Abstract
Human intestinal spirochetosis (HIS) is associated with overgrowth of the large intestine by spirochetes of the genus Brachyspira. The microbiological diagnosis of HIS is hampered by the fastidious nature and slow growth of Brachyspira spp. In clinical practice, HIS is diagnosed histopathologically, and a significant portion of cases may be missed. Fluorescence in situ hybridization (FISH) is a molecular method that allows the visualization and identification of single bacteria within tissue sections. In this study, we analyzed intestinal biopsy samples from five patients with possible HIS. All specimens yielded positive results by histopathological techniques. PCR amplification and sequencing of the 16S rRNA gene were performed. Sequences of two isolates clustered in the group of Brachyspira aalborgi, whereas in three cases, the sequences were highly similar to that of Brachyspira pilosicoli. Three phylotypes showed mismatches at distinct nucleotide positions with Brachyspira sp. sequences published previously. In addition, culture for Brachyspira was successful in three cases. On the basis of these data, we designed and evaluated a Brachyspira genus-specific 16S rRNA-directed FISH probe that detects all of the Brachyspira spp. published to date. FISH of biopsy samples resulted in strong, unequivocal signals of brush-like formations at the crypt surfaces. This technique allowed simultaneous visualization of single spirochetes and their identification as Brachyspira spp. In conclusion, FISH provides a fast and accurate technique for the visualization and identification of intestinal spirochetes in tissue sections. It therefore represents a valuable tool for routine diagnosis of HIS.
Human intestinal spirochetosis (HIS) is a histologically defined condition of the human distal intestinal tract characterized by helical microorganisms attached at one end to the surface epithelium of the colonic mucosa. This association forms a so-called “false brush border” (13). While certain Brachyspira spp. are recognized as the causative agents of swine dysentery and porcine intestinal spirochetosis (3, 11), their clinical significance and pathogenic potential for humans remain unclear. Various studies have reported on the association of these bacteria with intestinal disorders such as chronic watery diarrhea (8, 12) and on clinical improvement following antimicrobial therapy (29). In contrast, others have suggested that intestinal spirochetes are harmless commensals in humans (7). The prevalence of HIS ranges from 1.2% (23) to >40% (17, 34), depending on presumable patient risk factors, such as origin from developing countries, immunodeficiency, or homosexuality. Recently, Peruzzi et al. (30) discovered a prevalence of 12% in a selected population, indicating that HIS is an important differential diagnosis for patients with chronic gastrointestinal disorders and risk factors.
Two intestinal spirochetes have been identified in humans so far: Brachyspira aalborgi (15) and Brachyspira pilosicoli (36). Both species require selective media, and B. aalborgi is an extremely slow growing, fastidious microorganism that requires anaerobic incubation for as long as 4 weeks (4, 34). For this reason, HIS is primarily diagnosed histopathologically. The fuzzy basophilic fringe, 4 to 7 μm thick, on the epithelial layer of the colonic mucosa is visible in hematoxylin-and-eosin (HE)-stained histological sections and is considered pathognomonic for HIS. Tissue morphology usually remains unaltered, and no inflammatory reaction is observed (20).
However, diagnosis of HIS on the basis of HE staining requires experienced laboratory personnel and accurate interpretation, and silver staining is often needed to confirm the diagnosis (10). Therefore, a significant portion of cases may be missed, especially since B. pilosicoli might also colonize the epithelium without the characteristic end-on attachment, impeding identification by light microscopy at low magnification (24). Furthermore, histopathology does not provide information about the identity of the microorganisms, thereby precluding epidemiological studies. More importantly, the inability to identify the organism also hampers accurate therapy, since the intestinal spirochetes are suspected to differ in virulence, and therefore some cases of HIS may require antibiotic therapy more urgently than others (5, 29).
The genus Brachyspira currently comprises seven established species and several proposed species. Among some Brachyspira species, the high level of 16S rRNA gene conservation precludes interspecies differentiation by 16S rRNA gene methods and necessitates further molecular analyses. However, all known species isolated from humans can be identified and differentiated via their 16S rRNA genes. In line with the genetic variation discovered in Brachyspira species, such as Brachyspira hyodysenteriae (2) and Brachyspira innocens (9), recent molecular studies have also identified human Brachyspira strains genetically distinct from B. aalborgi and B. pilosicoli. This heterogeneity was confirmed by sequencing of 16S rRNA (14, 21) or NADH oxidase (25) genes, fluorescence in situ hybridization (FISH) (16, 17), or multilocus enzyme electrophoresis (33). Pettersson et al. (31) analyzed biopsy samples from two adults by 16S rRNA gene sequencing and consequently proposed to divide the B. aalborgi lineage into three phylogenetic clusters, including the type strain, B. aalborgi 513A, in the first cluster.
The extent of intraspecies genetic variation in human intestinal spirochetes is unclear and difficult to estimate, because few complete 16S rRNA gene sequences are available. Further epidemiologic and phylogenetic investigations are needed to elucidate spirochete genetic diversity and to facilitate the evaluation of the molecular diagnostic tools that are presently available.
FISH is a microscopic method that allows simultaneous visualization and identification of microorganisms. Jensen and colleagues (3, 16, 17) designed several genus- or species-specific oligonucleotide probes targeting the 16S or 23S rRNA of Brachyspira spp. and applied them successfully to porcine and human intestinal biopsy specimens. However, no genus-specific 16S rRNA-directed probe for diagnostic use targeting all Brachyspira spp. known so far has been developed.
In the present study, intestinal biopsy specimens from five patients with possible HIS were analyzed histopathologically and by culture, FISH, PCR amplification, and 16S rRNA gene sequencing. Biopsy specimens from a healthy control group were analyzed retrospectively by histopathology and FISH. The purpose was (i) to acquire further information about the phylogenetic structure of the Brachyspira spp. associated with HIS, (ii) to design a FISH probe covering all Brachyspira spp. based on the currently available sequence data, and (iii) to evaluate FISH as a fast and robust diagnostic screening tool for HIS.
MATERIALS AND METHODS
Patients.
Five patients (HIS1 to HIS5), four men and one woman (ages, 37 to 84 years), were included in this study. These patients suffered from chronic intestinal disorders of unknown etiology. Intestinal biopsies were performed for further differential diagnosis including HIS (Table 1). The patients had been admitted to four different German hospitals: HIS1 to the University Hospital Hamburg-Eppendorf, HIS2 to the Königin Elisabeth Herzberge Hospital Berlin, HIS3 and HIS5 to the Charité University Hospital, Campus Benjamin Franklin, and HIS4 to the Hospital Augsburg. All patients underwent colonoscopies; four biopsy specimens were obtained from the colon, and one biopsy specimen each was obtained from the ileum and the sigmoid. Clinical development and response to treatment were followed up. Additional intestinal biopsy specimens were collected from patients HIS1, HIS3, and HIS5, who underwent control colonoscopies after the cessation of treatment. Risk factors for HIS (immunosuppression, homosexuality, and contact with animals) were assessed for all patients.
TABLE 1.
HIS patients diagnosed in this study by histopathology, FISH and 16S rRNA gene sequencing
| Patient | Gender | Age at time of diagnosis (yr) | Clinical feature(s) | Treatment, response | Biopsy specimen location | Result of:
|
Results of sequencing of the 16S rRNA genea | |
|---|---|---|---|---|---|---|---|---|
| Histopathological diagnosis | FISH with EUB338 and BRACHY | |||||||
| HIS1 | Female | 37 | Chronic diarrhea | Metronidazole, partial recovery | Colon | Positive | Positive | B. aalborgi, 99.8% homology with AF200693 (1,307 bp, 2 MM) |
| HIS2 | Male | 84 | Bloody stools, colitis | No treatment | Colon | Positive | Positive | B. aalborgi, 100% homology with AF200693 (1,414 bp, 0 MM) |
| HIS3 | Male | 49 | Chronic diarrhea, HIV infection | Metronidazole, complete recovery | Sigmoid | Positive | Positive | B. pilosicoli-like, 99.4% homology with U14927 (1,412 bp, 7 MM) |
| HIS4 | Male | 38 | Chronic diarrhea | Metronidazole, complete recovery | Ileum, colon | Positive | Positive | B. pilosicoli-like, 99.5% homology with U14927 (1,465 bp, 7 MM) |
| HIS5 | Male | 65 | Chronic diarrhea, HIV infection | Metronidazole, partial recovery | Colon | Positive | Positive | B. pilosicoli-like, 99.4% homology with U14927 (1,421 bp, 8 MM) |
Given as the organism identified, the percentage of homology with the closest match in EMBL/GenBank, the accession number of the closest match (the length of the sequence, the number of mismatches [MM]).
To provide negative controls for the biopsy specimens, seven biopsy samples from healthy patients (four females, 56 to 68 years old, and three males, 63 to 65 years old) screened for colorectal cancer at Charité University Hospital, Campus Benjamin Franklin, were included in the study. These samples were found to be normal by histopathology and to be HIS negative by FISH with EUB338 and BRACHY.
Culture.
Biopsy samples were plated onto brain heart infusion agar supplemented with 10% bovine blood, 400 μg spectinomycin ml−1, 25 μg colistin ml−1, and 12.5 μg rifampin (rifampicin) ml−1 (4). Plates were incubated at 37°C in anaerobic jars under an atmosphere of 80% N2-10% H2-10% CO2 for 6 weeks.
Histopathological diagnosis.
Intestinal biopsy samples were fixed in formalin, embedded in paraffin wax, sectioned (thickness, 4 μm), and stained with HE. The appearance of the typical hematoxyphilic fringe on the brush border of the surface epithelium under light microscopy, confirmed by Warthin-Starry silver staining, was classified as “HIS positive”.
Specimen processing for FISH.
Biopsy samples were fixed in 3.7% (vol/vol) formaldehyde in phosphate-buffered saline (pH 7.4) containing 50% (vol/vol) ethanol and were stored at 4°C for 24 h. The embedding procedure using cold polymerizing resin and the sectioning technique were performed as described elsewhere (26). For FISH, a prewarmed hybridization solution (20 μl) containing 0.9 M NaCl, 20 mM Tris HCl (pH 7.3), and 0.01% sodium dodecyl sulfate was mixed with 20 pmol of the respective oligonucleotide probe and carefully applied to the tissue sections. Probes were synthesized commercially and 5′ end labeled with a fluorochrome, either Cy3 (indocarbocyanine) or Cy5 (indodicarbocyanine) (both from Biomers, Ulm, Germany). After incubation in a dark humid chamber at 50°C for 2 h, slides were rinsed with sterile double-distilled water, air dried, and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) containing DAPI (4′,6-diamidino-2-phenylindole). For microscopy, an epifluorescence microscope (Axioplan 2; Carl Zeiss, Jena, Germany) equipped with narrow band filter sets (AHF Analysentechnik, Tübingen, Germany) was used.
Oligonucleotide probes.
The 16S rRNA-directed bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) (1) and the nonspecific nucleic acid stain DAPI were used to screen for bacterial colonization. To selectively identify intestinal spirochetes, we designed a 16S rRNA-directed FISH probe named BRACHY (5′-ATTAGTCCATGTTTCCAT-3′; corresponding to Escherichia coli positions 153 to 170) (6) that is specific for Brachyspira spp. The probe was evaluated by comparison to all available sequences in the EMBL and GenBank databases. A clinical isolate of B. pilosicoli and the nearest phylogenetic neighbors at the probe binding site, Enterococcus faecium (ATCC 19434) and Spirochaeta halophila (DSM 10522), with two mismatches each, were used as positive and negative controls, respectively, and were included throughout the study. Furthermore, the probe was tested against other cultivable spirochetes, i.e., Borrelia garinii (tick isolate; R. Ackermann, University Hospital of Cologne, Cologne, Germany), the oral treponeme Treponema denticola (ATCC 33521), and Leptospira biflexa and Leptospira interrogans (provided by V. Sambri, Section of Microbiology, St. Orsola Hospital, University of Bologna, Bologna, Italy). The identities of all strains were confirmed by PCR and 16S rRNA gene sequencing. Bacterial strains for positive and negative controls were fixed as described elsewhere (27).
DNA extraction.
For DNA extraction, a commercially available respiratory specimen preparation kit (Amplicor; Roche Molecular Systems Inc., Branchburg, NJ) was used on the isolates (for HIS3, HIS4, and HIS5) or on 20 sections from Technovit-embedded biopsy samples (HIS1 and HIS2) as recommended by the manufacturer.
PCR amplification and sequencing.
The 16S rRNA gene was amplified using bacterial primers TPU1 (AGA GTT TGA TCM TGG CTC AG; corresponding to Escherichia coli positions 8 to 27) and RTU8 (AAG GAG GTG ATC CAK CCR CA; corresponding to E. coli positions 1541 to 1522) (38).
Amplicons were analyzed in an automated capillary DNA sequencer (CEQ 8000, Beckman Coulter, Krefeld, Germany). The sequences obtained were compared with currently available data from the public databases (EMBL and GenBank) using BLAST and FASTA in the sequence analysis program Husar, version 4.1. (Deutsches Krebsforschungszentrum, Heidelberg, Germany).
Phylogenetic analysis.
The 16S rRNA gene sequences obtained from the biopsy specimens were aligned and compared with previously published Brachyspira sp. sequences available from GenBank by using Husar, version 4.1. The type strains of B. aalborgi, B. pilosicoli, B. innocens, Brachyspira murdochii, Brachyspira intermedia, B. hyodysenteriae, Brachyspira alvinipulli, and “Brachyspira canis,” as well as 28 other B. aalborgi, B. pilosicoli, “Brachyspira ibaraki,” and “Brachyspira suanatina” strains (Table 2), were included. A neighbor-joining phylogenetic tree (32) was constructed using PAUP, version 4.1b, on the basis of a distance matrix corrected by the two-parameter model of Kimura (19).
TABLE 2.
Strain designations, EMBL/GenBank accession numbers, and references of the Brachyspira strains included in this study
| Strain/phylotype | Brachyspira sp. | Origin | EMBL/GenBank accession no. | Reference as given in GenBank |
|---|---|---|---|---|
| 513AT (NCTC 11492T) | B. aalborgi | Human | Z22781 | Hookey et al. (2003) |
| W1 | B. aalborgi | Human | AF200693 | Kraaz et al. (2000) |
| 719-00 | B. aalborgi | Human | AM039526 | Klitgaard et al. (2005) |
| HIS/ML15/5/02 | B. aalborgi | Human | AB177983 | Nakamura et al. (2005) |
| HISM28/5/02 | B. aalborgi | Human | AB120022 | Hirane et al. (2004) |
| HISM27/5/02 | B. aalborgi | Human | AB120021 | Hirane et al. (2004) |
| HISM26/5/02 | B. aalborgi | Human | AB120020 | Hirane et al. (2004) |
| HISM16/5/02 | B. aalborgi | Human | AB120010 | Hirane et al. (2004) |
| HISM15/5/02 | B. aalborgi | Human | AB120009 | Hirane et al. (2004) |
| W3b | B. aalborgi | Human | AY349949 | Raasbaeck et al. (2004) |
| W2f | B. aalborgi | Human | AY349948 | Raasbaeck et al. (2004) |
| W2b | B. aalborgi | Human | AY349947 | Raasbaeck et al. (2004) |
| Hca20 | B. aalborgi | Human | AF228807 | Pettersson et al. (2004) |
| Tvb03 | B. aalborgi | Human | AF228819 | Pettersson et al. (2004) |
| Tvb40 | B. aalborgi | Human | AF228821 | Pettersson et al. (2004) |
| HIS24/11/99 | “B. ibaraki” | Human | AB079583 | Adachi (2003) |
| OmanN26 | B. pilosicoli | Human | AY187057 | Mikosza et al. (2005) |
| Br1622 | B. pilosicoli | Porcine | AY514024 | Fossi et al. (2004) |
| P43T (ATCC 51139T) | B. pilosicoli | Porcine | U14927 | Fellstroem et al. (2007) |
| C162 | B. pilosicoli | Porcine | U14928 | Fellstroem et al. (2007) |
| AN916:90 | B. pilosicoli | Porcine | U14929 | Fellstroem et al. (2007) |
| CF8 | B. pilosicoli | Canine | AB120008 | Manabe et al. (2004) |
| CD2S | B. pilosicoli | Canine | AB120007 | Manabe et al. (2004) |
| CD1S | B. pilosicoli | Canine | AB120006 | Manabe et al. (2004) |
| H98-5 | B. pilosicoli | Canine | AY349946 | Johansson et al. (2004) |
| CN 140 | B. pilosicoli | Canine | AY349945 | Johansson et al. (2004) |
| Dog 17 | B. pilosicoli | Canine | AY349944 | Johansson et al. (2004) |
| 24072-93a | B. pilosicoli | Canine | AY349943 | Johansson et al. (2004) |
| AN2608/97 | B. pilosicoli | Canine | AF245123 | Johansson et al. (2007) |
| A3077 | B. pilosicoli | Canine | AF245120 | Johansson et al. (2007) |
| B256T (ATCC 29790T) | B. innocens | Porcine | U14920 | Fellstroem et al. (1994) |
| 56-150T | B. murdochii | Porcine | AY312492 | Johansson et al. (2003) |
| PSW/AT (ATCC 51140T) | B. intermedia | Porcine | U23033 | Stanton et al. (2001) |
| B78T (ATCC 51933T) | B. hyodysenteriae | Porcine | U14930 | Harris et al. (1972) |
| C1T (ACTT 51933T) | B. alvinipulli | Avian | U23030 | Stanton et al. (2006) |
| A2T | “B. canis” | Canine | AY349936 | Johansson et al. (2004) |
| AN3949:2/02 | “B. suanatina” | Avian | AY352290 | Jansson et al. (2004) |
| AN1418:2/01 | “B. suanatina” | Avian | AY352282 | Jansson et al. (2004) |
| HIS1 | B. aalborgi | Human | FM178385 | This study |
| HIS2 | B. aalborgi | Human | FM178386 | This study |
| HIS3 (isolate) | B. pilosicoli-like | Human | FM178387 | This study |
| HIS4 (isolate) | B. pilosicoli-like | Human | FM178388 | This study |
| HIS5 (isolate) | B. pilosicoli-like | Human | FM178389 | This study |
Nucleotide sequence accession numbers.
Five almost complete 16S rRNA gene sequences obtained from the tissue isolates were submitted to the EMBL database with the accession numbers listed in Table 2.
RESULTS
Response to treatment and follow-up.
Two patients had no known risk factors for HIS, whereas patient HIS4 was homosexual, and patients HIS3 and HIS5 were infected with human immunodeficiency virus (HIV). Four patients were treated with metronidazole. Upon therapy, the diarrhea of patients HIS3 and HIS4 resolved, while patients HIS1 and HIS5 showed partial improvement of symptoms. Patient HIS2 improved without treatment.
Histopathological findings.
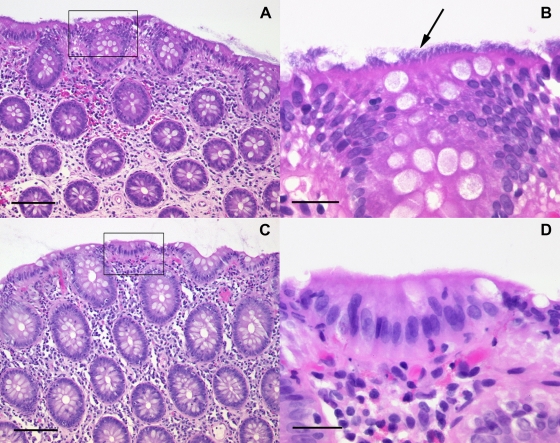
In all cases, the typical hematoxyphilic band on the brush border of the epithelial layer of the mucosa could be detected in HE-stained sections (Fig. 1). In addition, the tissue sections of patient HIS2 showed signs of chronic ulcerative colitis.
FIG. 1.
Spirochetosis of the colon. Shown are HE-stained sections of colon biopsy specimens from patient HIS3 (A and B) and control patient 5 (C and D). (A) Low-magnification image showing a “fringed” blue line along the surface epithelium. Bar, 40 μm. (B) A high-magnification image of the area boxed in panel A displays numerous hematoxyphilic organisms (arrow) at the luminal border of the colonic mucosa. Bar, 20 μm. (C and D) Normal tissue from a control patient at low and high magnification, respectively.
For patients HIS1, HIS3, and HIS5, biopsy samples from control colonoscopies were available for histological follow-up. In the follow-up biopsy samples of patients HIS1 and HIS3, minimal residual spirochetosis was suspected on the basis of the Warthin-Starry silver staining results, whereas patient HIS5 was completely negative.
Biopsy specimens from the control group were HIS negative (Fig. 1).
Culture.
After 6 weeks of anaerobic incubation, a thin haze of pinpoint-like colonies was observed on agar plates inoculated with biopsy material from patients HIS3, HIS4, and HIS5. Single colonies with motile spirochetes as verified by dark-field microscopy were subcultured. The identities of the strains were confirmed by PCR and 16S rRNA gene sequencing, showing the highest homology with the B. pilosicoli type strain P43 (U14927).
Cultures of the control biopsy samples of patients HIS1 and HIS5 that were collected after treatment remained negative for Brachyspira spp. For patients HIS1 and HIS2, no bacterial isolates could be obtained, because we received only formalin-fixed specimens.
FISH.
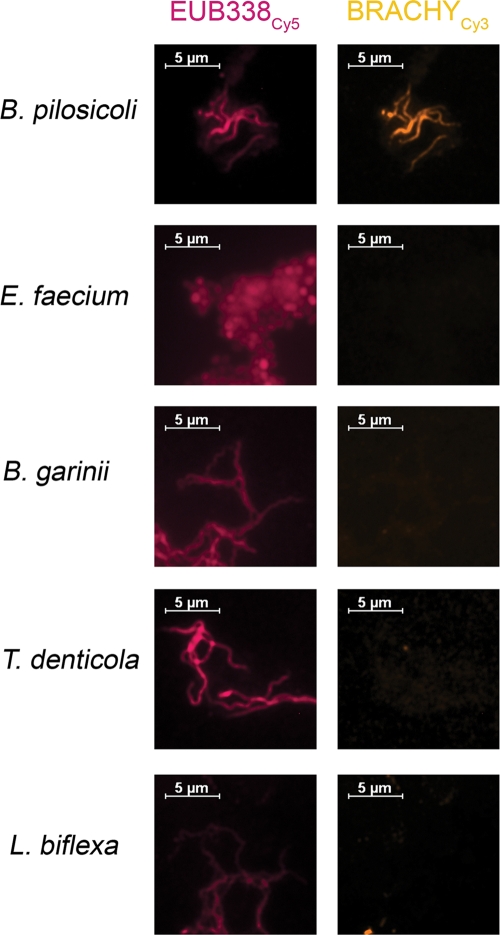
The oligonucleotide probe BRACHY, specific for the genus Brachyspira, was designed and evaluated prior to application to tissue sections. Compared to all sequences from EMBL and GenBank available as of January 2007, probe BRACHY showed 100% homology solely with the Brachyspira sp. 16S rRNA gene. Stringent hybridization conditions were adjusted using B. pilosicoli as a positive control and E. faecium and S. halophila as negative controls, with two mismatches each to the probe sequence. All other cultivable spirochetes investigated yielded no signal with this probe (Fig. 2).
FIG. 2.
Evaluation of probe BRACHY, specific for Brachyspira spp. Fixed bacteria from cultures of B. pilosicoli, E. faecium, B. garinii, T. denticola, and L. biflexa were simultaneously hybridized with BRACHYCy3 (orange) and EUB338Cy5 (magenta). Left and right panels in each row show identical microscopic fields with filter sets for Cy3 and Cy5, respectively. BRACHY showed specific hybridization, giving a positive signal solely with B. pilosicoli.
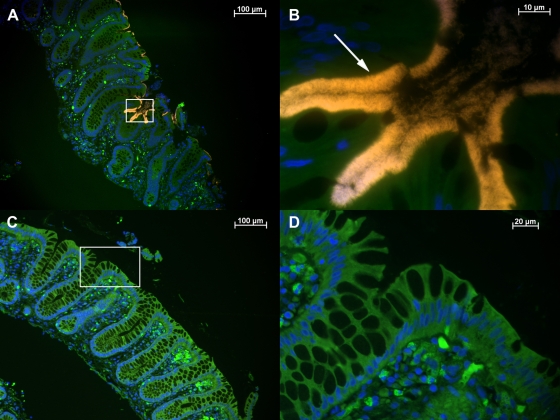
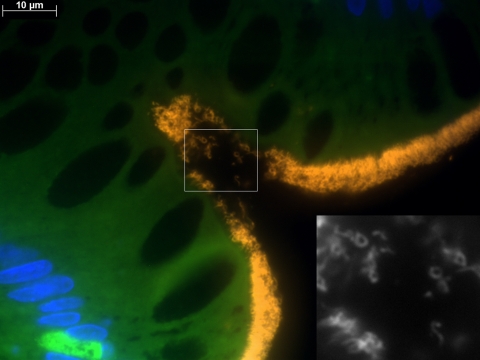
Tissue sections from all five patients showed positive signals after hybridization with both probes BRACHY and EUB338. At low magnification (×100), this positive reaction could be easily visualized as a conspicuous bright fringe coating the surface epithelium (Fig. 3A). Higher magnification (×1,000) revealed densely packed spirochetes attached at one end to the mucosa (Fig. 3B). In addition to the usual helical shape, microorganisms with different morphologies were detected, yielding positive signals with both EUB338 and BRACHY (Fig. 4).
FIG. 3.
Visualization and identification of Brachyspira spp. in a colon biopsy specimen by FISH. Sections of colon biopsy specimens from patient HIS4 (A and B) and control patient 1 (C and D) were hybridized with BRACHYCy3 (orange) and stained with DAPI (blue). (A) Overview. An overlay of the Cy3, fluorescein isothiocyanate, and DAPI filter set results in a bright orange fringe covering the surface epithelium of the crypts and contrasting with the green background fluorescence of the tissue. (B) A higher magnification of the microscopic field boxed in panel A reveals the characteristic end-on attachment (arrow) as well as single Brachyspira sp. organisms in the lumen. (C and D) Absence of spirochetes in normal tissue from a control patient, shown at low and high magnification, respectively.
FIG. 4.
Identification of different Brachyspira morphologies by FISH with BRACHY. A section of a colon biopsy specimen from patient HIS4 was hybridized with BRACHYCy3 (orange) and stained with DAPI (blue). In addition to the typical presentation of spirochetes in a helical shape, attached by one end to the surface epithelium, microorganisms with different morphologies can be visualized. Yielding a positive signal with BRACHYCy3, these microorganisms are identified as Brachyspira spp., as shown in the black-and-white image (inset) that was taken with the Cy3 filter set only.
No spirochetes could be detected in control biopsy samples collected from patients HIS1, HIS3, and HIS5 after treatment. The absence of a positive reaction with BRACHY was confirmed by the lack of detection of spirochetal morphotypes with EUB338 or DAPI, both at low and at high magnification. However, in the samples from patient HIS1, various rods and cocci in the intestinal lumen stained positive with EUB338 and DAPI.
No spirochetes were detected in the biopsy specimens from the control group (Fig. 3C and D).
PCR and analysis of sequence data.
For all five HIS cases, almost complete sequences of the 16S rRNA gene, ranging from 1,299 to 1,436 bp, were obtained. In comparison with currently available data from EMBL and GenBank, the sequences had the highest homology to previously published Brachyspira spp. Sequences from patients HIS1 and HIS2 could be identified as B. aalborgi; the HIS1 sequence yielded 99.8% homology with the sequence of accession number AF200693, and the HIS2 sequence was identical with the AF200693 sequence (22). Sequences from patients HIS3, HIS4, and HIS5 showed 99.4%, 99.5%, and 99.4% homology with the B. pilosicoli type strain, P43 (U14927), differing in seven, seven, and eight nucleotide positions, respectively. Although these three isolates were from different patients living in two geographically distant towns in Germany, they were closely related, exhibiting only three to five nucleotide mismatches. At three nucleotide positions at the end of the 16S rRNA gene, the isolates were identical to each other but different from B. pilosicoli P43T.
Accordingly, the HIS1 and HIS2 sequences clustered in the B. aalborgi group of a phylogenetic tree based on an alignment comprising 1,264 nucleotide positions (Fig. 5). Sequences HIS1 and HIS2 clustered together close to the B. aalborgi type strain, 513A (accession number Z22781), and were included in cluster 1 of the three phylogenetic clusters defined by Pettersson et al. (31). The HIS3, HIS4, and HIS5 sequences fell into the B. pilosicoli lineage. They formed a separate branch within this group, although a bootstrap percentage of 63% indicated only moderate stability for the branching of this node.
FIG. 5.
Neighbor-joining tree showing the phylogenetic relationships among different Brachyspira spp. The evolutionary tree, based on 16S rRNA gene sequence alignment, comprises 1,264 nucleotide positions. B. hyodysenteriae serves as the outgroup. The scale bar represents 0.001 substitution per nucleotide position. The stability of the branching order is represented by the bootstrap percentages, obtained from 1,000 resamplings of the data and placed at the major nodes. Colors differentiate sequences from human (orange), porcine (red), canine (green), and avian (blue) strains.
DISCUSSION
The clinical relevance of intestinal spirochetes for humans is still unresolved (37). Here we present data from five patients with possible HIS, suffering from symptoms of chronic intestinal disorders. After confirmation of the diagnosis, four patients underwent antimicrobial chemotherapy, resulting in clinical improvement and significant reductions in the numbers of spirochetes in control biopsy samples. The parallel course of positive FISH signals for symptomatic patients strongly suggests that the intestinal spirochetes were potentially pathogenic and may have been the causative agent of the intestinal disorders of our patients. This conclusion conflicts with reports of healthy patients with histologically diagnosed HIS (28, 31). The discrepancy might be explained by differences in virulence between and within Brachyspira spp., as well as by differences in the immune status of the patients, since HIV infection is a well-described risk factor for HIS. Brooke et al. (5) have suggested that B. aalborgi may be a commensal human-adapted species that is able to overgrow and to cause symptoms under certain conditions, while B. pilosicoli could have greater pathogenic potential in humans. This discussion reveals the insufficiency of present diagnostic tools and the need for identification of the intestinal spirochetes in addition to visualization. In order not to miss a case of HIS, we designed a 16S rRNA-targeting probe specific for all members of the genus Brachyspira, which yielded a positive signal in all five cases. The positive FISH signals were obvious even at low magnification and could be easily detected. Furthermore, FISH permitted the detection and identification of microorganisms as spirochetes even if they were detached from the surface epithelium and altered in their morphology (Fig. 4). Since FISH is simple, rapid, and inexpensive, it can easily be included in routine diagnostic procedures in addition to traditional histopathological techniques.
In contrast to histology using light microscopy, FISH allows identification of intestinal spirochetes on a genus-specific, species-specific, and even intraspecies-specific level (17). However, this requires a thorough inventory of the species involved. A challenging aspect of the diagnosis of HIS is the genetic heterogeneity of the intestinal spirochetes. The sequencing of the 16S rRNA gene revealed 99.8% and 100% homology with B. aalborgi for two cases (HIS1 and HIS2). In three cases, the sequences differed from those of previously published Brachyspira spp. and formed a distinct branch in the phylogenetic tree. Although the HIS3, HIS4, and HIS5 isolates were obtained from three different patients from two geographically distant German areas, they clustered together in the B. pilosicoli group, leading to the speculation that they might be epidemiologically and clinically relevant. These sequences, highly identical with the B. pilosicoli type strain, P43, in the first 800 nucleotides, showed distinct variations at the end of the 16S rRNA gene. This phenomenon demonstrates the importance of complete 16S rRNA gene sequencing to avoid underestimation of the variations and to enlarge the databases needed for critical evaluation and careful optimization of the present diagnostic techniques. Therefore, clinicians should consider sending samples from potential HIS patients to a specialized center for confirmation of the diagnosis, Brachyspira culture, and further molecular analysis.
In conclusion, intestinal spirochetes are genetically heterogeneous microorganisms with pathogenic potential that can be reliably visualized and identified by FISH. Since it is inexpensive and rapid, FISH can be included in routine diagnostic procedures. Furthermore, the use of this method can facilitate further epidemiological studies to determine the clinical significance of Brachyspira spp. and to investigate the extent of intraspecies genetic variation.
Acknowledgments
We thank Y. Gräser for access to PAUP, version 4.1b, and help with the phylogenetic analysis. We are grateful to G. Fiedler, A. Pohlisch, and J. Imlau for excellent technical assistance. Thanks are due to G. Jechart for providing biopsy samples from the patient in Augsburg and to U. Lippert for providing biopsy material from the patient in Hamburg. We thank D. Ramsey for critical reading of the manuscript.
This work was supported by the Sonnenfeld-Stiftung, Berlin, Germany; by a grant from Charité—Universitätsmedizin Berlin, Berlin, Germany, to D.S.; and by a Rahel-Hirsch grant from Charité—Universitätsmedizin to A.M.
There is no conflict of interest for any of the authors.
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 561919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atyeo, R. F., S. L. Oxberry, and D. J. Hampson. 1999. Analysis of Serpulina hyodysenteriae strain variation and its molecular epidemiology using pulsed-field gel electrophoresis. Epidemiol. Infect. 123133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boye, M., T. K. Jensen, K. Møller, T. D. Leser, and S. E. Jorsal. 1998. Specific detection of the genus Serpulina, S. hyodysenteriae and S. pilosicoli in porcine intestines by fluorescent rRNA in situ hybridization. Mol. Cell. Probes 12323-330. [DOI] [PubMed] [Google Scholar]
- 4.Brooke, C. J., T. V. Riley, and D. J. Hampson. 2003. Evaluation of selective media for the isolation of Brachyspira aalborgi from human faeces. J. Med. Microbiol. 52509-513. [DOI] [PubMed] [Google Scholar]
- 5.Brooke, C. J., T. V. Riley, and D. J. Hampson. 2006. Comparison of prevalence and risk factors for faecal carriage of the intestinal spirochaetes Brachyspira aalborgi and Brachyspira pilosicoli in four Australian populations. Epidemiol. Infect. 134627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius, J., M. L. Palmer, J. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 754801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie, J. D. 2003. Intestinal spirochetes. Organisms in search of a disease? Am. J. Clin. Pathol. 120820-821. [DOI] [PubMed] [Google Scholar]
- 8.Douglas, J. G., and V. Crucioli. 1981. Spirochaetosis: a remediable cause of diarrhoea and rectal bleeding? Br. Med. J. (Clin. Res. Ed.) 2831362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duhamel, G. E., D. J. Trott, N. Muniappa, M. R. Mathiesen, K. Tarasiuk, J. I. Lee, and D. J. Hampson. 1998. Canine intestinal spirochetes consist of Serpulina pilosicoli and a newly identified group provisionally designated “Serpunlina canis” sp. nov. J. Clin. Microbiol. 362264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteve, M., A. Salas, F. Fernández-Bañares, J. Lloreta, M. Mariné, C. I. Gonzalez, M. Forné, J. Casalots, R. Santaolalla, J. C. Espinós, M. A. Munshi, D. J. Hampson, and J. M. Viver. 2006. Intestinal spirochetosis and chronic watery diarrhea: clinical and histological response to treatment and long-term follow up. J. Gastroenterol. Hepatol. 211326-1333. [DOI] [PubMed] [Google Scholar]
- 11.Fellström, C., B. Pettersson, J. Thomson, A. Gunnarsson, M. Persson, and K. E. Johansson. 1997. Identification of Serpulina species associated with porcine colitis by biochemical analysis and PCR. J. Clin. Microbiol. 35462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gad, A., R. Willén, K. Furugård, B. Fors, and M. Hradsky. 1977. Intestinal spirochaetosis as a cause of longstanding diarrhoea. Uppsala J. Med. Sci. 8249-54. [DOI] [PubMed] [Google Scholar]
- 13.Harland, W. A., and F. D. Lee. 1967. Intestinal spirochaetosis. Br. Med. J. 3718-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hookey, J. V., S. P. Barrett, C. S. Reed, and P. Barber. 1994. Phylogeny of human intestinal spirochaetes inferred from 16S rDNA sequence comparisons. FEMS Microbiol. Lett. 117345-350. [DOI] [PubMed] [Google Scholar]
- 15.Hovind-Hougen, K., A. Birch-Andersen, R. Henrik-Nielsen, M. Orholm, J. O. Pedersen, P. S. Teglbjærg, and E. H. Thaysen. 1982. Intestinal spirochetosis: morphological characterization and cultivation of the spirochete Brachyspira aalborgi gen. nov., sp. nov. J. Clin. Microbiol. 161127-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, T. K., M. Boye, P. Ahrens, B. Korsager, P. S. Teglbjærg, C. F. Lindboe, and K. Møller. 2001. Diagnostic examination of human intestinal spirochetosis by fluorescent in situ hybridization for Brachyspira aalborgi, Brachyspira pilosicoli, and other species of the genus Brachyspira (Serpulina). J. Clin. Microbiol. 394111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, T. K., P. S. Teglbjærg, C. F. Lindboe, and M. Boye. 2004. Demonstration of Brachyspira aalborgi lineages 2 and 3 in human colonic biopsies with intestinal spirochaetosis by specific fluorescent in situ hybridization. J. Med. Microbiol. 53341-343. [DOI] [PubMed] [Google Scholar]
- 18.Käsbohrer, A., H. R. Gelderblom, K. Arasteh, W. Heise, G. Grosse, M. L'age, A. Schönberg, M. A. Koch, and G. Pauli. 1990. Intestinal spirochetosis in HIV infection: prevalence, isolation and morphology of spirochetes. Dtsch. Med. Wochenschr. 1151499-1506. (In German.) [DOI] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16111-120. [DOI] [PubMed] [Google Scholar]
- 20.Körner, M., and J. O. Gebbers. 2003. Clinical significance of human intestinal spirochetosis—a morphologic approach. Infection 31341-349. [DOI] [PubMed] [Google Scholar]
- 21.Kraatz, W., U. Thunberg, B. Pettersson, and C. Fellström. 2001. Human intestinal spirochetosis diagnosed with colonoscopy and analysis of partial 16S rDNA sequences of involved spirochetes. Anim. Health Res. Rev. 2111-116. [PubMed] [Google Scholar]
- 22.Kraaz, W., B. Pettersson, U. Thunberg, L. Engstrand, and C. Fellström. 2000. Brachyspira aalborgi infection diagnosed by culture and 16S ribosomal DNA sequencing using human colonic biopsy specimens. J. Clin. Microbiol. 383555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. I., and D. J. Hampson. 1992. Intestinal spirochaetes colonising Aboriginals from communities in the remote north of Western Australia. Epidemiol. Infect. 109133-141. [PMC free article] [PubMed] [Google Scholar]
- 24.Mikosza, A. S., and D. J. Hampson. 2001. Human intestinal spirochetosis: Brachyspira aalborgi and/or Brachyspira pilosicoli? Anim. Health Res. Rev. 2101-110. [PubMed] [Google Scholar]
- 25.Mikosza, A. S., M. A. Munshi, and D. J. Hampson. 2004. Analysis of genetic variation in Brachyspira aalborgi and related spirochaetes determined by partial sequencing of the 16S rDNA and NADH oxidase genes. J. Med. Microbiol. 53333-339. [DOI] [PubMed] [Google Scholar]
- 26.Moter, A., G. Leist, R. Rudolph, K. Schrank, B. K. Choi, M. Wagner, and U. B. Göbel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 1442459-2467. [DOI] [PubMed] [Google Scholar]
- 27.Moter, A., and U. B. Göbel. 2000. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 4185-112. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen, R. H., M. Orholm, J. O. Pedersen, K. Hovind-Hougen, P. S. Teglbjærg, and E. H. Thaysen. 1983. Colorectal spirochaetosis: clinical significance of the disease. Gastroenterology 8562-67. [PubMed] [Google Scholar]
- 29.Peghini, P. L., J. G. Guccion, and A. Sharma. 2000. Improvement of chronic diarrhea after treatment for intestinal spirochetosis. Dig. Dis. Sci. 451006-1010. [DOI] [PubMed] [Google Scholar]
- 30.Peruzzi, S., C. Gorrini, G. Piccolo, A. Calderaro, G. Dettori, and C. Chezzi. 2007. Human intestinal spirochaetosis in Parma: a focus on a selected population during 2002-2005. Acta Biomed. 78128-132. [PubMed] [Google Scholar]
- 31.Pettersson, B., M. Wang, C. Fellström, M. Uhlén, G. Molin, B. Jeppsson, and S. Ahrné. 2000. Phylogenetic evidence for novel and genetically different intestinal spirochetes resembling Brachyspira aalborgi in the mucosa of the human colon as revealed by 16S rDNA analysis. Syst. Appl. Microbiol. 23355-363. [DOI] [PubMed] [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 33.Stanton, T. B., D. J. Trott, J. I. Lee, A. J. McLaren, D. J. Hampson, B. J. Paster, and N. S. Jensen. 1996. Differentiation of intestinal spirochaetes by multilocus enzyme electrophoresis analysis and 16S rRNA sequence comparisons. FEMS Microbiol. Lett. 136181-186. [DOI] [PubMed] [Google Scholar]
- 34.Tompkins, D. S., M. A. Waugh, and E. M. Cooke. 1981. Isolation of intestinal spirochaetes from homosexuals. J. Clin. Pathol. 341385-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trivett-Moore, N. L., G. L. Gilbert, C. L. H. Law, D. J. Trott, and D. J. Hampson. 1998. Isolation of Serpulina pilosicoli from rectal biopsy specimens showing evidence of intestinal spirochetosis. J. Clin. Microbiol. 36261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott, D. J., T. B. Stanton, N. S. Jensen, G. E. Duhamel, J. L. Johnson, and D. J. Hampson. 1996. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int. J. Syst. Bacteriol. 46206-215. [DOI] [PubMed] [Google Scholar]
- 37.van Mook, W. N. K. A., G. H. Koek, A. J. A. M. van der Ven, T. L. Ceelen, and R. P. Bos. 2004. Human intestinal spirochaetosis: any clinical significance? Eur. J. Gastroenterol. Hepatol. 1683-87. [DOI] [PubMed] [Google Scholar]
- 38.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]