Abstract
Polyomavirus BK (BKV) establishes latent infection in various human tissues, including the kidney. Reactivation following renal transplantation (RT) may cause BKV-associated nephropathy, leading to graft loss. BKV reactivation is often associated with extensive rearrangements in the BKV noncoding regulatory region (NCRR). We explored the formation and predominance of the rearrangements versus the diversity of the rearrangements by cloning and characterizing PCR-amplified NCRR sequences from six Israeli RT patients. We found a high frequency and a high degree of diversity of rearrangements: NCRRs that contained major rearrangements (mrNCRRs), including large insertions and deletions, were detected in 0 to 100% of the clones from individual samples (mean, 50% and 67% in plasma and urine, respectively). In addition, we found a high frequency of mrNCRRs that contained single-nucleotide variations (snvNCRRs) among identical mrNCRRs and archetype clones. mrNCRRs were present in plasma and in concomitantly collected urine samples, but for each patient, only a subset of the mrNCRRs and snvNCRRs were present in both compartments at the same time and/or in subsequent samples from the same compartment. Some mrNCRRs were observed over several months, indicating the continuous replication of the viral genomes carrying them. Phylogenetic analysis based on the snvNCRR in the archetype clones grouped isolates from four of the patients into a new subgroup of genotype IV. Genotypes Ib-1 and Ib-2 were also found. Isolates from two patients had NCRRs from two genotypes, one concurrently with a RT and one after a second RT. Our study prompts further investigation of the functional consequences of NCRR rearrangements to assess their biological significance and their putative role in disease progression and prognosis.
A large proportion of the adult population has been infected by the human polyomaviruses BK virus (BKV) and/or JC virus (JCV) (39). Primary infection is usually asymptomatic and occurs at an early age (11). BKV and JCV then enter a latent state and persist in various organs, including the kidneys, ureter, brain, and spleen, in the absence of detectable viremia (9, 10, 31). Infection, reinfection, and reactivation may occur when individuals receive aggressive immunosuppressant drug therapy for renal, bone marrow, pancreas, and lung allogeneic transplants (4, 18, 23, 41) or chemotherapy for cancer (24) or when their immune systems have been compromised by infections with viruses such as human immunodeficiency virus (28, 32). Infection, reinfection, and reactivation in such individuals may lead to overt diseases that depend on both the virus and the organ involved. For example, the most common syndrome caused by BKV in kidney transplant patients is polyomavirus-associated nephropathy (PVAN) (for recent reviews, see references 1, 14, 20, 35, and 37), although some individuals develop ureteral stenosis (21), whereas hemorrhagic cystitis is the most common manifestation of BKV infection in bone marrow transplant recipients (5). In contrast, JCV has rarely been reported as the sole causative agent of PVAN in patients after renal transplantation (RT) (12, 20, 26) but is the main causative agent of progressive multifocal leukoencephalopathy in AIDS patients.
PVAN progression has been divided into three stages: stage A, an initial stage, is characterized by limited focal nonlytic viral activity accompanied by low but slowly increasing creatinine levels; stage B, a florid stage, is associated with elevated plasma creatinine levels and accumulating damage to the kidney that could lead to chronic kidney dysfunction and end-stage renal failure; and stage C is the final stage in which fibrotic sclerosing has become irreversible (13, 19, 35). During virus reactivation in such patients, the noncoding regulatory region (NCRR), which contains the origin of DNA replication as well as promoter and enhancer elements necessary for the transcription and translation of early and late viral genes, undergoes extensive rearrangements involving duplications and deletions (3, 37). Similar rearrangements were observed during JCV replication in patients with progressive multifocal leukoencephalopathy (2). These rearrangements may play an important role in virus replication by increasing or decreasing the number or the affinities of host transcription factor binding sites (22). The viral load is believed to reflect the total number of viral genomes in circulation but does not provide an indication of the proportion or the sequence structure of individual NCRR variants within this quasispecieslike population. Specific NCRR rearrangements in urine and/or blood might become the hallmark of a high risk for progression into PVAN if they can be linked to biological activities (34). The search for such variants has been limited by the fact that most of the sequencing studies have been conducted with uncloned viral populations (for a review, see the work of Moens and Van Ghelue [34]), and if such variants had existed among those populations, they may have been present at levels below the amount needed to appear above the background sequencing noise.
An additional parameter that may affect the biological properties of BKV and thus the risk for disease progression is the virus genotype. BKV has four antigenically differentiated serotypes, serotypes I to IV (27), and four corresponding genotypes based on the VP1 epitope region (25) and full-length genomic sequences (49). Genotype I is further divided into subgenotypes Ia, Ib-1, Ib-2, and Ic. Zheng et al. (49) and Nishimoto et al. (36) have shown by geographically associated phylogenetic analysis that genotypes I and IV are the most prevalent in Europe and throughout Asia but that only genotype I is prevalent in Africa. Furthermore, each subgenotype of genotype I was also the most prevalent in a particular geographic area.
In the current study, we have conducted an in-depth analysis of the NCRRs in reactivated BKV by cloning the NCRRs from urine and plasma samples from six RT patients with elevated plasma creatinine and PVAN-predictive BKV viruria levels to gain insight into the process of quasispeciation during BKV reactivation. NCRR structures from samples concurrently collected from the urogenital and blood compartments were compared, and the genomic subtypes were identified by phylogenetic comparisons with sequences from the DDBJ/EMBL/GenBank nucleotide sequence database.
(Portions of this work were submitted to Bar Ilan University, Ramat Gan, Israel, by T. T. Perets in partial fulfillment of the degree of doctor of philosophy.)
MATERIALS AND METHODS
Clinical samples.
Concurrent plasma and urine samples with BKV loads near or above the level predictive of PVAN (19, 21, 35, 46) from four RT patients (patients A, B, C, and D) who developed high plasma creatinine levels were chosen for the study of BKV NCRR sequence reorganization. Additional pairs of concurrently collected urine and plasma samples were available for two of these four patients (patients A and C). BKV NCRR rearrangements were also analyzed in urine samples from two additional patients (patients E and F). Patient E was included after the development of high plasma creatinine levels and PVAN-predictive BKV loads after both a first and a second RT, while patient F was chosen for study after the development of a high plasma creatinine level and PVAN-predictive BKV viruria in the absence of detectable viremia. Concurrent kidney biopsy reports were available for five of the six patients (patients A to D and F). While the reports were not as rigorously detailed as is now recommended for the definitive diagnosis of PVAN (19, 47), the findings were consistent with PVAN. Patient profiles, plasma creatinine levels, and BKV loads are presented in Table 1. The study was approved by the Institutional Review Board of the Chaim Sheba Medical Center, Tel Hashomer, Israel, and complied with Israeli laws and regulations.
TABLE 1.
BKV loads in urine and plasma, plasma creatinine levels, and biopsy results for Israeli RT patients A to F
| Patienta | Time (mo) after RT | Viral load (no. of copies per ml)b
|
Plasma creatinine level (mg/dl) | Biopsy reportd | |
|---|---|---|---|---|---|
| Urine | Blood | ||||
| A (M, 53) | 3 | 2.37 × 105 | 1.26× 105 | 1.8 | Suspicion of rejection (tubular interstitial nephritis, tubular atrophy); suspicion of viral infection (visible inclusion bodies, infiltrate in medulla) |
| 8 | 8.02 × 106 | 1.73 × 105 | 2.7 | ||
| B (M, 74y | 10 | 3.95 × 107 | 2.48 × 105 | 3.2 | Viral infection (tubular changes with focal tubular atrophy, inclusion bodies with viral particles [determined by electron microscopy]) |
| C (M, 60) | 24 | 3.13 × 108 | 5.46 × 105 | 6.1 | Viral infection (decoy cells in urine, infiltrate in medulla); tubular changes (medula and glomerulus, BKV-positive inclusion bodies) |
| 29 | 3.57 × 108 | 1.02 × 106 | 7.5 | ||
| 34 | 4.14 × 108 | 1.64 × 106 | 7.8 | ||
| D (F, 14) | 84 | 4.67 × 108 | 5.34 × 106 | 7.8 | No concurrent biopsy available |
| E (F, 55)c | 3 | 1.65 × 104 | 1.1 | Support (virus-associated chronic tubulointerstitial nephritis); mild acute cellular rejection; grade 1A focal proliferative glomerulonephritis; focal atrophy of tubules; hyperchromasia of nuclei | |
| 13 | 1.69 × 105 | 1.6 | |||
| 16 | 3.06 × 108 | 6.1 | |||
| 2/51e | 3.60 × 107 | 2.1 | |||
| F (M, 52) | 2 | 2.70 × 109 | Below detection | 8.5 | Acute tubular necrosis |
The information in parentheses represents the patient's sex (F, female; M, male), the patient's age (in years) at the time of the first RT.
Viral loads (number of copies per ml) were determined by qPCR. Titers in boldface indicate loads near or above the PVAN-predictive thresholds for urine and plasma (19, 35).
No concurrent plasma samples were available for patient E.
Biopsy report from a renal biopsy taken at the time of the first patient sample. All of the text refers to that biopsy.
2/51, 51 months after the first transplant and 2 months after the second transplant.
Viral DNA extraction and purification.
Urine (1.2 ml) was ultracentrifuged at 4°C for 1 h at an average speed of 287,582 × g (90,000 rpm; TLA100.2 rotor; Optima TL ultracentrifuge; Beckman Coulter GmbH, Krefeld, Germany). The pellet was dissolved in 200 μl of lysis buffer (1 M urea, 200 mM Tris, 20 mM NaCl, 200 mM EDTA, pH 7.4) for 1 h at 55°C. Plasma was prepared from whole blood by centrifugation at 2,500 rpm (1,000 × g) for 10 min. Viral DNA was extracted from the treated urine pellet and plasma by use of a High Pure viral nucleic acid kit (Roche Applied Science, Panzberg, Germany), according to the manufacturer's instructions. For patient F, urine was clarified and virus was concentrated by centrifugation at 4,600 × g for 10 min above an Amicon ultrafilter (Millipore Corporation, Billerica, MA). The urine above the filter containing the concentrated virus was replaced by buffer A (100 mM Tris, 150 mM NaCl, 12.5 mM EDTA, pH 7.4) (29) by using three to four cycles of dilution with 12 to 15 ml of buffer A and recentrifugation at 4,600 × g for 10 min. The supernatant from the last centrifugation was treated with RQ1 RNase-free DNase (Promega, Madison, WI) for 10 min at 37°C to remove any nonencapsulated DNA. Phenol-chloroform extraction of the encapsulated DNA was performed as described by Le et al. (29) by using buffer B (100 mM Tris, 150 mM NaCl, and 12.5 mM EDTA, pH 7.4, containing 2% sodium dodecyl sulfate) and proteinase K. The extracted viral DNA was linearized at a unique restriction site with EcoRI (Fermentas, Glen Burnie, MD), according to the manufacturer's instructions, and separated by electrophoresis on a 1% agarose gel. The DNA band at 5100 nucleotides (nt), equivalent in size to the linearized BKV genome, was extracted from the gel with an Invisorb spin DNA extraction kit (Invitek, Berlin, Germany), according to the manufacturer's instructions.
PCR and real-time quantitative PCR (qPCR).
DNA was analyzed by PCR for the presence of polyomavirus DNA with panpolyomavirus primers PEP-1 and PEP-2 (3). Differentiation between BKV and JCV was done by heminested PCR with primer PEP-2 and primer BEP-1 or primer JEP-1 (3), respectively. Conditions for amplification were 1 cycle of denaturation at 93°C for 5 min, annealing at 60°C for 2 min, and elongation at 72°C for 2 min; 45 cycles of denaturation, annealing, and elongation at 93°C for 45 s, 55°C for 45 s, and 72°C for 90 s, respectively; and a final elongation at 72°C for 7 min. BKV-specific PCR products were identified by ethidium bromide staining of the amplification products electrophoresed on 2% agarose gels.
Real-time qPCR (activation at 95°C for 15 min, followed by 60 cycles of denaturation at 95°C for 15 s and elongation at 60°C for 1 min) was performed on ABI 7000 or 7700 real-time PCR machines (Applied Biosystems Inc., Foster City, CA) with primers PEP-1 and PEP-2 and with 6-carboxyfluorescein-labeled BEP-1 as a probe (3). Quantitation was done by automatic comparison to a BKV DNA plasmid preparation calibrated with an ABI Amplicheck human BKV quantitated viral DNA control (Advanced Biotechnologies Inc., Columbia, MD).
Cloning of NCRRs.
The BKV NCRR template in each clinical sample was amplified with pureTaq Ready-to-Go beads (Amersham Biosciences Europe GmbH, Freiburg, Germany) and primers BRR1 (5′-CAG GGT GAA ATT CCT TAC ACT TCC-3′) (42) and BKVP2R (5′-TGG CAA CTA GGT CCC CCA A-3′). ECOS 9-5 competent Escherichia coli cells (Yeastern Biotech Co, Taipei, Taiwan) were transformed with pGEM-T-Easy plasmids ligated to the amplified BKV NCRR template (pGEM-T easy vector kit; Promega), according to the manufacturer's instructions. Screening for white versus blue colonies identified colonies potentially containing NCRR inserts. Approximately 10 white colonies were randomly selected for each clinical sample and grown overnight in LB-ampicillin or super optimal broth with catabolite repression-ampicillin (SOC-ampicillin) liquid cultures. Plasmid DNA was purified with an UltraClean miniplasmid preparation kit, according to the manufacturer's instructions (MO BIO Laboratories Inc., Solana Beach, CA). The presence of a BKV NCRR template was confirmed by PCR with primers BKVP2R and BRR1, as described above. For evaluation of Taq enzyme amplification fidelity, the BKV NCRR insert from a single plasmid was amplified with BRR1 and BKVP2R, the PCR amplification products were cloned, 10 BKV-containing plasmids were identified as described above, and the sequences of the entire amplicon of the original and the recloned templates obtained with primers BRR1 and BKVP2R were compared. In addition, the precloning amplification of DNA extracted from the urine from one patient and the postcloning reamplification of the NCRRs from 12 clones for sequencing were performed with Ex Taq enzyme (Takara Bio Inc., Japan), which has efficient 3′-5′ exonuclease activity and a mutation rate 4.5 times lower than the rates of standard Taq DNA polymerases.
Sequence and phylogenetic analyses of NCRRs.
The NCRR template in each plasmid was amplified by PCR with primers BKVP2R and BRR1, as described above. Both strands of gel-purified (QIAquick gel extraction kit; Qiagen Inc., Valencia, CA) NCRRs were sequenced with an ABI Prism 3700 DNA analyzer, a BigDye Terminator (version 3.0) ready reaction cycle sequencing kit (Applied Biosystems, Foster City, CA), and sequencing-quality primers BKVP2R and BRR-1.
NCRR sequences that formed an ungapped alignment with the NCRR sequence of prototype strain WW (GenBank accession number AB211371) when they were analyzed with the Sequencher program (Gencodes Corporation, Ann Arbor, MI) were designated archetype NCRR sequences. The most likely origin of each nucleotide in nonarchetype NCRRs, e.g., NCRRs that contained major insertions and/or deletion rearrangements (mrNCRRs), was also determined with the aid of the Sequencher program. Specifically, the NCRR sequence from each mrNCRR clone was subfragmented in silico until all nucleotides in each subfragment formed a completely overlapping, nongapped contig with individual P, Q, R, and S segments of strain WW. Single-nucleotide variants among the cloned mrNCRRs with the same pattern of rearrangements (snvNCRRs) were also identified by using the Sequencher program.
Phylogenetic comparisons were performed by neighbor joining of aligned sequence data bootstrapped 1,000 times with the ClustalX program (44). Phylogenetic analyses included sequences from all Israeli archetypelike snvNCRRs; prototype strain WW; those isolates with archetype NCRRs from among the full-length sequences analyzed by Zheng et al. (49); three isolates from a study by Sharma et al. (40); and six additional complete sequences of genotype IV isolates deposited in the EMBL database by Y. Yogo, H. Zheng, and Y. Nishimoto. Phylogenetic trees were visualized with the njplot and unrooted programs (M. Gouy, Laboratoire de Biometrie et Biologie Evolutive, Université Lyon, CNRS, Lyon, France [http://pbiol.univlyon1.fr/software/njplot.html]).
In silico reconstruction of archetypelike sequences from rearranged clones.
The supplemental material describes the in silico reconstruction of archetypelike sequences from rearranged clones.
Nucleotide sequence accession numbers.
All Israeli archetype and mrNCRR sequences and their snvNCRR sequences were submitted to the EMBL database and were assigned accession numbers FM995409 to FM995423, FM995424 to FM995436, FM995437 to FM995446, FM995447 to FM995459, FM995460 to FM995483, and FM995484 to FM995487 for patients A to F, respectively.
RESULTS
Demonstration of concomitant presence of more than one mrNCRR and/or minor snvNCRR BKV variant in clinical samples.
qPCR with primers targeting the BKV T antigen indicated the presence of high viral loads in each clinical sample (Table 1) without indicating whether the BKV NCRRs of viruses in the sample were rearranged or contained one (16) or more than one (34) mrNCRR. To characterize the BKV NCRRs in each clinical sample, each strand of gel-purified PCR-amplified DNA was sequenced with primers BKVP2R and BRR1. As the sequence from each of the clinical samples progressed into the NCRR from either direction, there was an abrupt change in the graphic component of the sequencing results. Strings of equidistant unique peaks, e.g., nucleotide positions where >90% of all BKV NCRR DNA templates had the same nucleotide at each nucleotide position, were followed by a series of ambiguous nucleotide peaks with the progressive loss of interpeak periodicity. This pattern is indicative of the presence of significant numbers (>10%) of templates with major differences in their sequences and lengths.
To confirm this conclusion, PCR-amplified NCRR sequences derived from the total DNA of each individual clinical sample were cloned as described in Materials and Methods, and for each clinical sample, the NCRRs of 8 to 21 clones were sequenced. In contrast to the profile for total DNA, the sequence profile of each individual BKV NCRR clone consisted of a pattern of unique peaks throughout the entire length of the NCRR. The reason for the sequencing of at least eight clones for each clinical sample was to look for diversity among the NCRRs and to gain a rough indication of the relative frequencies of archetype and nonarchetype NCRRs. The NCRR sequence from each clone was compared to the NCRR sequence of BKV archetype strain WW by using the convention described in the review by Moens et al. (33). Specifically, the 233 nt of the archetype WW NCRR sequence was arbitrarily divided into four segments, segments P, Q, R, and S, with lengths of 68, 39, 63, and 63 nt, respectively, to more easily describe the major rearrangements within the NCRR. We have classified a cloned NCRR as WW-like archetype 1 (WWL1) if its sequence consisted of a single copy of complete P, Q, R, and S segments arranged in the correct order. For some patients, a slightly altered NCRR archetype was isolated. It consisted of a single copy of complete P, Q, R, and S segments arranged in the correct order, except that the S segment contained a single thymidine nucleotide inserted between nucleotide positions 49 and 50. A cloned NCRR with this archetypelike structure has been designated WW-like archetype 2 (WWL2) to distinguish it from the former WWL1 archetype. For each patient, each unique pattern in which partial or complete segments were deleted from and/or inserted into either of these archetypelike sequences was designated an mrNCRR and was assigned a unique mrNCRR variant name (V1, V2, etc.). Mixtures of archetype and mrNCRRs were found, confirming the simultaneous presence of templates with major sequence differences.
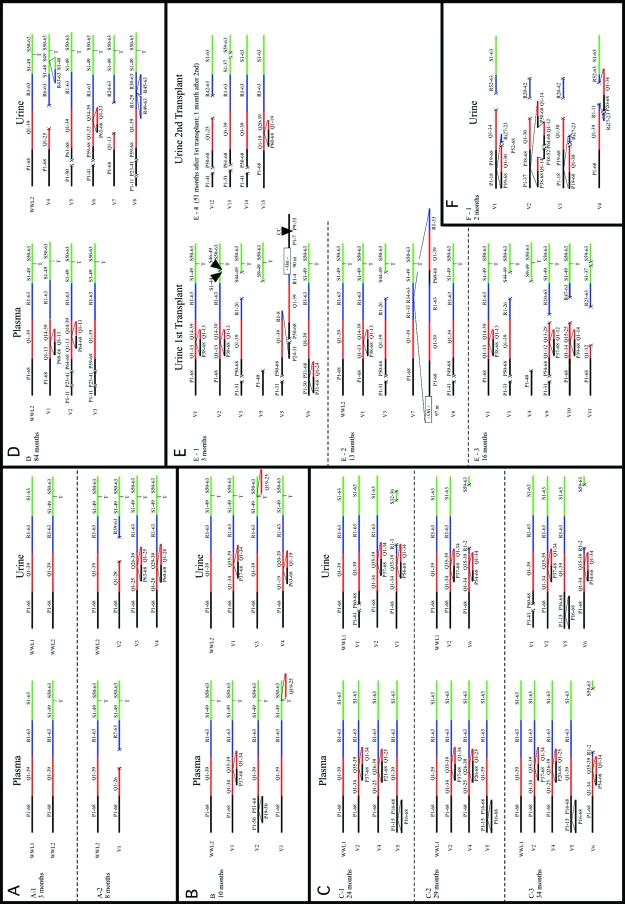
The pattern of each unique segment arrangement for each clinical sample from each patient is shown in Fig. 1. Some unique mrNCRR patterns appeared in more than one clone from different clinical samples from the same patient, including samples taken months apart (patients C and E). This is an indication of the continuous replication rather than the casual appearance and disappearance of such variants.
FIG. 1.
Schematic representation of all of the mrNCRRs from the urine and plasma of the RT patients. Each archetypelike NCRR or mrNCRR is represented by a single line of joined black, red, green, and blue lines representing segments P (68 nt), Q (39 nt), R (63 nt), and S (63 nt), respectively (34). Above each colored line is a letter confirming the name of the segment, followed by a pair of numbers to indicate the range of nucleotides present. The S segment of the WWL2 archetypes and the mrNCRRs derived from them are represented by a T below the line between nucleotide positions 49 and 50. The letter at the top left of each box identifies the patient. The name of each unique major mrNCRR variant appears at the left of each linear representation. Discontinuous lines separate samples from the same patient taken at different times after RT. The relative distribution of mrNCRR variants and the presence of individual snvNCRR variants for each NCRR for each urine and plasma sample are indicated in Table 2.
The relative distribution of archetypelike clones and/or mrNCRRs in paired plasma and urine samples from each patient were not identical, and some mrNCRR variants were unique to either one of these compartments (Table 2). For example, in patient A, the ratios of the WWL1 archetype to the WWL2 archetype were 85:15 in plasma and 20:80 in urine 3 months after transplantation; and in patient D, major variants V1, V2, and V3 were isolated exclusively from plasma, whereas five other variants, variants V4 to V8, were isolated exclusively from urine. Interestingly, when the sequences from all plasma and urine samples from patients A through D were analyzed separately, segment Q and an adjacent region of segment P were overrepresented as a consequence of insertions, while segment R was underrepresented as a consequence of deletions (Fig. 2.). The patterns for samples from each compartment were similar but not identical. For example, portions of segments S and P were underrepresented in plasma isolates but not urine isolates. The reason for these differences is not understood at present. The overall frequencies of mrNCRRs among plasma- and urine-derived clones from all patients were 50% and 67%, respectively.
TABLE 2.
Distribution of BKV mrNCRR and snvNCRR variants in urine samples or paired plasma and urine samples of RT patients
| Patient-sample no./mo after RT | Plasma clones
|
Urine clones
|
||||||
|---|---|---|---|---|---|---|---|---|
| mrNCRRa
|
snvNCRR variantsb | mrNCRR
|
snvNCRR variants | |||||
| Name | No. | % | Name | No. | % | |||
| A-1/3 | WWL1 | 11 | 85 | 1, 2 | WWL1 | 2 | 20 | 1 |
| WWL2 | 2 | 15 | 3 | WWL2 | 8 | 80 | 1, 2, 3 | |
| A-2/8 | WWL2 | 8 | 80 | 1, 4, 5, 6 | WWL2 | 20 | 84 | 1, 5, 7, 8, 9 |
| V1 | 2 | 20 | 1 | V2 | 1 | 4 | 1 | |
| V3 | 2 | 8 | 1 | |||||
| V4 | 1 | 4 | 1 | |||||
| B-1/10 | WWL2 | 9 | 56 | 1, 2, 3, 4, 5 | WWL2 | 3 | 33 | 1 |
| V1 | 4 | 25 | 1, 2 | V1 | 4 | 44 | 1, 3 | |
| V2 | 2 | 13 | 1, 2 | V3 | 1 | 11 | 2 | |
| V3 | 1 | 6 | 1 | V4 | 1 | 11 | 1 | |
| C-1/24 | WWL1 | 3 | 33 | 1 | WWL1 | 6 | 67 | 1, 2 |
| V2 | 3 | 33 | 1, 2 | V1 | 1 | 11 | 1 | |
| V4 | 2 | 23 | 1 | V2 | 1 | 11 | 3 | |
| V5 | 1 | 11 | 1 | V3 | 1 | 11 | 1 | |
| C-2/29 | WWL1 | 5 | 46 | 1 | WWL1 | 8 | 67 | 1, 2 |
| V2 | 2 | 18 | 1 | V2 | 3 | 25 | 2 | |
| V4 | 3 | 27 | 1 | V6 | 1 | 8 | 1 | |
| V5 | 1 | 9 | 1 | |||||
| C-3/34 | WWL1 | 2 | 18 | 1 | ||||
| V2 | 4 | 36 | 1 | V1 | 2 | 22 | 1 | |
| V4 | 1 | 9 | 1 | V2 | 4 | 44 | 1 | |
| V5 | 3 | 27 | 1 | V5 | 1 | 12 | 1 | |
| V6 | 1 | 9 | 1 | V6 | 2 | 22 | 1 | |
| D-1/84 | WWL2 | 2 | 20 | 1, 2 | WWL2 | 3 | 39 | 1, 3, 4 |
| V1 | 1 | 10 | 1 | V4 | 1 | 12 | 1 | |
| V2 | 2 | 20 | 1 | V5 | 1 | 12 | 1 | |
| V3 | 5 | 50 | 1, 2 | V6 | 1 | 12 | 1 | |
| V7 | 1 | 12 | 1 | |||||
| V8 | 1 | 12 | 1 | |||||
| E-1/3 | V1 | 6 | 55 | 1, 2, 3 | ||||
| V2 | 1 | 9 | 1 | |||||
| V3 | 1 | 9 | 1 | |||||
| V4 | 1 | 9 | 1 | |||||
| V5 | 1 | 9 | 1 | |||||
| V6 | 1 | 9 | 1 | |||||
| E-2/13 | WWL2 | 1 | 10 | 1 | ||||
| V1 | 4 | 40 | 2, 4 | |||||
| V3 | 3 | 30 | 1 | |||||
| V7 | 1 | 10 | 1 | |||||
| V8 | 1 | 10 | 1 | |||||
| E-3/16 | V1 | 6 | 55 | 2 | ||||
| V3 | 1 | 9 | 2 | |||||
| V4 | 1 | 9 | 1 | |||||
| V9 | 1 | 9 | 1 | |||||
| V10 | 1 | 9 | 1 | |||||
| V11 | 1 | 9 | 1 | |||||
| E-4/1 (51)c | V12 | 1 | 6 | 1 | ||||
| V13 | 12 | 75 | 1, 2, 3, 4, 5 | |||||
| V14 | 2 | 13 | 1 | |||||
| V15 | 1 | 6 | 1 | |||||
| F-1/2 | V1 | 5 | 42 | 1 | ||||
| V2 | 1 | 8 | 2 | |||||
| V3 | 1 | 8 | 1 | |||||
| V4 | 5 | 42 | 1 | |||||
The name for each NCRR clone indicates whether it is one of the two archetypes (WWL1 and WWL2) or an mrNCRR clone with major deletions and/or insertions. mrNCRR clones for each individual patient are named V1 to Vn (as shown in Fig. 1), where n is a serial number given to each new unique pattern for that patient only and does not indicate the identity of patterns between patients. The percent column indicates the frequency of each mrNCRR clone among all of the clones derived from the specific clinical sample.
Numbers in this column refer to the names of clones with point mutations (single-nucleotide variants) which were found among the particular archetype or mrNCRR clone shown at the left.
Fifty-one months after the first transplant and 1 month after the second transplant.
FIG. 2.
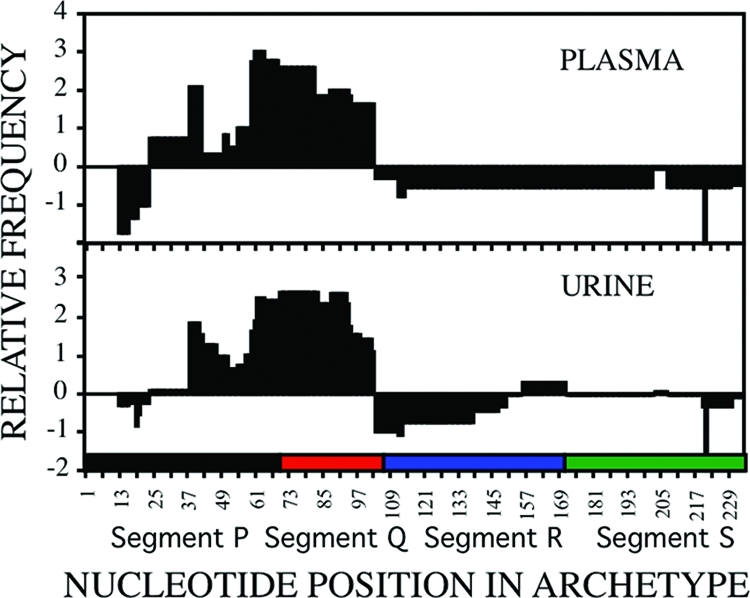
Overrepresentation and underrepresentation of segments of the NCRR in plasma and urine samples as a result of insertions and deletions. For each clinical sample, the insertions for each cloned mrNCRRs were removed in silico, and all that remained of each sequence was aligned to its respective WWL1 or WWL2 archetypes, allowing for large gaps (Sequencher program). The partial or complete segments contained in the insertions that had been removed in silico were then added to the alignment. The number of nucleotides at each position was determined and then normalized as a function of the number of clones that were analyzed for that clinical sample. The shaded areas represent the average normalized value for each nucleotide position for all plasma samples and all urine samples. A value of >0 indicates overrepresentation, a value <0 indicates underrepresentation, and a value of 0 indicates that either there were no insertions or deletions at this position or that the number of insertions was compensated for by the number of deletions.
Unexpectedly for a DNA virus, sequence analyses using eukaryotic DNA polymerase for replication also revealed snvNCRRs among clones with identical WWL1 or WWL2 archetypelike or mrNCRR architectures. The Taq DNA polymerase used in the PCR amplification prior to cloning did not have proofreading ability; however, its estimated misincorporation rate is less than 1 base per 10,000 bases amplified (30, 45; http://www.vivo.colostate.edu/hbooks/genetics/biotech/enzymes/hotpolys.html). It is highly unlikely that Taq polymerase infidelity produced the snvNCRRs, since the entire cloned region amplified with primers BRR1 and BKVP2R that included the NCRR averaged approximately 781 nt in length. Moreover, the same point mutations appeared in mrNCRRs derived from samples taken long periods of time apart. To experimentally exclude the possibility that the large number of snvNCRRs observed was due to Taq polymerase infidelity, we reamplified and subcloned the NCRR from one archetype clone from patient D using the same Taq polymerase and cloning protocol. Analysis of the entire 738-nt sequences (the sequences of the amplicons minus the sequences of the primers) from 10 randomly selected subclones indicated that the snvNCRRs most likely represent true variations in sequences since the sequence of each of the 10 clones was identical to that of the original clone. Further confirmation that the T residue inserted between positions 49 and 50 of segment S of the WWL2-like archetypes and the existence of snvNCRRs were not artifacts was provided by an experiment in which the PCR amplifications of DNA before cloning and after cloning of NCRR DNA from the urine sample taken from patient A 8 months after RT were performed with the Takara Ex Taq enzyme, which has an efficient 3′-5′ exonuclease activity and a mutation rate 4.5 times lower than that of the standard Taq DNA polymerase. Eleven of the 12 clones analyzed had a WWL2-like archetype; i.e., all had a T residue inserted between positions 49 and 50 of the S segment. In addition to an snvNCRR previously found only in plasma, WWL2-5, there were two new snvNCRRs among these 11 archetypes, WWL2-8 and WWL2-9. The T residue was also present in the S segment of the 12th clone, a new mrNCRR, variant V4.
Analyses of clones derived from the 19 clinical samples revealed the simultaneous presence of multiple mrNCRRs and multiple snvNCRR variants in 17. The relative distribution of archetype and unique mrNCRR clones and the number of snvNCRRs for each is presented in Table 2. For patients B, C, and D, all mrNCRR variants were exclusively reorganizations of the specific WW-like NCRR archetype isolated from the respective patients, while for patients A and E, both WW-like archetypes were isolated. For patient A, the WWL1 and WWL2 archetypes were present in the same sample; however, all mrNCRR variants in a subsequent sample were exclusively derived from the WWL2 archetype. In contrast, for patient E, the WWL2 archetype and WWL2-derived mrNCRRs were isolated after the first transplantation, while WWL1-like mrNCRRs were exclusively isolated 1 month after a second RT, more than 3 years later.
The procedure used to amplify and clone NCRRs from urine by PCR with primers BRR1 and BKVP2R (patients A to E) might have amplified products from subgenomic DNA trapped in cells pelleted during ultracentrifugation. The inclusion of unencapsulated subgenomic DNA fragments was much less likely for BKV DNA isolated from cell-free plasma (patients A to D). This raised the question of whether the mrNCRR clones were subgenomic replication intermediates that were never encapsulated and thus irrelevant for the virus phenotype, although the reappearance of the same mrNCRR over long periods of time, when such samples were available, suggested that they were derived from replicating virus. To resolve this question experimentally, in samples taken from patient F, whose urine had a very high viral load, unencapsulated DNA was removed before extraction from intact virions by addition of a DNase digestion step before DNA extraction, and only linearized full-length DNA extracted from the gels after electrophoresis was used for PCR amplification and cloning of the amplified NCRRs. Highly rearranged mrNCRRs but no WWL clones were found in the encapsulated virions, clearly demonstrating that mrNCRRs can be found in intact viral particles.
Phylogenetic analysis of Israeli archetypelike snvNCRRs.
Twenty-two archetypelike NCRR variants were cloned from 7 of 12 urine samples and 6 of 7 plasma samples. Different snvNCRRs were represented among the archetypelike clones from the same patient and even the same clinical sample (Table 2).
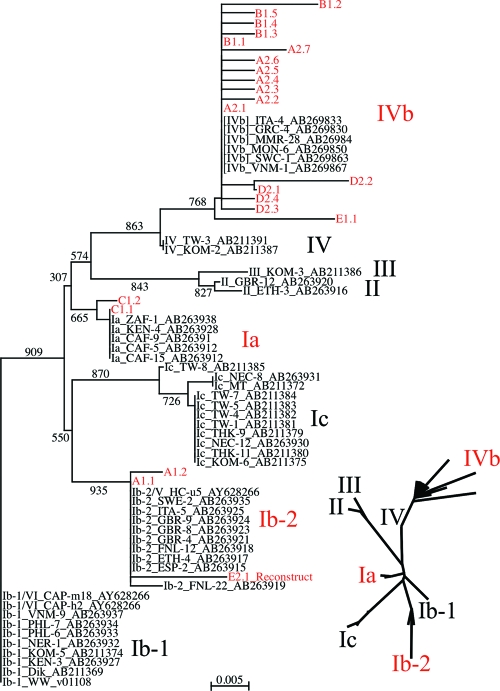
Zheng et al. (49) showed that the full-length BKV genomic sequences in the DDJB/EMBL/GenBank sequence database separated into four major BKV genomic subtypes corresponding to the four serotypes. Their phylogenetic analysis was based on comparison of BKV genomic sequences from which the NCRRs had been deleted in silico, since some sequences had archetypelike NCRRs, while others had mrNCRRs. To determine the genomic subtypes on the basis of the NCRR sequences, we first needed to demonstrate that the archetype NCRR sequences excluded were representative of the portion of the genomes from which they had been excluded. Two phylogenetic trees were generated from the subset of sequences analyzed by Zheng and colleagues (49) that had archetypelike NCRRs and from isolates purported to represent new genotypes, tentatively designated V and VI by Sharma et al. (40). The first tree was generated from full-length sequences from which the NCRRs were excluded in silico (see Fig. S1 in the supplemental material) to re-create a tree like the one generated by Zheng et al. (49), while the second one was generated from the NCRR sequences that had been excluded (see Fig. S2 in the supplemental material). Strain WW sequences were used as the outgroup reference for each of these and subsequent phylogenetic trees. The BKV sequences in both phylogenetic trees segregated into four genotypes. Moreover, comparison of the two trees indicates that all of the BKV isolates that clustered together on the basis of the full-length genomic sequences from which the NCRR sequences had been removed also clustered together when genotypes were formed exclusively on the basis of the archetype NCRR sequences. The tentatively designated type V and VI isolates of Sharma et al. (40) mapped to the previously designated subgroups Ib-2 and Ib-1 (49), respectively. The phylogenetic information is also presented as an unrooted tree in Fig. S1 and S2 in the supplemental material as an insert at the bottom of each figure in unrooted format. The overall branch topography of the NCRR tree was similar but not identical to that of the tree generated from complete genomes with NCRR sequences excised, and the bootstrap values were lower (see Fig. S1 and S2, respectively, in the supplemental material). However, the clear separation between genotypes and even subgenotypes appeared to be sufficient to allow the use of NCRR archetypelike sequences for genotyping. Therefore, a new neighbor-joining phylogenetic tree for archetypal NCRR sequences was prepared; the new tree included the sequences of all snvNCRR variants isolated from the Israeli patients A to E. The in silico reconstruction of archetypelike NCRRs from samples E-4 and F-1 are described in detail in the supplemental material. Seventeen of the Israeli archetype-2-like snvNCRRs clustered into a new genomic subtype most closely related to but well separated from members of genotype IV (data not shown). The DDBJ/EMBL/GenBank database was then searched by using one of these outlying Israeli sequences, and six highly similar archetypelike NCRR sequences that had not yet been assigned a genotype were found. The NCRR archetype tree was then re-created by including all of the sequences in Fig. S2 in the supplemental material and the Israeli NCRR sequences with the six closely related untyped sequences (Fig. 3). We have tentatively assigned the name subgenotype IVb (assuming that the original genotype IV would be redesignated IVa) to this new lineage, since the two archetype-1-like sequence variants tentatively designated V and VI by Sharma et al. (40) clustered within subgenotypes Ia and Ib-2, respectively.
FIG. 3.
Genotyping of Israeli BKV isolates from RT patients on the basis of their archetype NCRR sequences. To form the tree, nonarchetype NCRR sequences were excised in silico from the complete BKV sequences of all isolates, and when necessary, the archetype was reconstructed (see the supplemental material for details). Six additional complete BKV sequences were downloaded from the EMBL database. All archetype NCRR sequences were aligned, the data set was bootstrapped 1,000 times, and a nearest-neighbor phylogenetic tree was generated with the ClustalX program. The tree was visualized by using the njplot program (large image at the bottom right). The WW NCRR sequence was used as the reference outgroup. An unrooted tree was also visualized by using the unrooted program (inserts at the bottom right). For visual clarity, only the names of the genotypes and subgenotypes appear for the unrooted tree. Isolates are designated by their genotype and subgentoype, as described by Zheng et al., (49); the isolate name; and then the EMBL accession number. Israeli archetype isolates are designated by a letter to indicate the patient; the number 1 or 2 to indicate whether the isolate was WWL1 or WWL2, respectively; a period; and a number to indicate the snvNCRR variant. The name subgenotype IVb has been introduced to describe the new subgroup of genotype IV, assuming that the original genotype IV will be renamed IVa.
The WWL1 and WWL2 archetypes were isolated from patients A and E. In patient A, genomic subtypes V (WWL2) and Ib-2 (WWL1) cocirculated, while in patient E, subtype IVb (WWL2) was isolated after the first kidney transplantation and mrNCRR variants of subtype Ib-1 (WWL1) were isolated after the second kidney transplantation.
DISCUSSION
The natural history of BKV infection and reactivation in transplant recipients is poorly understood. Early observations in RT patients with PVAN revealed frequent rearrangements in the NCRR genomic region in BKV isolates recovered from various body compartments, including the urogenital compartment (urine, kidney biopsy specimens) and circulatory compartment (blood, serum, plasma) (34). Highly effective immunosuppressive treatments are probably the main cause for the virus's ability to replicate freely (48). However, early gene expression, replication capacity, cytopathology, and disease progression are influenced by various viral factors, such as the virus genotype and major sequence modifications. Duplications, deletions, and/or modifications that arise during reactivation of BKV may affect regulatory protein binding motifs in the region encompassing the NCRR and the origin of replication (16, 34, 42). These rearrangements may also allow the virus to adapt to ongoing changes within the host cell environment (22).
The six Israeli patients were either at stage B or at stage C of PVAN, as judged by their serum creatinine levels, viral loads, and biopsy results and the loss of the first transplant in patient E (19, 35).
Gosert et al. (16) isolated BKV DNA from clinical samples of RT patients and characterized the genomic sequences of the NCRRs under conditions of sensitivity supposedly able to detect mixtures containing at least 5% of a minority species. They did not find any minority species in their samples. In contrast to the results of Gosert et al. (16), direct sequencing of PCR-amplified NCRRs from total DNA extracted from clinical samples in our study yielded results that indicated the presence of multiple templates of various sizes and sequences. This was proven by cloning, which revealed the presence of BKVs with unique major NCRR rearrangements as well as with archetypelike sequences, as reported by others (42). Altogether, 221 clones from six patients were characterized. Importantly, some mrNCRR and snvNCRR variants appeared in more than one clinical sample from a given individual taken at intervals of several months, suggesting continuous replication over time. In addition, in all patients for whom paired plasma and urine samples were available, some mrNCRR variants appeared in the paired urine and plasma samples, while other variants were unique to each of these different compartments and the frequencies of overrepresented and underrepresented segments of the NCRR were similar but not identical (Table 2; Fig. 2). This observation may indicate either different sites of virus replication in patients with PVAN or a selective transfer of virus from the affected kidney's tissue into the bloodstream. The clinical or biological significance of our observations are currently unclear. Indeed, Gosert et al. (16) and others (34) did not find a correlation of specific rearrangement patterns with disease progression, although Gosert et al. (16) demonstrated an increased replicative capacity and an increase in cytopathology for all of their mrNCRR variants. Our mrNCRR clones will enable us to further study the biological properties of the rearranged and WWL viruses in a tissue culture and/or an animal model system. In this context, it should be noted that a cloned mrNCRR variant obtained after bone marrow transplantation (6) exhibited similarity to an mrNCRR rearrangement found in one of our RT patients.
A large and varying repertoire of BKV variants present in a host at the same time is reminiscent of quasispeciation in RNA viruses, which also employ strategies of recombination and single-nucleotide substitution (15, 17). The surprising number of single-nucleotide-substitution variants among the archetypelike and the mrNCRR variants suggests that the proofreading ability of the eukaryotic host DNA polymerase and/or the excision and repair mechanism became degraded during BKV infection after RT. Support for this generalized degradation comes from two directions. The first is that single-nucleotide variations are not restricted to the NCRR and have been observed in coding regions as well (38, 40; T. T. Perets et al., unpublished data). The second is from studies correlating agnoprotein expression with impaired double-strand-break repair activity, increased chromosome fragmentation, and reduced expression of the Ku70 and Ku80 eukaryotic DNA repair proteins in cells and inhibition of double-strand-break repair activity in cell extracts (8). Polyomaviruses replicate according to the onion skin model, in which the origin of replication (Ori) can be activated multiple times during S phase. DNA synthesis moves bidirectionally away from the Ori, and the separation of daughter molecules is a slow step involving replicative intermediates with at least three chains, nascent DNA and two daughter strands (7). At the initial stage of replication, the three Ori genes are in very close proximity, and this may facilitate major rearrangements through self-recombination. Indeed, a second Ori was found in two of the clones from patient E.
Given the high frequency of single-nucleotide substitutions in the BKV genome (38, 40; Perets et al., unpublished; this report), it is not surprising that several genotypes and subgenotypes have already been reported (for example, see the work of Zheng et al. [49]). A two-step process was required in order to assign the sequences of the Israeli isolates to published BKV genotypes. In the first step, we demonstrated that archetype NCRR-based phylogenetic analysis provided results equivalent to those obtained by phylogenetic analyses based on VP1 or whole-genome sequences (40, 49). Once this had been established (see the supplemental material), we then performed a phylogenetic analysis of the Israeli archetypical NCRR sequences and published genotypes to identify the genotypes and subgenotypes of the Israeli isolates. The phylogenetic analysis of all Israeli archetypelike minor variants revealed that most of the Israel BKV archetypelike isolates did not belong to any of the previously recognized genotypes or subgenotypes reported by Zheng et al. (49) (Fig. 3). They did, however, cluster with a group of six untyped sequences from other countries that had been deposited in the EMBL database after the analysis of Zheng et al. (49) was published. We have tentatively designated this new genetic cluster subgenotype IVb.
The presence of a thymidine residue between positions 49 and 50 of segment S in Israeli WW-like archetype 2 was previously reported for isolates from Ethiopia, the United Kingdom, and Japan. The Israeli isolates were members of subgenotype IVb, whereas those from Ethiopia, the United Kingdom, and Japan belonged to genogroup II or III. Since ancestral sequences are unavailable, it is not possible to determine whether the presence of the T residue represents an insertion or whether its absence represents a deletion. It may be more likely that this single-nucleotide insertion or deletion occurred independently in different genotypes than it is that these isolates from such different genotypes shared a common ancestor in the distant past.
The data of Zheng et al. (49) suggested geographical clustering of BKV genotypes and subgenotypes. There is a growing trend for prospective transplant patients to travel to other countries to procure kidneys and other solid organs due to insufficient local pools of organ donors. Thus, when the risk for disease progression in an individual with a depressed immune system is assessed, it may be necessary to take into account the fact that the patient will be exposed to a previously unencountered genomic subtype rather than to a reactivated genotype that the patient's immune system encountered before it was suppressed. Furthermore, clonal sequence analysis of virus from such patients may shed light on the yet unresolved question of whether reactivation involves a virus already present in the patient, a virus introduced during transplantation, and/or a virus reinfection after transplantation. With this respect, it is important to further assess the association between serotypes and genotypes.
In conclusion, sequence analysis of cloned NCRRs, in contrast to uncloned NCRRs, provided detailed information about the behavior of BKV during reactivation in different body compartments. A high frequency and diversity of replicating variants with NCRR rearrangements were found over long periods of time, which is in contrast to the finding of a predominance of a single mrNCRR reported by Gosert et al. (16). Sequence analysis also revealed a new BKV subgenotype with members in Israel and elsewhere. In addition to the biological importance of our findings, it may become clinically useful if specific subtypes or rearrangements can be linked to a particular disease state, as suggested by Stoner and Hubner (43). Once specific NCRR rearrangements become a hallmark for a high risk of developing PVAN, it may be possible to develop specific tests to detect such rearrangements.
Supplementary Material
Acknowledgments
This research was funded by the Israel Ministry of Health.
Footnotes
Published ahead of print on 4 March 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Acott, P. D., and H. H. Hirsch. 2007. BK virus infection, replication, and diseases in pediatric kidney transplantation. Pediatr. Nephrol. 221243-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agostini, H. T., C. F. Ryschkewitsch, E. J. Singer, and G. L. Stoner. 1997. JC virus regulatory region rearrangements and genotypes in progressive multifocal leukoencephalopathy: two independent aspects of virus variation. J. Gen. Virol. 78(Pt 3)659-664. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, R. R., S. Dagostin, and K. V. Shah. 1989. Detection of BK virus and JC virus in urine and brain tissue by the polymerase chain reaction. J. Clin. Microbiol. 271174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, R. R., K. V. Shah, P. Charache, and R. Saral. 1988. BK and JC virus infections in recipients of bone marrow transplants. J. Infect. Dis. 158563-569. [DOI] [PubMed] [Google Scholar]
- 5.Azzi, A., S. Cesaro, D. Laszlo, K. Zakrzewska, S. Ciappi, R. De Santis, R. Fanci, G. Pesavento, E. Calore, and A. Bosi. 1999. Human polyomavirus BK (BKV) load and haemorrhagic cystitis in bone marrow transplantation patients. J. Clin. Virol. 1479-86. [DOI] [PubMed] [Google Scholar]
- 6.Carr, M. J., G. P. McCormack, K. J. Mutton, and B. Crowley. 2006. Unique BK virus non-coding control region (NCCR) variants in hematopoietic stem cell transplant recipients with and without hemorrhagic cystitis. J. Med. Virol. 78485-493. [DOI] [PubMed] [Google Scholar]
- 7.Cole, C., and S. Conzen. 2001. Polyomaviridae: the viruses and their replication, p. 2141-2174. In D. Knipe, P. Howley, D. Griffin, R. Lamb, M. Martin, B. Roizman, and S. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 8.Darbinyan, A., K. M. Siddiqui, D. Slonina, N. Darbinian, S. Amini, M. K. White, and K. Khalili. 2004. Role of JC virus agnoprotein in DNA repair. J. Virol. 788593-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolei, A., V. Pietropaolo, E. Gomes, C. Di Taranto, M. Ziccheddu, M. A. Spanu, C. Lavorino, M. Manca, and A. M. Degener. 2000. Polyomavirus persistence in lymphocytes: prevalence in lymphocytes from blood donors and healthy personnel of a blood transfusion centre. J. Gen. Virol. 811967-1973. [DOI] [PubMed] [Google Scholar]
- 10.Dorries, K. 2001. Latent and persistent polyomavirua infection, p. 561-584. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss Inc., New York, NY.
- 11.Dorries, K. 1998. Molecular biology and pathogenesis of human polyomavirus infections. Dev. Biol. Stand. 9471-79. [PubMed] [Google Scholar]
- 12.Drachenberg, C. B., H. H. Hirsch, J. C. Papadimitriou, R. Gosert, R. K. Wali, R. Munivenkatappa, J. Nogueira, C. B. Cangro, A. Haririan, S. Mendley, and E. Ramos. 2007. Polyomavirus BK versus JC replication and nephropathy in renal transplant recipients: a prospective evaluation. Transplantation 84323-330. [DOI] [PubMed] [Google Scholar]
- 13.Drachenberg, C. B., J. C. Papadimitriou, H. H. Hirsch, R. Wali, C. Crowder, J. Nogueira, C. B. Cangro, S. Mendley, A. Mian, and E. Ramos. 2004. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am. J. Transplant. 42082-2092. [DOI] [PubMed] [Google Scholar]
- 14.Drachenberg, C. B., J. C. Papadimitriou, and E. Ramos. 2006. Histologic versus molecular diagnosis of BK polyomavirus-associated nephropathy: a shifting paradigm? Clin. J. Am. Soc. Nephrol. 1374-379. [DOI] [PubMed] [Google Scholar]
- 15.Eigen, M. 1993. Viral quasispecies. Sci. Am. 26942-49. [DOI] [PubMed] [Google Scholar]
- 16.Gosert, R., C. H. Rinaldo, G. A. Funk, A. Egli, E. Ramos, C. B. Drachenberg, and H. H. Hirsch. 2008. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 205841-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 748434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haririan, A., E. R. Ramos, C. B. Drachenberg, M. R. Weir, and D. K. Klassen. 2002. Polyomavirus nephropathy in native kidneys of a solitary pancreas transplant recipient. Transplantation 731350-1353. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch, H. H., D. C. Brennan, C. B. Drachenberg, F. Ginevri, J. Gordon, A. P. Limaye, M. J. Mihatsch, V. Nickeleit, E. Ramos, P. Randhawa, R. Shapiro, J. Steiger, M. Suthanthiran, and J. Trofe. 2005. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation 791277-1286. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch, H. H., C. B. Drachenberg, J. Steiger, and E. Ramos. 2006. Polyomavirus-associated nephropathy in renal transplantation: critical issues of screening and management. Adv. Exp. Med. Biol. 577160-173. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch, H. H., W. Knowles, M. Dickenmann, J. Passweg, T. Klimkait, M. J. Mihatsch, and J. Steiger. 2002. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N. Engl. J. Med. 347488-496. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch, H. H., and J. Steiger. 2003. Polyomavirus BK. Lancet Infect. Dis. 3611-623. [DOI] [PubMed] [Google Scholar]
- 23.Hogan, T. F., E. C. Borden, J. A. McBain, B. L. Padgett, and D. L. Walker. 1980. Human polyomavirus infections with JC virus and BK virus in renal transplant patients. Ann. Intern. Med. 92373-378. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, T. F., B. L. Padgett, D. L. Walker, E. C. Borden, and Z. Frias. 1983. Survey of human polyomavirus (JCV, BKV) infections in 139 patients with lung cancer, breast cancer, melanoma, or lymphoma. Prog. Clin. Biol. Res. 105311-324. [PubMed] [Google Scholar]
- 25.Jin, L., P. E. Gibson, W. A. Knowles, and J. P. Clewley. 1993. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J. Med. Virol. 3950-56. [DOI] [PubMed] [Google Scholar]
- 26.Kazory, A., D. Ducloux, J. M. Chalopin, R. Angonin, B. Fontaniere, and H. Moret. 2003. The first case of JC virus allograft nephropathy. Transplantation 761653-1655. [DOI] [PubMed] [Google Scholar]
- 27.Knowles, W. 2001. The epidemiology of BK virus and the occurrence of antigenic and genomic subtypes, p. 527-559. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss Inc., New York, NY.
- 28.Knowles, W. A., D. Pillay, M. A. Johnson, J. F. Hand, and D. W. Brown. 1999. Prevalence of long-term BK and JC excretion in HIV-infected adults and lack of correlation with serological markers. J. Med. Virol. 59474-479. [PubMed] [Google Scholar]
- 29.Le, C. T., G. C. Gray, and S. K. Poddar. 2001. A modified rapid method of nucleic acid isolation from suspension of matured virus: applied in restriction analysis of DNA from an adenovirus prototype strain and a patient isolate. J. Med. Microbiol. 50571-574. [DOI] [PubMed] [Google Scholar]
- 30.Lundberg, K. S., D. D. Shoemaker, M. W. Adams, J. M. Short, J. A. Sorge, and E. J. Mathur. 1991. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene 1081-6. [DOI] [PubMed] [Google Scholar]
- 31.Major, E. 2001. Human polyomaviruses, p. 2175-2196. In D. Knipe, P. Howley, D. Griffin, R. Lamb, M. Martin, B. Roizman, and S. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 32.Markowitz, R. B., H. C. Thompson, J. F. Mueller, J. A. Cohen, and W. S. Dynan. 1993. Incidence of BK virus and JC virus viruria in human immunodeficiency virus-infected and -uninfected subjects. J. Infect. Dis. 16713-20. [DOI] [PubMed] [Google Scholar]
- 33.Moens, U., T. Johansen, J. I. Johnsen, O. M. Seternes, and T. Traavik. 1995. Noncoding control region of naturally occurring BK virus variants: sequence comparison and functional analysis. Virus Genes 10261-275. [DOI] [PubMed] [Google Scholar]
- 34.Moens, U., and M. Van Ghelue. 2005. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology 331209-231. [DOI] [PubMed] [Google Scholar]
- 35.Nickeleit, V., and M. J. Mihatsch. 2006. Polyomavirus nephropathy in native kidneys and renal allografts: an update on an escalating threat. Transplant Int. 19960-973. [DOI] [PubMed] [Google Scholar]
- 36.Nishimoto, Y., H. Y. Zheng, S. Zhong, H. Ikegaya, Q. Chen, C. Sugimoto, T. Kitamura, and Y. Yogo. 2007. An Asian origin for subtype IV BK virus based on phylogenetic analysis. J. Mol. Evol. 65103-111. [DOI] [PubMed] [Google Scholar]
- 37.Randhawa, P., and D. C. Brennan. 2006. BK virus infection in transplant recipients: an overview and update. Am. J. Transplant. 62000-2005. [DOI] [PubMed] [Google Scholar]
- 38.Randhawa, P. S., K. Khaleel-Ur-Rehman, P. A. Swalsky, A. Vats, V. Scantlebury, R. Shapiro, and S. Finkelstein. 2002. DNA sequencing of viral capsid protein VP-1 region in patients with BK virus interstitial nephritis. Transplantation 731090-1094. [DOI] [PubMed] [Google Scholar]
- 39.Shah, K. V. 1996. Polyomavirses, p. 2027-2043. In D. M. Knipe, B. Roizman, P. M. Howley, T. P. Monath, S. E. Straus, R. M. Chanock, J. L. Melnick, and B. N. Fields (ed.), Fields virology, vol. 2, 3rd ed. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 40.Sharma, P. M., G. Gupta, A. Vats, R. Shapiro, and P. Randhawa. 2006. Phylogenetic analysis of polyomavirus BK sequences. J. Virol. 808869-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shitrit, D., L. Nirit, S. I. Shiran, G. Izbicki, D. Sofer, M. Eldad, and M. R. Kramer. 2003. Progressive multifocal leukoencephalopathy in a lung transplant recipient. J. Heart Lung Transplant. 22946-950. [DOI] [PubMed] [Google Scholar]
- 42.Stoner, G. L., R. Alappan, D. V. Jobes, C. F. Ryschkewitsch, and M. L. Landry. 2002. BK virus regulatory region rearrangements in brain and cerebrospinal fluid from a leukemia patient with tubulointerstitial nephritis and meningoencephalitis. Am. J. Kidney Dis. 391102-1112. [DOI] [PubMed] [Google Scholar]
- 43.Stoner, G. L., and R. Hubner. 2001. The human polyomaviruses: past, present, and future, p. 611-663. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. Wiley-Liss Inc., New York, NY.
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tindall, K. R., and T. A. Kunkel. 1988. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry 276008-6013. [DOI] [PubMed] [Google Scholar]
- 46.Vats, A., R. Shapiro, P. Singh Randhawa, V. Scantlebury, A. Tuzuner, M. Saxena, M. L. Moritz, T. J. Beattie, T. Gonwa, M. D. Green, and D. Ellis. 2003. Quantitative viral load monitoring and cidofovir therapy for the management of BK virus-associated nephropathy in children and adults. Transplantation 75105-112. [DOI] [PubMed] [Google Scholar]
- 47.Wadei, H. M., A. D. Rule, M. Lewin, A. S. Mahale, H. A. Khamash, T. R. Schwab, J. M. Gloor, S. C. Textor, M. E. Fidler, D. J. Lager, T. S. Larson, M. D. Stegall, F. G. Cosio, and M. D. Griffin. 2006. Kidney transplant function and histological clearance of virus following diagnosis of polyomavirus-associated nephropathy (PVAN). Am. J. Transplant. 61025-1032. [DOI] [PubMed] [Google Scholar]
- 48.Wu, C., P. Randhawa, and J. McCauley. 2006. Transplantation: polyomavirus nephropathy and the risk of specific immunosuppression regimens. Sci. World J., p.512-528. [DOI] [PMC free article] [PubMed]
- 49.Zheng, H. Y., Y. Nishimoto, Q. Chen, M. Hasegawa, S. Zhong, H. Ikegaya, N. Ohno, C. Sugimoto, T. Takasaka, T. Kitamura, and Y. Yogo. 2007. Relationships between BK virus lineages and human populations. Microbes Infect. 9204-213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.