Abstract
Burkholderia gladioli, primarily known as a plant pathogen, is involved in human infections, especially in patients with cystic fibrosis (CF). In the present study, the first respiratory isolates recovered from 14 French patients with CF and 4 French patients without CF, identified by 16S rRNA gene analysis, were tested for growth on B. cepacia selective media, for identification by commercial systems, and for their antimicrobial susceptibilities, and were compared by pulsed-field gel electrophoresis (PFGE). Patients' data were collected. All 18 isolates grew on oxidation-fermentation-polymyxin B-bacitracin-lactose medium and Pseudomonas cepacia agar, but only 13 grew on Burkholderia cepacia selective agar. API 20NE strips did not differentiate B. gladioli from B. cepacia, whereas Vitek 2 GN cards correctly identified 15 isolates. All isolates were susceptible to piperacillin, imipenem, aminoglycosides, and ciprofloxacin and were far less resistant to ticarcillin than B. cepacia complex organisms. Fifteen PFGE types were observed among the 18 isolates, but shared types were not identified among epidemiologically related patients. The microbiological follow-up of CF patients showed that colonization was persistent in 3 of 13 documented cases; B. gladioli was isolated from posttransplantation cultures of blood from 1 patient. Among the patients without CF, B. gladioli was associated with intubation (three cases) or bronchiectasis (one case). In summary, the inclusion of B. gladioli in the databases of commercial identification systems should improve the diagnostic capabilities of those systems. In CF patients, this organism is more frequently involved in transient infections than in chronic infections, but it may be responsible for complications posttransplantation; patient-to-patient transmission has not been demonstrated to date. Lastly, B. gladioli appears to be naturally susceptible to aminoglycosides and ciprofloxacin, although resistant isolates may emerge in the course of chronic infections.
Burkholderia gladioli is mainly known as a plant pathogen; and three phytopathogenic pathovars have been characterized: B. gladioli pathovar gladioli (formerly Pseudomonas marginata), which causes a leaf and corn disease in gladioli and irises; B. gladioli pathovar alliicola (formerly Pseudomonas alliicola), which causes onion bulb rot; and B. gladioli pathovar agaricicola, which causes soft rot in the mushroom Agaricus bitorquis (1, 20). A fourth pathovar, B. gladioli pathovar cocovenenans, has recently been proposed for the former species Burkholderia (Pseudomonas) cocovenenans, previously shown to be a junior synonym of B. gladioli (9), on the basis of its production of lethal toxins responsible for severe food poisoning in China (14). Lastly, Pseudomonas antimicrobica, isolated from the mealy bug and antagonistic to fungi pathogenic for plants, such as Botrytis cinerea, has also been reclassified as B. gladioli (8).
The first cases of respiratory tract infections due to B. gladioli were reported in 1989 in patients with cystic fibrosis (CF) (5). This organism was further described in patients with chronic granulomatous disease (23) or underlying immunocompromise (12, 25). The spectrum of infections caused by B. gladioli includes respiratory tract infections (5, 12, 23), septicemia (12, 23, 25), abscesses (15), osteomyelitis (4), keratitis (19, 22), and adenitis (12). In contrast to the closely related Burkholderia cepacia complex species, B. gladioli was not initially considered to be virulent in patients with CF (5); but it was later associated with acute respiratory tract infections (2), abscesses (15), posttransplantation septicemia (16, 18), and lymph node infections (17). A recent review of 30 colonized CF patients in North Carolina hospitals demonstrated that infection was transient in 60% of cases (17). To date, person-to-person transmission of B. gladioli has not been reported and the presumed cross-infection of six patients with the same strain described by Wilsher et al. in New Zealand in 1997 (32) was later attributed to the ET12 strain of Burkholderia cenocepacia (7). These findings highlight the risk of misidentification when phenotypic methods are used. Molecular identification of B. gladioli is mainly based on rRNA gene-specific signatures and can be performed by means of species-specific PCR (3, 6, 31), amplified rRNA gene restriction analysis (ARDRA) (24), or sequencing of the 16S rRNA gene (11). The aim of the present study was to assess widely used microbiological tools for the recovery and identification of B. gladioli from respiratory specimens, to test the antimicrobial susceptibilities and genetic diversity of clinical isolates, and to analyze the epidemiology of respiratory colonization with this species.
MATERIALS AND METHODS
Bacterial strains.
The present study included 18 Burkholderia primary clinical isolates recovered from sputum samples and sent to the Observatoire Burkholderia cepacia for analysis between 1996 and 2005, as well as the 4 following reference strains: the pathovar reference strains of B. gladioli pathovar gladioli (ATCC 10248) and pathovar alliicola (ATCC 19302), which were obtained from the Collection Française des Bactéries Phytopathogènes (Angers, France), and the strains deposited as the type strains of Burkholderia cocovenenans (ATCC 33664) and Pseudomonas antimicrobica (ATCC 49839), both of which have been transferred to the species B. gladioli and which were purchased from the Laboratorium voor Microbiologie, Ghent, Belgium.
ARDRA identification.
ARDRA was performed as described previously (24) with five restriction enzymes, i.e., AluI, CfoI, DdeI, MspI, and XmnI.
Growth tests on Burkholderia cepacia selective media.
Three selective media were tested: oxidation-fermentation-polymyxin B-bacitracin-lactose (OFPBL) (30); Pseudomonas cepacia agar (PCA), purchased from AES, Combourg, France; and Burkholderia cepacia selective agar (BCSA), kindly provided by bioMérieux (Marcy l'Etoile, France). Growth was checked after 24, 48, and 72 h at 37°C.
Biochemical testing.
Two commercial identification systems, i.e., API 20NE strips and Vitek 2 GN cards, were used. Inoculation was performed according to the manufacturer's recommendations (bioMérieux).
Antimicrobial susceptibility testing.
The antibiotic susceptibilities of the 18 clinical isolates were determined by the disk diffusion test, according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM). The inoculum suspension was swabbed on Mueller-Hinton agar (bioMérieux). Disks (Bio-Rad, Marnes-la-Coquette, France) containing the following antibiotics were tested: ticarcillin (75 μg), ticarcillin-clavulanic acid (75/10 μg), piperacillin (75 μg), piperacillin-tazobactam (75/10 μg), ceftazidime (30 μg), cefepime (30 μg), aztreonam (30 μg), imipenem (10 μg), pefloxacin (5 μg), ciprofloxacin (5 μg), gentamicin (15 μg), tobramycin (10 μg), amikacin (30 μg), and colistin (50 μg). In addition, the MICs of ticarcillin, ticarcillin-clavulanic acid, piperacillin, ceftazidime, and gentamicin were measured by the agar dilution technique with Mueller-Hinton agar and a Steers-Foltz replicator, which delivered 104 CFU per spot. Escherichia coli CIP 76.24 and Pseudomonas aeruginosa CIP 76.110 were included as quality controls (10). As there are no specific breakpoints for B. gladioli in the CA-SFM guidelines, general recommendations or specific recommendations for B. cepacia, when they existed, were applied to the interpretation of the susceptibility tests.
PFGE typing.
Genotyping was performed with the 18 clinical isolates. Isolates recovered from sputum and a culture of blood from one patient were compared. The macrorestriction pattern of total DNA was determined by PFGE (CHEF-DRIII apparatus; Bio-Rad, Ivry sur Seine, France) with XbaI, as described previously (27). GelCompar software (Applied Maths, Kortrijk, Belgium) was used to establish a DNA similarity matrix on the basis of the Dice coefficient (two-by-two strain comparisons). The dendrogram was constructed by using the unweighted-pair-group method of arithmetic averages clustering algorithm with the Dice coefficient. To ensure that the gels were comparable, Staphylococcus aureus NCTC 8325 was included as a reference strain. Isolates with indistinguishable PFGE patterns were assigned to the same clone and clonal variant. Strains that differed by up to (and including) six bands were considered to belong to different clonal variants; strains that differed by more than six bands were considered to belong to different clones (29). Clones were designated by letters, and clonal variants were designated by suffix numbers.
Epidemiological data.
The sex, age, and sputum microbiology at the time of isolate acquisition, as well as the available clinical data, were recorded for each colonized CF patient and each patient without CF. The persistence of infection in the CF patients was evaluated from the annual surveillance of Burkholderia infections by the Observatoire Burkholderia cepacia: patients were considered persistently infected if they were declared B. gladioli positive for at least for 3 subsequent years and with at least four positive cultures per year, transiently infected if they were declared B. gladioli positive only once and culture negative thereafter (minimal duration of follow-up after the primary colonization, 2 years), and intermittently infected if they were declared B. gladioli positive at least twice with zero to two positive cultures per year. In all cases, sputum was cultured at least four times a year.
RESULTS
Identification by ARDRA.
Two ARDRA profiles that differed by their DdeI restriction patterns were observed among the 4 reference strains and 17 of the 18 clinical isolates (Table 1). The last clinical isolate, which differed by its MspI restriction pattern, was confirmed to be B. gladioli by Peter Vandamme (Laboratorium voor Microbiologie). These profiles are easily distinguishable from the profiles observed within the B. cepacia complex (24).
TABLE 1.
ARDRA profiles of 4 reference strains and 18 clinical isolates of B. gladioli
| Reference strain(s) (species) exhibiting each ARDRA profilea | No. of clinical isolates exhibiting each ARDRA profile/total no. tested | ARDRA profile
|
||||
|---|---|---|---|---|---|---|
| AluI | CfoI | DdeI | MspI | XmnI | ||
| CFBP 2427 (ATCC 10248) (B. gladioli pathovar gladioli), LMG 11626 (ATCC 33664) (“Pseudomonas cocovenenans”), and LMG 18920 (ATCC 49839) (“Pseudomonas antimicrobica”) | 9/18 | B | B | B | B | A |
| CFBP 2422 (ATCC 19302) (B. gladioli pathovar alliicola) | 8/18 | B | B | F | B | A |
| No reference strain | 1/18 | B | B | F | Z | A |
CFBP, Collection Française de Bactéries Phytopathogènes (Angers, France); LMG, Laboratorium voor Microbiologie (Ghent, Belgium); ATCC, American Type Culture Collection (Manassas, VA).
Growth tests on Burkholderia cepacia selective media and biochemical testing.
Good growth on OFPBL medium and PCA was observed for the 18 clinical isolates and the 4 reference strains after 24 or 48 h at 37°C. There was poor growth of B. gladioli on BCSA after 72 h at 37°C for 13 of the 18 clinical isolates and no growth on BCSA for 5 clinical isolates and the 4 reference strains tested. Eighteen of the 22 strains were identified as B. cepacia by use of the API 20NE system (likelihood, ≥98%) with the mention “possible B. gladioli”; 3 clinical isolates were unidentified, and 1 reference strain was misidentified as Pseudomonas fluorescens (likelihood, 80.7%). The Vitek 2 GN cards provided an accurate identification to the species level for 15 of the 18 clinical isolates and 3 of the 4 reference strains; 3 clinical isolates were misidentified: 2 as P. fluorescens (likelihoods, 96.63 and 94.69%, respectively) and 1 as Pseudomonas oryzhihabitans (likelihood, 87.14%); for 1 reference strain, supplemental testing was required to differentiate B. gladioli from P. fluorescens.
Antimicrobial susceptibility.
By use of the disk diffusion test, all isolates were demonstrated to be susceptible to piperacillin, imipenem, ciprofloxacin, and aminoglycosides and resistant to colistin. Of the 18 clinical isolates, 9 were susceptible and 9 were intermediately susceptible to ticarcillin, whereas all isolates were susceptible to ticarcillin-clavulanic acid. Eight isolates were resistant to ceftazidime, 10 were resistant to cefepime, and 13 were resistant to aztreonam. The MICs of ticarcillin, ticarcillin-clavulanic acid, piperacillin, ceftazidime, and gentamicin ranged from 8 to 64 mg/liter, 4/2 to 32/2 mg/liter, <2 to 4 mg/liter, and < 0.5 to 2 mg/liter, respectively. Among the β-lactams tested, piperacillin exhibited the best antimicrobial activity, since all clinical isolates were inhibited at a concentration of 4 mg/liter, whereas ceftazidime exhibited moderate and variable activity. The distribution of the MICs for the clinical isolates is shown in Fig. 1.
FIG. 1.
Distribution of the MICs of three ß-lactams and gentamicin among the 18 clinical isolates. The antibiotic susceptibility breakpoints (in mg/liter) of the 18 clinical isolates are according to the recommendations of CA-SFM. BC, specific guidelines for B. cepacia, when they exist; S, susceptible; I, intermediate; R, resistant.
PFGE typing.
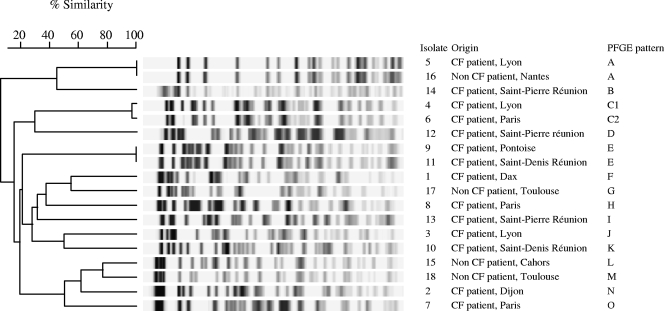
Fifteen PFGE types were observed among the 18 clinical isolates (Fig. 2). Three pairs of isolates recovered from epidemiologically unrelated patients (i.e., isolates 5 and 16, isolates 4 and 6, and isolates 9 and 11) shared the same PFGE types (types A, C, and E, respectively), whereas isolates recovered from epidemiologically related patients (i.e., isolates 3 and 4; isolates 6 and 7; isolates 10 and 11; and isolates 12, 13, and 14) were genetically unrelated.
FIG. 2.
PFGE dendrogram with the 18 clinical isolates constructed by using the unweighted-pair-group method of arithmetic averages algorithm.
Epidemiological data.
The clinical isolates were recovered from the sputum of 14 patients with CF (8 males, 6 females) and 4 patients without CF (2 males, 2 females) (Table 2). Among the 14 CF patients, the mean age at the time of acquisition of B. gladioli was 14 ± 10 years (range, 5 to 42 years). Complete microbiological data were available for 13 of the 14 CF patients; B. gladioli was the sole pathogen isolated from 2 patients, whereas the other patients were coinfected with S. aureus (nine cases) and/or other gram-negative nonfermenters (B. cenocepacia, one case; Stenotrophomonas maltophilia and Achromobacter xylosoxidans, one case; P. aeruginosa, one case). It was possible to evaluate the persistence of infection in only 13 of the 14 patients, since patient 1, who was chronically colonized with B. cepacia, died 1 year after the first recovery of B. gladioli. Colonization was transient in 8 of 13 patients, intermittent in 2 patients, and persistent in 3 patients. The three persistently infected patients had at least five positive cultures per year for at least 4 years. One patient who was transiently colonized with B. gladioli became chronically colonized with B. multivorans. Lastly, patient 6, who was persistently colonized with B. gladioli from 1996 at the age of 12 years, underwent a first bilateral lung transplantation in 2002. Sputum specimens obtained posttransplantation were B. gladioli negative, except for once at month 38. Due to the occurrence of bronchiolitis obliterans syndrome, a second transplantation was performed in 2007, at month 51. Fourteen days after a difficult immediate postoperative course, the patient developed bilateral pneumonia, and B. gladioli was recovered from a bronchoalveolar lavage fluid specimen and from two blood cultures on days 18 and 24. Central venous and arterial catheters were removed, and the bloodstream infection was successfully treated with a combination of piperacillin-tazobactam and tobramycin. Nevertheless, bronchial aspirates and bronchoalveolar lavage samples persistently grew B. gladioli. At month 3.5 posttransplantation, fiberoptic bronchoscopy demonstrated ulceronecrotic infiltration of the right anterior tracheal wall. Computed tomography of the chest disclosed an increasing and then compressive mediastinal mass which was shown by mediastinoscopy to be an abscess, which was culture positive for B. gladioli. Blood cultures remained negative. Transient clinical improvement was obtained during antibiotic treatment that included temocillin, piperacillin-tazobactam, tobramycin, rifampin (rifampicin), and co-trimoxazole; but the patient finally died at month 6 posttransplantation from uncontrolled pulmonary and mediastinal B. gladioli infection. Two respiratory isolates recovered in 2002 before the first transplantation were compared to the blood culture isolate recovered in 2007 by means of PFGE. The two respiratory isolates exhibited different PFGE types, and the PFGE type of the blood culture isolate was identical to the PFGE type of one of those isolates.
TABLE 2.
Epidemiological characteristics of 18 cases of respiratory tract colonization with B. gladioli
| Patient | Origin in France | Sexa | Clinical background | First recovery
|
|
|---|---|---|---|---|---|
| Age (yr) | Associated pathogen(s) | ||||
| 1 | Centre Hospitalier, Dax | F | CF | 8 | B. cenocepacia |
| 2 | Hôpital du Bocage, Dijon | M | CF | 8 | S. maltophilia, A. xylosoxidans |
| 3 | Hôpital Debrousse, Lyon | M | CF | 16 | S. aureus, P. aeruginosa |
| 4 | Hôpital Debrousse, Lyon | M | CF | 8 | S. aureus, Haemophilus influenzae |
| 5 | Hôpital Lyon Sud, Pierre-Bénite | F | CF | 28 | S. aureus |
| 6 | Centre Hélio-Marin, Saint-Trojan Hôpital Robert-Debré, Paris Hôpital Foch, Suresnes | F | CF | 12 | S. aureus |
| 7 | Hôpital Robert-Debré, Paris | F | CF | 14 | None |
| 8 | Hôpital Cochin, Paris | M | CF | 42 | None |
| 9 | Hôpital René-Dubos, Pontoise | F | CF | 9 | NDb |
| 10 | Hôpital d'Enfants, Saint-Denis de la Réunion | M | CF | 5 | S. aureus |
| 11 | Hôpital d'Enfants, Saint-Denis de la Réunion | M | CF | 19 | S. aureus, Aspergillus fumigatus |
| 12 | Centre Hospitalier, Saint-Pierre de la Réunion | M | CF | 7 | S. aureus |
| 13 | Centre Hospitalier, Saint-Pierre de la Réunion | M | CF | 7 | S. aureus |
| 14 | Centre Hospitalier, Saint-Pierre de la Réunion | F | CF | 12 | S. aureus |
| 15 | Centre Hospitalier, Cahors | M | No CF, intubation | 71 | None |
| 16 | Centre Hospitalier, Nantes | F | No CF, bronchiectasis | 75 | None |
| 17 | Hôpital Rangueil, Toulouse | F | No CF, intubation | 77 | None |
| 18 | Hôpital Rangueil, Toulouse | M | No CF, intubation | 37 | None |
F, female; M, male.
ND, not documented.
The mean age of the four patients without CF was 65 ± 18 years (age range, 37 to 77 years), and B. gladioli was the sole pathogen isolated. Three patients (patients 15, 17, and 18) were intubated; and one presented with bronchiectasis. For patient 15, who had a severe traumatic thorax injury with hemoperitoneum, the recovery of B. gladioli from bronchial aspirates was associated with marked respiratory symptoms, fever, and an elevated C-reactive protein level and white blood cell count. On the contrary, in patients 17 and 18, colonization with B. gladioli was asymptomatic.
DISCUSSION
Burkholderia gladioli is a colistin-resistant organism mainly isolated from the respiratory tract, especially in patients with CF. In these patients, it may be responsible for acute respiratory infections and posttransplantation bloodstream infections, but in contrast to B. cepacia complex organisms, it has so far not been involved in patient-to-patient transmission. It is thus important for the clinical microbiology laboratory to ensure the efficient recovery as well as the accurate identification of B. gladioli.
In the present study of 18 clinical isolates and 4 reference strains, we showed that B. gladioli grew poorly on BCSA but grew well on PCA and OFPBL medium. This is in agreement with the findings of a previous study of Henry et al., in which clinical specimens were inoculated on the same media: it was shown that BCSA allowed the growth of B. gladioli from specimens from only one of the five positive cases (13). The gentamicin susceptibilities of the 18 clinical isolates tested in the present study (maximal MIC, 2 mg/liter) probably explains their low level of recovery on BCSA, which contains gentamicin (10 mg/liter) as an inhibitory agent. The good performance of PCA observed in our study is more surprising, since this medium contains ticarcillin (100 mg/liter), which was shown to be moderately active against B. gladioli, with MICs ranging from 8 to 64 mg/liter. Moreover, PCA did not perform better than BCSA in the study of Henry et al. (13). In conclusion, the use of BCSA for the recovery of B. cepacia complex organisms from respiratory specimens from patients with CF might lead to an underestimation of the rate of colonization with B. gladioli; OFPBL can be considered the best medium for the recovery of B. gladioli, and further tests based on the inoculation of a larger number of B. gladioli-positive sputum specimens are required to validate the use of PCA for the recovery of this organism.
Molecular identification of B. gladioli mostly relies on widely used 16S rRNA gene-based methods (ARDRA or sequencing), which are sufficient to distinguish this species from members of the B. cepacia complex, given the differences in their rrs gene sequences (28). However, these methods are not available in all clinical microbiology laboratories, and if they are available, they are generally used as second-line techniques, when the phenotypic identification is unsatisfactory. The biochemical tests included in the API 20NE strip do not allow the differentiation of B. cepacia and B. gladioli, and 18 of the 22 strains tested were identified as B. cepacia. On the contrary, the Vitek 2 system readily identified 18 strains (15 of 18 clinical isolates and 3 of 4 reference strains) to the species level, with no misidentifications as B. cepacia occurring. The most common misidentification was P. fluorescens (in one case with the API 20NE system and in two cases with Vitek 2 GN cards), but colistin resistance eliminated this misidentification. In contrast to the Vitek 2 system, the Phoenix (BD Diagnostic Systems, Sparks, MD) and MicroScan WalkAway (Dade Behring, West Sacramento, CA) automated microbiology systems do not include B. gladioli in their databases to date; Snyder et al. reported the identification of one strain of B. gladioli as Acinetobacter baumannii by the MicroScan system and a Burkholderia or a Ralstonia sp. by the Phoenix system (26). In conclusion, the Vitek 2 GN card is efficient for the routine identification of B. gladioli, provided that coherence with the antibiotic susceptibility profile is checked.
The 18 primary isolates of B. gladioli tested in the present study were far less resistant to antibiotic compounds than B. cepacia complex isolates. Ticarcillin was shown to be moderately active, and the combination of ticarcillin and clavulanic acid displayed slightly decreased MICs. All isolates were found to be susceptible to piperacillin, imipenem, aminoglycosides, and ciprofloxacin; but at least 50% of the isolates were resistant to ceftazidime, cefepime, or aztreonam. Nevertheless, analysis of the antimicrobial susceptibilities of sequential isolates recovered from the three persistently infected CF patients showed the appearance of aminoglycoside resistance in all three patients and ciprofloxacin resistance in two of the three patients (data not shown). Genotyping of serial isolates from the same patient would be necessary to determine whether resistant and susceptible isolates belong to the same clone. Interestingly, in patient 6, two different respiratory isolates were identified by PFGE, but the susceptible blood isolate was identical to a resistant respiratory isolate. Jones et al. (15) previously reported that the strain involved in multiple abscesses in a CF patient who received several intravenous courses of antibiotics, including tobramycin, became resistant. These observations suggest that in contrast to B. cepacia complex organisms, B. gladioli is naturally susceptible to aminoglycosides but that acquired resistance may occur.
Few reports of the genotypic analysis of multiple isolates recovered in the same care unit have been published. Four CF isolates from the same children's CF center compared by randomly amplified polymorphic DNA analysis (13) exhibited unique patterns, as did 17 isolates from patients with CF and 1 isolate from a patient without CF in the same pulmonary medicine department compared by using XbaI and SpeI macrorestriction analysis by PFGE (17). Conversely, isolates recovered from two patients with corneal ulcers in the same department of ophthalmology displayed identical SpeI macrorestriction profiles, but the patients were unrelated and a common source of infection could not be identified (19). In the present study, 18 respiratory isolates recovered from patients with CF and patients without CF in 12 different care units were compared by XbaI macrorestriction analysis. In four of the participating CF centers, two to three patients were colonized with B. gladioli, but the strains from patients within the same CF center were genetically distinct. Thus, colonization of CF patients seems to be due to independent acquisition from the natural environment. It is interesting to note that of our 14 CF patients, 5 lived on Reunion Island, which suggests that B. gladioli might be more abundant in the natural environment of this overseas department that on the European continent. On the contrary, strains with identical profiles were observed in geographically unrelated patients, even in one patient from Reunion Island and one patient from the continent. The genotypic diversity within the species is unknown so far, and our results suggest that identical genotypic fingerprints do not constitute unchallengeable evidence of epidemiological relatedness.
Comparison of our national data to those reported by Kennedy et al. (17) in a CF and lung transplantation center suggests the similar epidemiology of B. gladioli in French and North American CF patients with regard to the mean age at the time of primary colonization (14 ± 10 and 15.4 ± 6 years, respectively), the high prevalence of S. aureus cocolonization (9/13 patients [69%] and 30/33 patients [91%], respectively), and the transient feature of most colonizations (8/13 patients [61.5%] and 18/30 patients [60%], respectively). Nevertheless, only 1 of our 13 documented cases (7.7%) was infected with P. aeruginosa at the time of B. gladioli acquisition, whereas Kennedy et al. (17) reported coinfection in 21 of 33 patients (64%).
The performance of lung transplantation in CF patients harboring B. gladioli is debated, due to the risk of complications posttransplantation, such as abscesses, bacteremia, or mediastinal lymph node infection (16, 17, 18). Kanj et al. (16) and Kennedy et al. (17) reported the occurrence of infectious complications in two of two and one of three B. gladioli-colonized CF lung transplant recipients, respectively; but these infections were successfully treated. In our study, the only lung transplant recipient developed a bloodstream infection on day 18 posttransplantation and died from progressive ongoing B. gladioli mediastinal and pulmonary infection at month 6 posttransplantation. Finally, in a study of a large cohort of Burkholderia-infected patients, Murray et al. (21) demonstrated that the 14 B. gladioli-positive patients had higher rates of mortality posttransplantation than B. multivorans-positive patients or uninfected patients, but the cause of death was not specified.
Respiratory tract infections with B. gladioli have also rarely been reported in patients without CF: two patients with chronic granulomatous disease (23), one patient with primary ciliary dyskinesia, and one mechanically ventilated patient (17). In the present study, this organism was recovered from three intubated patients and one patient with bronchiectasis. Thus, mechanical ventilation and chronic pulmonary diseases might be risk factors for respiratory infection with B. gladioli. The occurrence of such infections is infrequent, but they may be increasingly recognized, due to improvements in the methods of microbiological diagnosis.
In summary, the inclusion of B. gladioli in the databases of commercial identification systems, such as the Vitek 2 system, will probably improve the reliability of the microbiological diagnosis of infections with this species. Nevertheless, efficient recovery from the sputum of patients with CF requires the use of a gentamicin-free selective medium, due to the usual susceptibility of primary isolates to aminoglycosides. The antibiotic susceptibility pattern of B. gladioli, with the exception of colistin resistance, is quite different from that of B. cepacia complex organisms; and the susceptibilities to aminoglycosides, imipenem, and ticarcillin-clavulanic acid are useful characteristics for use in the differentiation of the two species. Nevertheless, aminoglycoside-resistant isolates may appear in the course of chronic infections. In agreement with the findings of the previous study of Kennedy et al. (17) in a North American pulmonary medicine department, the present national French study demonstrated that most, but not all, colonizations with B. gladioli in CF patients are transient and that in contrast to strains belonging to the closely related B. cepacia complex, this organism does not seem to be acquired by patient-to-patient transmission. The risk of bloodstream infection and/or abscesses posttransplantation was confirmed in the present study, with bacteremia and a mediastinal abscess occurring in the only lung transplant recipient in our cohort. Finally, B. gladioli may also be involved in respiratory infections in patients without CF, especially in those who are intubated.
Acknowledgments
We thank the following individuals from the indicated cities in France for sending clinical B. gladioli isolates: E. Bingen, Paris; J. Caillon, Nantes; J. Carrere, Giens; M. de Montclos, Lyon; C. Denoix, Saint-Denis Réunion; J.-M. Duez, Dijon; A.-M. Freydière, Lyon; P. Honderlick, Suresnes; J.-P. Lafargue, Dax; F. Landais, Saint-Trojan; G. Paul, Paris; S. Picot, Saint-Pierre Réunion; and M. Thibault, Pontoise. We acknowledge the Observatoire Burkholderia cepacia network for providing clinical and biological information. We thank Nicola Coley for revising the manuscript.
We are indebted to Vaincre la Mucoviscidose (Paris, France) for financial support.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Ballard, R., N. Palleroni, M. Doudoroff, R. Stanier, and M. Mandel. 1970. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J. Gen. Microbiol. 60199-214. [DOI] [PubMed] [Google Scholar]
- 2.Barker, P., R. Wood, and P. Gilligan. 1997. Lung infection with Burkholderia gladioli in a child with cystic fibrosis: acute clinical and spirometric deterioration. Pediatr. Pulmonol. 23123-125. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1998. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J. Clin. Microbiol. 362748-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyanton, B., L. Noroski, H. Reddy, M. Dishop, M. Hicks, J. Versalovic, and E. Moylett. 2005. Burkholderia gladioli osteomyelitis in association with chronic granulomatous disease: case report and review. Pediatr. Infect. Dis. J. 24837-839. [DOI] [PubMed] [Google Scholar]
- 5.Christenson, J. C., D. F. Welch, G. Mukwaya, M. J. Muszynski, R. E. Weaver, and D. J. Brenner. 1989. Recovery of Pseudomonas gladioli from respiratory tract specimens of patients with cystic fibrosis. J. Clin. Microbiol. 27270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clode, F., M. Kaufmann, H. Malnick, and T. Pitt. 1999. Evaluation of three oligonucleotide primer sets in PCR for the identification of Burkholderia cepacia and their differentiation from Burkholderia gladioli. J. Clin. Pathol. 52173-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clode, F. E., L. A. Metherell, and T. L. Pitt. 1999. Nosocomial acquisition of Burkholderia gladioli in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 160374-375. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., M. Gillis, and P. Vandamme. 2000. Pseudomonas antimicrobica Attafuah and Bradbury 1990 is a junior synonym of Burkholderia gladioli (Severini 1913) Yabuuchi et al. 1993. Int. J. Syst. Evol. Microbiol. 502135-2139. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., B. Holmes, K. Kersters, J. R. W. Govan, and P. Vandamme. 1999. Burkholderia cocovenenans (van Damme et al. 1960) Gillis et al. 1995 and Burkholderia vandii Urakami et al. 1993 are junior synonyms of Burkholderia gladioli (Severini 1913) and Burkholderia plantarii (Azegami et al. 1987) Urakami et al. 1994, respectively. Int. J. Syst. Bacteriol. 4937-42. [DOI] [PubMed] [Google Scholar]
- 10.Fass, R., and J. Barnishan. 1979. Minimal inhibitory concentrations of 34 antimicrobial agents for control strains Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853. Antimicrob. Agents Chemother. 16622-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferroni, A., I. Sermet-Gaudelus, E. Abachin, G. Quesne, G. Lenoir, P. Berche, and J.-L. Gaillard. 2002. Use of 16S rRNA gene sequencing for identification of nonfermenting gram-negative bacilli recovered from patients attending a single cystic fibrosis center. J. Clin. Microbiol. 403793-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves, M., T. Robin, A. M. Chipman, J. Wong, S. Khashe, and J. M. Janda. 1997. Four additional cases of Burkholderia gladioli infection with microbiological correlates and review. Clin. Infect. Dis. 25838-842. [DOI] [PubMed] [Google Scholar]
- 13.Henry, D., M. Campbell, C. McGimpsey, A. Clarke, L. Louden, J. L. Burns, M. H. Roe, P. Vandamme, and D. Speert. 1999. Comparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosis. J. Clin. Microbiol. 371004-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao, Z., Y. Kawamura, N. Mishima, R. Yang, N. Li, X. Liu, and T. Ezaki. 2003. Need to differentiate lethal toxin-producing strains of Burkholderia gladioli, which cause severe food poisoning: description of B. gladioli pathovar cocovenenans and an emended description of B. gladioli. Microbiol. Immunol. 7915-925. [DOI] [PubMed] [Google Scholar]
- 15.Jones, A., T. Stanbridge, B. Isalska, M. Dodd, and A. Webb. 2001. Burkholderia gladioli: recurrent abscesses in a patient with cystic fibrosis. J. Infect. 4269-71. [DOI] [PubMed] [Google Scholar]
- 16.Kanj, S. S., V. Tapson, R. D. Davis, J. Madden, and I. Browning. 1997. Infections in patients with cystic fibrosis following lung transplantation. Chest 112924-930. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy, M., R. Coakley, S. Donaldson, R. Aris, K. Hohneker, J. Wedd, M. Knowles, P. Gilligan, and J. Yankaskas. 2007. Burkholderia gladioli: five year experience in a cystic fibrosis and lung transplantation center. J. Cyst. Fibros. 6267-273. [DOI] [PubMed] [Google Scholar]
- 18.Khan, S., S. Gordon, P. Stillwell, T. Kirby, and A. Arroliga. 1996. Empyema and bloodstream infection caused by Burkholderia gladioli in a patient with cystic fibrosis after lung transplantation. Pediatr. Infect. Dis. J. 15637-639. [DOI] [PubMed] [Google Scholar]
- 19.Lestin, F., R. Kraak, and A. Podbielski. 2008. Two cases of keratitis and corneal ulcers caused by Burkholderia gladioli. J. Clin. Microbiol. 462445-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lincoln, S., T. Fermor, D. Stead, and J. Sellwood. 1991. Bacterial soft rot of Agaricus bitorquis. Plant Pathol. 40136-144. [Google Scholar]
- 21.Murray, S., J. Charbeneau, B. C. Marshall, and J. J. LiPuma. 2008. Impact of Burkholderia infection on lung transplantation in cystic fibrosis. Am. J. Respir. Crit. Care Med. 178363-371. [DOI] [PubMed] [Google Scholar]
- 22.Ritterband, D., M. Shah, K. Cohen, J. Lawrence, and J. Seedor. 2002. Burkholderia gladioli keratitis associated with consecutive recurrent endophthalmitis. Cornea 21602-603. [DOI] [PubMed] [Google Scholar]
- 23.Ross, J. P., S. M. Holland, V. J. Gill, E. S. DeCarlo, and J. I. Gallin. 1995. Severe Burkholderia (Pseudomonas) gladioli infection in chronic granulomatous disease: report of two successfully treated cases. Clin. Infect. Dis. 211291-1293. [DOI] [PubMed] [Google Scholar]
- 24.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 372201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin, J., S. Kim, M. Shin, S. Suh, D. Ryang, and M. Jeong. 1997. Bacteremia due to Burkholderia gladioli: case report. Clin. Infect. Dis. 251264-1265. [DOI] [PubMed] [Google Scholar]
- 26.Snyder, J. W., G. K. Munier, and C. L. Johnson. 2008. Direct comparison of the BD Phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 462327-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talon, D., M. Dupont, J. Lesne, M. Thouverez, and Y. Michel-Briand. 1996. Pulsed-field gel electrophoresis as an epidemiological tool for clonal identification of Aeromonas hydrophila. J. Appl. Bacteriol. 80277-282. [DOI] [PubMed] [Google Scholar]
- 28.Tayeb, L. A., M. Lefevre, V. Passet, L. Diancourt, S. Brisse, and P. A. D. Grimont. 2008. Comparative phylogenies of Burkholderia, Ralstonia, Comamonas, Brevundimonas and related organisms derived from rpoB, gyrB and rrs gene sequences. Res. Microbiol. 159169-177. [DOI] [PubMed] [Google Scholar]
- 29.Tenover, F., R. Arbeit, R. Goering, et al. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18426-439. [DOI] [PubMed] [Google Scholar]
- 30.Welch, D., M. Muszynski, C. Pai, M. Marcon, M. Hribar, P. Gilligan, J. Matsen, P. Ahlin, B. Hilman, and S. Chartrand. 1987. Selective and differential medium for recovery of Pseudomonas cepacia from the respiratory tracts of patients with cystic fibrosis. J. Clin. Microbiol. 251730-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitby, P., K. Carter, K. Hatter, J. LiPuma, and T. Stull. 2000. Identification of members of the Burkholderia cepacia complex by species-specific PCR. J. Clin. Microbiol. 382962-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilsher, M., J. Kolbe, A. Morris, and D. Welch. 1997. Nosocomial acquisition of Burkholderia gladioli in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 1551436-1440. [DOI] [PubMed] [Google Scholar]