA | 12 |
| HSJ216 | 1997 | Portugal | 4 | 0 | 1 | 3 | 7 | 99 | 2 | 24 | G4 | C1 | t037 | 239 | 8 | IIIA | 10 |
| HU25 | 1993 | Brazil | 4 | 0 | 1 | 3 | 6 | 0 | 2 | 25 | G4 | C1 | t138 | 239 | 8 | IIIA | 59 |
| HU272 | 2002 | Hungary | 4 | 0 | 1 | 3 | 10 | 2 | 2 | 26 | G4 | R2 | t787 | 239 | 8 | III | 17 |
| TAW166 | 1998 | Taiwan | 4 | 0 | 1 | 3 | 10 | 8 | 2 | 27 | G4 | N2 | t036 | 254 | 8 | IV | 6, 7 |
| TAW97 | 1998 | Taiwan | 4 | 0 | 1 | 3 | 7 | 3 | 2 | 28 | G4 | C2 | t037 | 239 | 8 | IIIA | 6, 7 |
| BK1953 | 1995 | United States | 4 | 0 | 1 | 4 | 11 | 4 | 2 | 29 | G5 | F1 | t051 | 247 | 8 | IA | 52 |
| HGSA13 | 1998 | Portugal | 4 | 0 | 1 | 4 | 11 | 4 | 2 | 29 | G5 | N1 | t051 | 247 | 8 | IA | 12 |
| HUR97 | 1998 | Hungary | 4 | 0 | 1 | 4 | 11 | 4 | 2 | 29 | G5 | F8 | t051 | 247 | 8 | IA | 50 |
| COL | 1965 | United Kingdom | 4 | 0 | 1 | 4 | 10 | 6 | 2 | 30 | G5 | F6 | t008 | 250 | 8 | I | 51 |
| E2213 | 1965 | Denmark | 4 | 0 | 1 | 4 | 11 | 6 | 2 | 31 | G5 | F4 | t051 | 247 | 8 | I | 19 |
| E2453 | 1965 | Denmark | 4 | 0 | 1 | 4 | 11 | 6 | 2 | 31 | G5 | F2 | t051 | 247 | 8 | I | 19 |
| HPV107 | 1992 | Portugal | 4 | 0 | 1 | 3 | 11 | 4 | 2 | 32 | G5 | F1 | t051 | 247 | 8 | IA | 55 |
| GRE18 | 1998 | Greece | 4 | 0 | 99 | 2 | 7 | 2 | 2 | 33 | G6 | F5 | t037 | 239 | 8 | III | 7 |
| HGSA15 | 1994 | Portugal | 4 | 0 | 99 | 2 | 7 | 99 | 2 | 34 | G6 | C4 | t037 | 239 | 8 | IIIA | 12 |
| HAR22 | 1991 | United Kingdom | 2 | 3 | 0 | 3 | 15 | 12g | 2 | 35 | G7 | J1 | t022 | 22 | 22 | IV | 47 |
| HGSA128 | 2000 | Portugal | 2 | 3 | 0 | 3 | 16 | 12g | 2 | 36 | G7 | J2 | t032 | 79 | 22 | IV | 7, 12 |
| HU303 | 2003 | Portugal | 2 | 3 | 0 | 3 | 16 | 12g | 2 | 36 | G7 | J3 | t032 | 22 | 22 | IV | 17 |
| IPOP2 | 2001 | Portugal | 2 | 3 | 99 | 3 | 16 | 12g | 2 | 37 | G7 | J1 | t032 | 22 | 22 | IV | 7 |
| DEN4415 | 2001 | Denmark | 2 | 1 | 2 | 2 | 9 | 3 | 2 | 38 | G8 | K1 | t021 | 36 | 30 | II | 26 |
| HAR24 | 1993 | United Kingdom | 2 | 1 | 2 | 2 | 11 | 2 | 2 | 39 | G8 | K1 | t018 | 36 | 30 | II | 47 |
| HGSA202 | 2003 | Portugal | 2 | 1 | 2 | 2 | 11 | 3 | 2 | 40 | G8 | K3 | t018 | 30 | 30 | MSSA | 13 |
| DEN4358 | 2001 | Denmark | 0 | 0 | 0 | 3 | 10 | 1g | 2 | 41 | G9 | H8 | t116 | 45 | 45 | V | 26 |
| PLN49 | 1997 | Poland | 0 | 0 | 0 | 3 | 10 | 1g | 2 | 41 | G9 | H9 | t015 | 45 | 45 | IV | 7, 42 |
| CA04 | 1998 | United States | 0 | 0 | 1 | 3 | 8 | 1g | 2 | 42 | G9 | H6 | t124 | 45 | 45 | IV | 20 |
| CV81 | 1997 | Cape Verde | 0 | 0 | 1 | 3 | 11 | 1g | 2 | 43 | G9 | H3 | t861 | 508 | 45 | MSSA | 11 |
| HAR38 | 1996 | Germany | 0 | 0 | 1 | 3 | 9 | 1g | 2 | 44 | G9 | H1 | t004 | 45 | 45 | IV | 47 |
| HU281 | 2002 | Hungary | 0 | 0 | 1 | 3 | 9 | 1g | 2 | 44 | G9 | H5 | t038 | 45 | 45 | IV | 17 |
| HSA19 | 1992 | Portugal | 0 | 0 | 1 | 3 | 7 | 1g | 2 | 45 | G9 | H4 | t1072 | 45 | 45 | MSSA | 5 |
| IPO516 | 2006 | Portugal | 0 | 0 | 1 | 3 | 10 | 1g | 2 | 46 | G9 | H7 | t2429 | 45 | 45 | V | 2 |
| IPOP56 | 2001 | Portugal | 0 | 0 | 1 | 3 | 10 | 1g | 2 | 46 | G9 | H2 | t1538 | 45 | 45 | MSSA | 5 |
| HU109 | 1996 | Hungary | 1 | 0 | 1 | 3 | 8 | 1 | 2 | 47 | G9 | E5 | t984 | 239 | 8 | III | 50 |
| GRE14 | 1998 | Greece | 4 | 2 | 6 | 3 | 7 | 1g | 1 | 48 | G10 | L1 | t044 | 80 | 80 | IV | 4, 7 |
| HFF189 | 2005 | Portugal | 4 | 2 | 6 | 3 | 7 | 1g | 1 | 48 | G10 | L2 | t044 | 80 | 80 | IV | This study |
| CHL5 | 1997 | Chile | 2 | 0 | 1 | 3 | 3 | 2 | 1 | 49 | G11 | I1 | t535 | 83 | 5 | I | 7, 9 |
| JP82 | 1997 | Japan | 2 | 0 | 1 | 3 | 8 | 2g | 1 | 50 | G11 | G2 | t375 | 92 | 509 | IVA | 7, 8 |
| COB111 | 1998 | Colombia | 2 | 1 | 2 | 3 | 7 | 3 | 1 | 51 | G12 | G1 | t1572 | 84 | 509 | IV | 7, 31 |
| HFF202 | 2005 | Portugal | 2 | 1 | 2 | 3 | 7 | 2 | 1 | 52 | G12 | X | t1537 | 707 | S4 | MSSA | This study |
| MW2 | 1998 | United States | 1 | 2 | 2 | 3 | 8 | 5g | 3 | 53 | S1 | T3 | t128 | 1 | 1 | IV | 14 |
| IPOP58 | 2001 | Portugal | 1 | 2 | 3 | 3 | 6 | 2 | 3 | 54 | S2 | U1 | t189 | 188 | 1 | MSSA | 5 |
| HSJ109 | 1995 | Portugal | 4 | 2 | 4 | 2 | 11 | 9 | 3 | 55 | S3 | ZC | t1897 | 573 | 1 | MSSA | 5 |
| HDE1 | 1996 | Portugal | 2 | 3 | 99 | 3 | 9 | 12 | 1 | 56 | S4 | B1 | t311 | 5 | 5 | IV | 54 |
| HBA3 | 2006 | Portugal | 2 | 3 | 1 | 2 | 3 | 3 | 1 | 57 | S5 | A1 | t535 | 5 | 5 | IV | 2 |
| HDE288 | 1996 | Portugal | 2 | 3 | 1 | 3 | 9 | 11 | 1 | 58 | S6 | B1 | t311 | 5 | 5 | VI | 54 |
| NCTC8325 | 1943 | United Kingdom | 3 | 0 | 1 | 4 | 10 | 6 | 3 | 59 | S7 | F3 | t211 | 8 | 8 | MSSA | http://www.nctc.org.uk |
| ARG199 | 1996 | Argentina | 4 | 99 | 7 | 3 | 11 | 9 | 1 | 60 | S8 | D4 | t148 | 86 | 8 | II | 7, 9, 18 |
| GRE4 | 1998 | Greece | 99 | 0 | 99 | 99 | 7 | 3 | 2 | 61 | S9 | R1 | t037 | 239 | 8 | IIIA | 7 |
| HAR36 | 1993 | Germany | 4 | 0 | 1 | 3 | 13 | 6 | 2 | 62 | S10 | F7 | t009 | 254 | 8 | IV | 47 |
| DEN2946 | 2001 | Denmark | 2 | 1 | 1 | 2 | 7 | 5 | 2 | 63 | S11 | K4 | t975 | 30 | 30 | IV | 26 |
| CV11 | 1997 | Cape Verde | 2 | 1 | 2 | 2 | 7 | 4 | 2 | 64 | S12 | K2 | t942 | 30 | 30 | MSSA | 11 |
| HGSA256 | 2003 | Portugal | 2 | 1 | 2 | 2 | 12 | 1 | 1 | 65 | S13 | K5 | t166 | 34 | 30 | MSSA | 13 |
| IPOP24 | 2001 | Portugal | 2 | 1 | 2 | 2 | 5 | 3 | 1 | 66 | S14 | K6 | t1076 | 34 | 30 | MSSA | 5 |
| E1114 | 1960 | Denmark | 0 | 1 | 1 | 3 | 8 | 2 | 1 | 67 | S15 | W | t518 | 50 | 50 | MSSA | 32 |
| TAW214 | 1998 | Taiwan | 2 | 1 | 4 | 2 | 7 | 11g | 4 | 68 | S16 | ZG | t437 | 59 | 59 | IV | 6, 7 |
| IPOP50 | 2001 | Portugal | 2 | 2 | 1 | 3 | 9 | 1g | 3 | 69 | S17 | T4 | t359 | 97 | 97 | MSSA | 5 |
| IPOP51 | 2001 | Portugal | 2 | 4 | 2 | 2 | 9 | 99 | 2 | 70 | S18 | U2 | t1075 | 106 | 101 | MSSA | 5 |
| CV55 | 1997 | Cape Verde | 3 | 1 | 1 | 3 | 8 | 2 | 2 | 71 | S19 | ZB | t937 | 398 | 398 | MSSA | 11 |
| JP87 | 1997 | Japan | 2 | 1 | 1 | 3 | 8 | 9g | 1 | 72 | S20 | V | t375 | 89 | 509 | IIvar | 7, 8 |
| E260 | 1957 | Denmark | 2 | 1 | 1 | 0 | 10 | 2 | 3 | 73 | S21 | ZE | t1194 | 446 | 1021 | MSSA | 32 |
| E3373 | 1967 | Denmark | 3 | 1 | 4 | 3 | 8 | 10 | 2 | 74 | S22 | F10 | t164 | 447 | S2 | MSSA | 32 |
| CV87 | 1997 | Cape Verde | 3 | 2 | 2 | 2 | 99 | 9 | 4 | 75 | S23 | S2 | t941 | 668 | S3 | MSSA | 11 |
| DCC1185 | 1997 | Portugal | 2 | 2 | 2 | 3 | 7 | 2 | 2 | 76 | S24 | ZA | t1065 | 580 | S5 | MSSA | 5 |
| E216 | 1957 | Denmark | 1 | 2 | 7 | 2 | 11 | 11 | 1 | 77 | S25 | ZH | t1191 | 445 | S6 | MSSA | 32 |
| E691 | 1959 | Denmark | 2 | 1 | 1 | 4 | 13 | 2 | 2 | 78 | S26 | ZD | t1207 | 49 | ND1 | MSSA | 32 |
| DCC300 | 1996 | Portugal | 2 | 1 | 1 | 2 | 12 | 2 | 1 | 79 | S27 | Y | t166 | 10 | ND2 | MSSA | 5 |
| DEN2230 | 2001 | Denmark | 3 | 0 | 2 | 3 | 10 | 2 | 2 | 80 | S28 | Z | t355 | 152 | ND3 | V | 26 |
| CV161 | 1997 | Cape Verde | 3 | 4 | 2 | 3 | 9 | 1g | 3 | 81 | S29 | F9 | t359 | 669 | ND4 | MSSA | 11 |
0, the size of the amplicon obtained was shorter than that of a complete repeat; 99, no amplification was obtained.
Numerical nomenclature of SIRU profiles. New numbers are attributed to profiles that differ in at least one allele.
S, singletons, i.e., SIRU profiles that appear once and have no related profiles.
PFGE types and subtypes were determined using thresholds of 80% and 98% similarity, respectively.
CC assignments were assessed by eBURST v3 on 9 June 2008. S, singletons; ND, not determined. The sequence types are included in groups for which no founder could be determined.
Strains that do not have SCCmec are labeled as MSSA strains.
SIRU05 amplification using the SIRU05R2 reverse primer.
Strains N315, NCTC8325, COL, and MW2 were included for reproducibility and methodology control, since their genomes are fully sequenced and were used in the theoretical design of the SIRU method (38).
PFGE analysis.
PFGE patterns were analyzed in BioNumerics, version 4.61, software (Applied Maths, Sint-Martens-Latem, Belgium) as previously described (25), with minor modifications, including an optimization setting of 1.0% for band pattern comparisons and a 98% Dice coefficient similarity cutoff for PFGE subtypes.
spa typing and MLST analysis.
spa types were assigned through the Ridom web server (http://spaserver.ridom.de). Additionally, for one isolate previously characterized as nontypeable, the spa type was determined through sequencing of SIRU21 (see below). MLST alleles and STs were identified through the MLST database (http://www.mlst.net), and CCs were defined using the eBURST v3 algorithm (http://eburst.mlst.net).
SIRU typing.
DNA was extracted as previously reported (3). The SIRU method was performed as previously described (36, 38), with the following minor modifications: (i) 0.5 U of AmpliTaq DNA polymerase (Applied Biosystems, CA) was used per PCR in a final volume of 25 μl, (ii) an annealing temperature of 59°C was used with primer SIRU05R2 (see below), and (iii) PCR products (10 μl) were resolved in a 2.5% Seakem LE (Cambrex, Rockland, ME) agarose gel in 0.5% Tris-borate-EDTA buffer (Bio-Rad, Hercules, CA) at 5 V/cm for 2.5 h. The size of each amplicon was determined by visual inspection by comparison with a 50-bp ladder size marker and by computer analysis using Bionumerics, version 4.61, software, which facilitates marker-based normalization of the migration distances and therefore guarantees accurate length measurements. The number of repeats was calculated, taking into account the combined size of the repeat unit and the flanking regions of each locus, as follows: (i) for SIRU01, repeat unit of 55 bp + flanking regions of 184 bp = 239 bp for one-repeat-length amplicon; (ii) for SIRU05, 60 bp + 146 bp = 206 bp; for SIRU05 with primer SIRU05R2, 60 bp + 156 bp = 216 bp; (iii) for SIRU07, 56 bp + 191 bp = 247 bp; (iv) for SIRU13, 64 bp + 148 bp = 212 bp; (v) for SIRU15 131 bp + 212 bp = 343 bp; (vi) for SIRU16, 159 bp + 162 bp = 321 bp; and (vii) for SIRU21, 24 bp + 96 bp = 120 bp.
The primers used to amplify each of the seven loci were previously published (36), except for those for locus 16 (SIRU16) and an additional reverse primer for locus 5 (SIRU05). New SIRU16 primers, SIRU16_2F (5′-TGGTGTTAATTTAGCTTGC-3′) and SIRU16_2R (5′-AAACGCAACTTGAAGAAACG-3′), were designed through sequence alignments of the SIRU16 loci of the seven S. aureus genomes previously considered for the design of the primers for the remaining loci (38). The new SIRU05 locus reverse primer was designed specifically for strains for which there was no amplification with the previously published primers (SIRU05L and SIRU05R), namely, for strains belonging to STs 1, 22, 45, and 80. Therefore, primer SIRU05R2 (5′-AGTTGTAGTCATCTTACTGC-3′) was designed through sequence alignments of the available SIRU05 loci of MW2, MSSA476 (both ST1), and EMRSA-15 (ST22) (sequence from the EMRSA-15 genome sequencing project at the Wellcome Trust Sanger Institute [http://www.sanger.ac.uk/sequencing/Staphylococcus/aureus/EMRSA15]). All sequence alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), with default parameters.
eBURST v3 software was used to cluster SIRU profiles. Isolates sharing six of seven loci with at least one isolate (single-locus variants) were included in the same SIRU group. Singletons represent profiles that appear once and have no related profiles in the collection.
Comparison of typing methods.
SID and the respective confidence intervals were calculated as described previously (33, 57). The quantitative level of congruence between typing methods was assessed by a framework proposed by Carriço et al. (15), based on the adjusted Rand (AR) and Wallace (W) coefficients, available at http://www.comparingpartitions.info/. The AR coefficient quantifies the global agreement between two methods, whereas the W coefficient indicates the probability that two isolates classified as the same type by one method are also classified as the same type by another method (15).
RESULTS
SIRU typeability.
In a first approach, we tested the method with four completely sequenced strains (N315 [ST5-SCCmec II], NCTC8325 [ST8-MSSA], COL [ST250-SCCmec I], and MW2 [ST1-SCCmec IV]) for which theoretical SIRU profiles had previously been published (36, 38). Five of the seven loci were amplified from the four tested strains; SIRU16 was amplified from strain MW2 only, whereas SIRU05 was amplified from all strains except MW2. A BLAST search showed that the published SIRU16 forward primer (SIRU16_L) (36) has similarity with strain MW2 but not with the three remaining tested strains. The SIRU16_L primer was found to have similarity with strain MSSA476, which belongs to ST1, like MW2, and with strain MRSA252 (ST36-SCCmec II). The primer was tested on additional ST36 isolates, and it performed well. These preliminary results led us to design a new SIRU16 primer that anneals with all available genomes of S. aureus strains (see Materials and Methods). The SIRU typing method using the new SIRU16 primers was applied to the whole collection. A seven-digit profile was obtained for only 70 of 104 strains, indicating a typeability of 67% when isolates with at least one nonamplified locus were considered nontypeable.
Considering the typeability of each locus separately, SIRU07 was the unique locus that showed 100% typeability. SIRU05 showed the lowest typeability (72%), followed by SIRU15 (95%), while the remaining SIRUs showed a typeability of 99% (one isolate was nontypeable). The consistent nonamplification of SIRU05 from all strains belonging to particular CCs, i.e., CC22, CC45, and CC80, led us to perform a BLAST search with different S. aureus genomes which showed that the published SIRU05 reverse primer (SIRU05_R) (36) has no similarity with strains MW2, MSSA476, EMRSA-15 (ST22), and RF122 (ST151). Therefore, a new SIRU05R2 reverse primer was designed considering these particular strains (see Materials and Methods), and a new PCR was performed on all isolates for which this locus was not amplified with the previously published SIRU05R primer.
The sequential use of the SIRU05R2 reverse primer increased the typeability from 67% to 89.4%. Moreover, the typeability of the SIRU05 locus alone rose to 95%. Among the 11 nontypeable isolates, 9 isolates showed no amplification for a single locus, 1 isolate showed no amplification for two loci (SIRU05 and SIRU15), and another isolate showed no amplification for three loci (SIRUs 1, 15, and 16).
For further evaluation of the method, an arbitrarily chosen neutral number of repeats (99) was attributed to all nonamplified loci. However, this artifact creates a limitation because two isolates showing no amplification at the same locus are considered identical in this allele.
SIRU locus discriminatory power.
Considering the discriminatory power of each locus, SIRU07 showed four different allele numbers, SIRU16 showed five, SIRU01 and SIRU13 showed six, and SIRU15 showed eight. Loci 5 and 21 showed the highest variability among the seven loci, with 14 and 13 different allele numbers, respectively. The SIRU21 locus, the only one located in a coding region, the spa gene, had a direct correlation with the spa type. However, an identical number of repeat units may contain sequence variations which are not detected by SIRU typing.
SIRU typing clonal assignment.
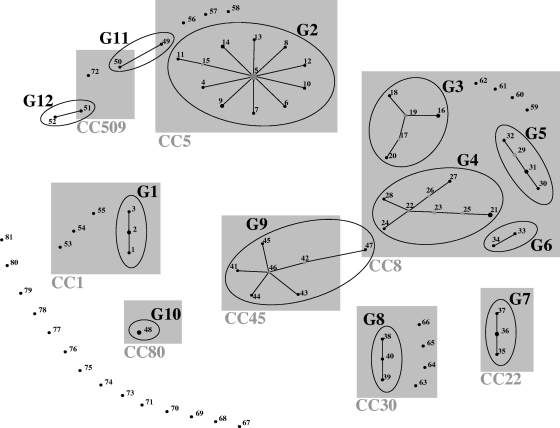
The 104 strains were distributed into 81 SIRU profiles (differing in at least one of the seven loci) that could be clustered into 12 groups and 29 singletons (Table 1). Figure 1 shows the assignment of groups when the eBURST v3 algorithm was applied to the collection. The largest SIRU group, G2 (n = 20), included the majority (19 of 23) of the strains belonging to CC5 (ST5, ST85, and ST100) and the related strain CC228-ST228. SIRU group G9 included the nine strains belonging to CC45 (ST45 and ST508). Similarly, G7 included the four CC22 strains (ST22 and ST79), and G10 included the two CC80-ST80 strains. SIRU group G8 included only three (ST30 and ST36) of the seven CC30 strains. The remaining four strains (ST30 and ST34) were classified as singletons. CC8 strains were divided into four SIRU groups (G3, G4, G5, and G6) and four singletons. Curiously, ST247 and ST250 strains were found in G5 only, while ST239 strains were distributed over G3, G4, and G6 but not in G5. SIRU group G1 included four of the seven CC1 strains.
FIG. 1.
Schematic representation of the SIRU groups identified by eBURST v3 analysis. The size of the dots is proportional to the number of isolates of each SIRU profile. Single-locus variants are linked by lines; CCs are highlighted in gray.
Unexpectedly, two isolates which were totally different by all other typing methods were grouped in the same SIRU group, G12. In the same way, one CC8 strain appeared to be a single-locus variant of a CC45 strain and was therefore included in the same G9 SIRU group, and a single CC5 strain was clustered with a CC509 strain in G11.
Three of the 12 SIRU groups included a single PFGE type, as follows: G7, PFGE J; G8, PFGE K; and G10, PFGE L. Eleven of the 13 singletons defined by PFGE were also defined as singletons by SIRU typing.
Concerning spa typing, all SIRU groups contained related spa types, except for (i) one G4 strain (TAW166, t036), (ii) one G9 strain (HU109, t984), (iii) two G11 strains (CHL5, t535; and JP82, t375), and (iv) the two G12 strains. Moreover, each spa type was associated with a single SIRU group, with the exception of t037, found in SIRU groups G4 and G6 belonging to the same CC, and t375, found in a G11 isolate and in singleton S20 (Table 1). Among the 11 different SCCmec types that characterized the MRSA collection (n = 78), with a few exceptions (Table 1), each SIRU profile was associated with a single SCCmec type.
Comparison of the discriminatory power of SIRU typing with that of other typing methods.
The SID values obtained for the different typing methods are presented in Table 2. Considering the 104 isolates, the SIRU method showed a very high discriminatory power at the profile level (SID = 99.23%), similar to that observed for PFGE at the subtype level (SID = 99.85%) and higher than that observed for spa typing, considering both the length and sequence variation (SID = 97.61%). Considering discrimination of the SIRU method at the group level (SID = 93.24%), it was higher than that of MLST (SID = 91.77%) but lower than that of PFGE at the type level (SID = 95.41%). However, since the confidence intervals of the methods overlap (Table 2), we cannot exclude the hypothesis that they have similar discriminatory powers at a 95% confidence level.
TABLE 2.
Number of types identified and SID for each typing method for the 104 S. aureus isolates
| Typing method | No. of types identified | SID (%) | 95% Confidence interval (%) |
|---|---|---|---|
| PFGE subtyping | 97 | 99.85 | 99.69-100.00 |
| SIRU profile typing | 81 | 99.23 | 98.66-99.81 |
| spa typing | 61 | 97.61 | 96.36-98.86 |
| PFGE typing | 33 | 95.41 | 94.11-96.70 |
| SIRU group typing | 41 | 93.24 | 90.62-95.87 |
| MLST | |||
| ST | 42 | 91.77 | 88.52-95.01 |
| CC | 25 | 84.37 | 79.68-89.06 |
Clustering concordance and directional agreement between SIRU typing and other typing methods.
The clustering concordance between SIRU typing and the remaining methods (PFGE, spa typing, MLST, and SCCmec typing) could be traced based on the calculation of AR coefficients for the whole collection (Table 3). The AR values obtained for the SIRU typing method indicated that the highest level of congruence was at the group level for the ST (ARSIRU group-ST = 0.5736), followed by the CC (ARSIRU group-CC = 0.4963). At the profile level, the highest congruence was with spa type (ARSIRU profile-spa type = 0.4313). Congruence between PFGE (type and subtype levels) and SIRU typing was shown to be particularly low (ARSIRU profile-PFGE type = 0.1067; ARSIRU profile-PFGE subtype = −0.0025).
TABLE 3.
AR and W values for the entire collection (n = 104)
| Typing method | AR coefficient
|
W coefficient
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SIRU group | SIRU profile | SIRU group | SIRU profile | PFGE type | PFGE subtype | spa type | MLST—ST | MLST—CC | |
| SIRU group typing | 0.1914 | 0.1113 | 0.2983 | 0.0138 | 0.2928 | 0.6713 | 0.9006 | ||
| SIRU profile typing | 0.1914 | 1.0000 | 0.4146 | 0.0000 | 0.9024 | 0.8293 | 1.0000 | ||
| PFGE typing | 0.3180 | 0.1067 | 0.4390 | 0.0691 | 0.0325 | 0.1870 | 0.4959 | 0.7724 | |
| PFGE subtyping | 0.0242 | −0.0025 | 0.6250 | 0.0000 | 1.0000 | 0.3750 | 1.0000 | 1.0000 | |
| spa typing | 0.4119 | 0.4313 | 0.8281 | 0.2891 | 0.3594 | 0.0234 | 0.8047 | 0.9453 | |
| MLST | |||||||||
| ST | 0.5736 | 0.1289 | 0.5510 | 0.0771 | 0.2766 | 0.0181 | 0.2336 | 1.0000 | |
| CC | 0.4963 | 0.0800 | 0.3895 | 0.0490 | 0.2270 | 0.0096 | 0.1446 | 0.5269 | |
The W coefficient was calculated to determine the capacity of SIRU typing to predict the classification achieved by other methods (Table 3). The results obtained showed that in this collection, two strains with the same SIRU profile have a 100% probability of belonging to the same CC, a 90% probability of sharing the same spa type, and an 83% probability of being classified in the same ST. The capacity of SIRU typing at the profile level to predict the PFGE type was low (WSIRU profile-PFGE type = 0.4146), and there was no correlation with the PFGE subtype (WSIRU profile-PFGE subtype = 0). On the other hand, the SIRU group showed a high correlation with the CC (WSIRU group-CC = 0.9006).
DISCUSSION
In the present study, we evaluated the performance of the SIRU typing method applied to a diverse collection of S. aureus isolates previously characterized by well-established typing methods. The SIRU method's typeability, i.e., the method's ability to assign a type to all isolates tested, was 89.4% when new primers designed for SIRU05 and SIRU16 locus amplification were used. The consistent nonamplification of SIRU05 from all strains belonging to ST1, CC22, CC45, and CC80 observed with our collection when we used the previously published SIRU05 primers is in agreement not only with previous studies focusing on SIRU typing of EMRSA-15 (CC22) isolates (29, 35-37) but also with the work of Ikawaty et al., who reported a reduced typeability for this specific locus on CC1, CC5, CC8, CC97, and CC228 isolates as well (39).
The ability of SIRU typing to assign a different type to two unrelated strains randomly sampled from the collection, i.e., its discriminatory power, was found to be very high (>99%) and was due mainly to the individual high levels of variability of SIRU05 and SIRU21 loci. High variability in SIRU21 is not surprising, since it is located in the known highly variable polymorphic region of the spa gene. The discriminatory power of SIRU typing at the profile level (SID = 99.23%) is similar to that of PFGE subtyping (SID = 99.85%) and spa typing (SID = 97.61%), considering the overlapping of the confidence intervals at 95%. Our results are concordant with a recent study, besides the fact that only six of the seven SIRU loci were taken into account (39). The observed high discriminatory power makes SIRU typing suitable for outbreak investigations, as also shown in studies by Hardy et al. where strains belonging to an outbreak or consecutively isolated from the same ward had the same or highly related SIRU profiles (35, 37). Moreover, in a study involving seven different outbreaks, variations in the number of SIRU repeats in strains belonging to the same outbreak were found to be rare (40).
The concordance between SIRU typing and well-established typing methods was measured by the calculation of the AR coefficient. We demonstrated that at the group level, SIRU typing showed the highest congruence with MLST (ST and CC), whereas at the profile level it showed the highest congruence with spa typing. The correspondence with PFGE was low, in contrast to the study of Ikawaty et al. on 50 S. aureus isolates, where the AR coefficient between the MLVA method and PFGE (AR = 0.599) was higher than that for spa typing (AR = 0.435) (39).
The major CCs defined by MLST (CC5, CC8, CC22, CC30, and CC45) were maintained when the eBURST v3 algorithm was applied to our SIRU profile data. The exception was CC8, known to include a high degree of variability in STs, which was divided into four well-distinguished groups. Moreover, a single CC8 strain was surprisingly clustered with CC45 isolates (G9). Considering that MLST is based on the variation in housekeeping genes that have a slow evolutionary clock and that the SIRU method looks into variable repeat regions that could evolve more rapidly by an introduction or deletion of a single repeat, clonal types might be affected by genomic rearrangements to different extents in the two methods. The SIRU loci in noncoding regions are less likely to be subject to natural selection that affects some VNTRs located on coding regions or promoters (43). Interestingly, among the seven SIRU loci, different evolutionary clocks could be observed, since SIRU01, SIRU07, SIRU13, SIRU15, and SIRU16 appeared to be generally monomorphic between strains from the same group and therefore more conserved during evolution. SIRU05 and SIRU21 add high levels of variability to the method and could be especially informative for recent levels of evolutionary divergence. Therefore, the SIRU typing method includes different scales of evolutionary divergence within the same system, making it suitable for studies with different purposes.
The predictive power between SIRU typing and other methods, translated by the W index, was maximum for CC and very high for spa typing and MLST in our collection. In opposition, the prediction of the PFGE results was low, as SIRU typing was able to distinguish among isolates with the same PFGE subtype and vice versa. Noller et al. (49) showed that during an investigation of an Escherichia coli O157:H7 outbreak, MLVA appeared to have a sensitivity equal to that of PFGE and a specificity that was even superior to that of PFGE. On another hand, SIRU typing showed the highest congruence with MLST results, i.e., ST and CC results. It is noteworthy that the predictive power of a method seems to be linked to the variability in the collection studied, as in a recent study involving two collections of S. aureus isolates, where for the first collection the MLVA method predicted the spa typing results but the reverse was not observed, while for the second collection both methods proved to be mutually predictive (39).
Our results show that SIRU typing analysis adds to the knowledge of the variability of the S. aureus genome and contributes to the understanding of genetic relationships among MRSA clones, as seen by Malachowa et al. (44).
In terms of convenience, the SIRU method was shown to be easy to perform, since it is based on single-locus PCRs, which could minimize the drawback of band size determination and even reduce the total time required for practical procedures if allied to the use of automated systems. Automated MLVA approaches were already proposed for S. aureus and Staphylococcus epidermidis genotyping through different PCR schemes (27, 28). In terms of cost, SIRU typing (11.2€/strain) is comparable to PFGE (13.2€/strain), but it is faster and technically easier to perform and much cheaper than spa typing (15.5€/strain) or MLST (80€/strain). Additionally, SIRU typing could be combined with spa typing, since the spa type may be determined by sequencing of the SIRU21 amplicons of nontypeable strains with conventional primers (1, 56).
In summary, as a PCR-based method, the SIRU typing method is relatively fast, accessible, and not expensive, which combined with its high discriminatory power makes it useful and reliable for short-term epidemiological investigations of S. aureus. In addition, its congruence with MLST results (at the CC and ST levels) makes it potentially valuable for evolutionary studies. In order for SIRU typing to be considered a useful tool in terms of epidemiological surveillance networks and evolutionary purposes, a public database similar to the databases available for other bacterial MLVA schemes (34) may be created for SIRU typing, allowing harmonization of S. aureus MLVA schemes and the interlaboratory exchange of data.
Acknowledgments
This work was supported by project POCTI/SAU-ESP/57841/2004 from Fundação para a Ciência e Tecnologia (FCT), Portugal, awarded to H. de Lencastre. T. Conceição was supported by grant SFRH/BD/21424/2005 from FCT.
We thank H. Westh for critical comments on the manuscript.
Footnotes
Published ahead of print on 4 March 2009.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires-de-Sousa, M., B. Correia, H. de Lencastre, et al. 2008. Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period.J. Clin. Microbiol. 462912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires-de-Sousa, M., C. E. Parente, O. Vieira-da-Motta, I. C. Bonna, D. A. Silva, and H. de Lencastre. 2007. Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Appl. Environ. Microbiol. 733845-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aires de Sousa, M., C. Bartzavali, I. Spiliopoulou, I. S. Sanches, M. I. Crisostomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 412027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aires de Sousa, M., T. Conceicao, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 435150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aires de Sousa, M., M. I. Crisostomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aires de Sousa, M., and H. de Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 413806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6253-258. [DOI] [PubMed] [Google Scholar]
- 9.Aires de Sousa, M., M. Miragaia, I. S. Sanches, S. Avila, I. Adamson, S. T. Casagrande, M. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America from 1996 to 1998. J. Clin. Microbiol. 392197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aires de Sousa, M., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 362590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aires De Sousa, M., I. Santos Sanches, M. L. Ferro, and H. De Lencastre. 2000. Epidemiological study of staphylococcal colonization and cross-infection in two West African hospitals. Microb. Drug Resist. 6133-141. [DOI] [PubMed] [Google Scholar]
- 12.Amorim, M. L., M. Aires de Sousa, I. S. Sanches, R. Sa-Leao, J. M. Cabeda, J. M. Amorim, and H. de Lencastre. 2002. Clonal and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus (MRSA) from a Portuguese hospital over time. Microb. Drug Resist 8301-309. [DOI] [PubMed] [Google Scholar]
- 13.Amorim, M. L., N. A. Faria, D. C. Oliveira, C. Vasconcelos, J. C. Cabeda, A. C. Mendes, E. Calado, A. P. Castro, M. H. Ramos, J. M. Amorim, and H. de Lencastre. 2007. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J. Clin. Microbiol. 452881-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 15.Carrico, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 442524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6189-198. [DOI] [PubMed] [Google Scholar]
- 17.Conceicao, T., M. Aires-de-Sousa, M. Fuzi, A. Toth, J. Paszti, E. Ungvari, W. B. van Leeuwen, A. van Belkum, H. Grundmann, and H. de Lencastre. 2007. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin. Microbiol. Infect. 13971-979. [DOI] [PubMed] [Google Scholar]
- 18.Corso, A., I. Santos Sanches, M. Aires de Sousa, A. Rossi, and H. de Lencastre. 1998. Spread of a methicillin-resistant and multiresistant epidemic clone of Staphylococcus aureus in Argentina. Microb. Drug Resist. 4277-288. [DOI] [PubMed] [Google Scholar]
- 19.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 989865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 1861344-1347. [DOI] [PubMed] [Google Scholar]
- 21.de Lencastre, H., I. Couto, I. Santos, J. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 1364-73. [DOI] [PubMed] [Google Scholar]
- 22.de Lencastre, H., A. de Lencastre, and A. Tomasz. 1996. Methicillin-resistant Staphylococcus aureus isolates recovered from a New York City hospital: analysis by molecular fingerprinting techniques. J. Clin. Microbiol. 342121-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lencastre, H., E. P. Severina, H. Milch, M. K. Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3289-296. [DOI] [PubMed] [Google Scholar]
- 24.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faria, N. A., J. A. Carrico, D. C. Oliveira, M. Ramirez, and H. de Lencastre. 2008. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 46136-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 431836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francois, P., A. Hochmann, A. Huyghe, E. J. Bonetti, G. Renzi, S. Harbarth, C. Klingenberg, D. Pittet, and J. Schrenzel. 2008. Rapid and high-throughput genotyping of Staphylococcus epidermidis isolates by automated multilocus variable-number of tandem repeats: a tool for real-time epidemiology. J. Microbiol. Methods 72296-305. [DOI] [PubMed] [Google Scholar]
- 28.Francois, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, B. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. van Leeuwen, A. van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 433346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghebremedhin, B., W. Konig, W. Witte, K. J. Hardy, P. M. Hawkey, and B. Konig. 2007. Subtyping of ST22-MRSA-IV (Barnim epidemic MRSA strain) at a university clinic in Germany from 2002 to 2005. J. Med. Microbiol. 56365-375. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert, F. B., A. Fromageau, L. Gelineau, and B. Poutrel. 2006. Differentiation of bovine Staphylococcus aureus isolates by use of polymorphic tandem repeat typing. Vet. Microbiol. 117297-303. [DOI] [PubMed] [Google Scholar]
- 31.Gomes, A. R., I. S. Sanches, M. Aires de Sousa, E. Castaneda, and H. de Lencastre. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Colombian hospitals: dominance of a single unique multidrug-resistant clone. Microb. Drug Resist. 723-32. [DOI] [PubMed] [Google Scholar]
- 32.Gomes, A. R., H. Westh, and H. de Lencastre. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 503237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 394190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guigon, G., J. Cheval, R. Cahuzac, and S. Brisse. 2008. MLVA-NET—a standardized web database for bacterial genotyping and surveillance. Euro Surveill. 13pii 18863. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=18863. [PubMed] [Google Scholar]
- 35.Hardy, K. J., S. Gossain, N. Henderson, C. Drugan, B. A. Oppenheim, F. Gao, and P. M. Hawkey. 2007. Rapid recontamination with MRSA of the environment of an intensive care unit after decontamination with hydrogen peroxide vapour. J. Hosp. Infect. 66360-368. [DOI] [PubMed] [Google Scholar]
- 36.Hardy, K. J., B. A. Oppenheim, S. Gossain, F. Gao, and P. M. Hawkey. 2006. Use of variations in staphylococcal interspersed repeat units for molecular typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 44271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy, K. J., A. Szczepura, R. Davies, A. Bradbury, N. Stallard, S. Gossain, P. Walley, and P. M. Hawkey. 2007. A study of the efficacy and cost-effectiveness of MRSA screening and monitoring on surgical wards using a new, rapid molecular test (EMMS). BMC Health Serv. Res. 7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardy, K. J., D. W. Ussery, B. A. Oppenheim, and P. M. Hawkey. 2004. Distribution and characterization of staphylococcal interspersed repeat units (SIRUs) and potential use for strain differentiation. Microbiology 1504045-4052. [DOI] [PubMed] [Google Scholar]
- 39.Ikawaty, R., R. J. Willems, A. T. Box, J. Verhoef, and A. C. Fluit. 2008. A novel multiple-locus variable-number tandem repeat (VNTR) analysis (MLVA) for rapid molecular typing of human Staphylococcus aureus. J. Clin. Microbiol. 463147-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kloos, W. E., and T. M. Bannerman. 1999. Staphylococcus and Micrococcus, p. 264-282. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, DC.
- 41.Kuwahara-Arai, K., N. Kondo, S. Hori, E. Tateda-Suzuki, and K. Hiramatsu. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob. Agents Chemother. 402680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leski, T., D. Oliveira, K. Trzcinski, I. S. Sanches, M. Aires de Sousa, W. Hryniewicz, and H. de Lencastre. 1998. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J. Clin. Microbiol. 363532-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 262567-2582. [DOI] [PubMed] [Google Scholar]
- 44.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 433095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 1141732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 513374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 411574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2902976-2984. [DOI] [PubMed] [Google Scholar]
- 49.Noller, A. C., M. C. McEllistrem, A. G. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 415389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira, D. C., I. Crisostomo, I. Santos-Sanches, P. Major, C. R. Alves, M. Aires-de-Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7349-361. [DOI] [PubMed] [Google Scholar]
- 52.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178164-171. [DOI] [PubMed] [Google Scholar]
- 53.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 411801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sa-Leao, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 371913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanches, I. S., M. Ramirez, H. Troni, M. Abecassis, M. Padua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J. Clin. Microbiol. 331243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 373556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simpson, E. H. 1949. Measurement of species diversity. Nature 163688. [Google Scholar]
- 58.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 22-11. [DOI] [PubMed] [Google Scholar]
- 59.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 332400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Belkum, A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 4922-27. [DOI] [PubMed] [Google Scholar]
- 61.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3)1-46. [DOI] [PubMed] [Google Scholar]
- 62.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Velazquez-Meza, M. E., M. Aires de Sousa, G. Echaniz-Aviles, F. Solorzano-Santos, G. Miranda-Novales, J. Silva-Sanchez, and H. de Lencastre. 2004. Surveillance of methicillin-resistant Staphylococcus aureus in a pediatric hospital in Mexico City during a 7-year period (1997 to 2003): clonal evolution and impact of infection control. J. Clin. Microbiol. 423877-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]