Abstract
Sialidase activity varies widely among strains and tends to correlate with strain virulence in the avian pathogen Mycoplasma synoviae. To characterize the forms of selection acting on enzymes required for sialic acid scavenging and catabolism, the ratios of nonsynonymous (Ka) to synonymous (Ks) mutation frequency were calculated for codons in the sialidase gene of 16 strains of M. synoviae and for its nearly identical homolog in four strains of Mycoplasma gallisepticum. The Ka/Ks (ω) values for the linked genes required for nutritive N-acetylneuraminate catabolism (nanA, nagC, nanE, nagA, and nagB) from nine strains of M. synoviae were also determined. To provide context, ω was determined for all corresponding genes of 26 strains of Clostridium perfringens and Streptococcus pneumoniae. Bayesian models of sequence evolution showed that only the sialidase of M. synoviae was under significant (P < 0.001) diversifying selection, while the M. synoviae genes for N-acetylneuraminate catabolism and all genes examined from M. gallisepticum, C. perfringens, and S. pneumoniae were under neutral to stabilizing selection. Diversifying selection acting on the sialidase of M. synoviae, but not on the sialidase of M. gallisepticum or the sialidases or other enzymes essential for sialic acid scavenging in other Firmicutes, is evidence that variation in specific activity of the enzyme is perpetuated by a nonnutritive function in M. synoviae that is influenced by the genomic context of the organism.
Mycoplasma synoviae and Mycoplasma gallisepticum are major pathogens of commercial poultry and are each independently associated with respiratory, reproductive, and joint diseases. The severity of clinical signs of mycoplasmosis ranges from unapparent to severe and is often influenced by the presence of secondary pathogens during infection. Numerous aspects of M. gallisepticum pathogenesis have been studied extensively, including primary (11, 34) and secondary (16, 23, 27) mechanisms for cytadherence, antigenic variation (23, 31), and enzymatic or metabolic factors (2, 12, 17) that contribute to its virulence.
The basis for virulence of M. synoviae is far less characterized (22). The best-understood contributing factor is a multigene system encoding variable lipoprotein hemagglutinins (vlhA genes), which are thought to constitute the primary mechanism of M. synoviae cytadherence to sialylated host cell receptors. Antigenic variation results from site-specific recombination among a large assemblage of vlhA pseudogenes present in a single 69-kb locus. Allele switching driven in vivo by the selective pressure of anti-VlhA antibodies is a proposed mechanism for M. synoviae to evade the adaptive immune system of the host (31, 32). In contrast, independently transcribed vlhA genes are distributed in several discrete clusters throughout the M. gallisepticum genome, and these are of secondary importance to the principal adhesin gene, gapA, and its accessory protein gene, crmA, in mediating adherence to sialylated receptors (34).
Adjacent to the vlhA locus, an M. synoviae gene encodes a cell-associated sialidase that displays essentially continuous quantitative variation in specific activity among strains. A nearly identical homolog (consensus 94.5% amino acid identity and 96.5% amino acid similarity) exists in M. gallisepticum, even though M. gallisepticum does not possess the enzymes that M. synoviae does for catabolizing liberated sialic acid (N-acetylneuraminate) for glycolysis. Because receptor desialylation reduces or abolishes cytadherence by both M. synoviae and M. gallisepticum, a balance between variable sialidase activity and avidity of adherence to sialylated receptors is predictable (25), but the selective pressures responsible for the observed correlation of sialidase activity with M. synoviae strain virulence remain unknown (3, 4, 26).
The rates of nonsynonymous (amino acid-altering; Ka or dN [15]) and synonymous (silent; Ks or dS) mutations within a gene can reveal the forms of selective pressure acting on the corresponding protein among strains or species (33). A Ka/Ks (ω) value of ≈1 implies that the gene is not under strong selection of any type, while an ω value of <1 indicates that the gene is under stabilizing selection that removes divergent forms from a population. In rare instances, ω is >1, which indicates that the gene is under diversifying selection, and suggests that the species is at its fittest when substantial sequence diversity is perpetuated among strains (28, 46). We examined the sialidases of M. synoviae and M. gallisepticum and linked genes required for N-acetylneuraminate catabolism by M. synoviae for evidence of selection that might help to elucidate their roles in the ecology and virulence of these species.
MATERIALS AND METHODS
Strains and culture conditions.
M. synoviae strains K3344, K4907A, K5395B (referred to as K5599A in a preliminary report [26]), FMT, MS117, MS173, MS178, and WVU1853T were cultured in modified Frey's medium including 1% (wt/vol) glucose, 15% (vol/vol) porcine serum, 0.05% (wt/vol) l-cysteine, and 0.05% (wt/vol) NAD as previously described (26). M. gallisepticum strains F, JR-67, R, and S6 were cultured in American Type Culture Collection medium 988 (SP-4) containing 0.5% (wt/vol) glucose and 15% (vol/vol) fetal bovine serum at 37°C in 5% (vol/vol) CO2 without agitation.
Amplification and nucleotide sequencing.
Genomic DNA was extracted from all strains of M. synoviae and M. gallisepticum using EasyDNA reagents according to the manufacturer's specifications (Invitrogen, Carlsbad, CA). Amplification and sequencing of the sialidase and N-acetylneuraminate catabolism locus of M. synoviae were described previously (25). Amplification of the sialidase gene from M. gallisepticum strains was performed by initial denaturation at 94°C followed by 30 cycles at 94°C (20 s), 50°C (20 s), and 72°C (3 min). A 10-min final extension was performed at 72°C. Forward (5′-TCA GAT CAT TAA ACT AGC GCC TAA-3′) and reverse (5′-CGC ATG ATA CGA TAA CGA AAT G-3′) primers and Expand High Fidelity PCR reagents (Roche Applied Sciences, Indianapolis, IN) were used. All amplicons were sequenced by primer walking using four-dye fluorescent dideoxy labeling methods. Sequence reads were assembled into contigs using Sequencher 4.7 (Gene Codes, Ann Arbor, MI).
Selection analyses.
Natural variation in the sialidase genes from the eight strains of M. synoviae that we sequenced, plus eight others in GenBank (see Table S1 in the supplemental material), was analyzed using Bayesian models of sequence evolution in the Selecton v2.4 software suite (38). Separate analyses of the aligned sequences were performed using the M8 model (45) and the mechanistic-empirical combined (MEC) model (10). Briefly, M8 is a mechanistic model that uses maximum likelihood methods accounting for factors such as different probabilities for transitions and transversions, codon bias, and among-site rate variation to estimate the proportion of codons with ω values of <1 (the beta distribution p0) and the proportion with ω values of ≥1 (ωs). Sites in the p0 category reflect stabilizing selection, and sites in the ωs category reflect either neutral or diversifying selection. This allows site-specific as well as global inferences about the forms of selection acting on the protein. The MEC model further accounts for differing empirical amino acid mutation probabilities; a position with physicochemically radical amino acid changes in polarity or charge is assigned a higher site-specific Ka value than a position with less extreme mutations. Compared to the more conservative M8 model, a smaller proportion of sites with ω values substantially greater than 1 may be sufficient for the MEC model to indicate diversifying selection despite a low global ω value for the protein.
To focus specifically on the catalytic residues of the enzyme (8, 19), nucleotide sequences bearing (i) the signature Trp-Arg-Ile-Pro motif, (ii) Arg37, Asp62, Asp100, and Glu230 equivalents (numbering of the Clostridium perfringens sialidase NanI [GenBank accession no. P10481]), and (iii) the signature “Asp box” (Ser/Thr-X-Asp-X-Gly-X-Thr-Trp/Phe) motifs were concatemerized in silico and analyzed using the M8 and MEC models. The M8 model was also used to analyze the M. synoviae sialidase gene homologs in M. gallisepticum strains F, JR-67, R, and S6 (GenBank accession no. FJ659840 to FJ659843) as well as the genes encoding the canonical N-acetylneuraminate catabolism pathway (N-acetylneuraminate lyase gene nanA, N-acetylmannosamine kinase gene nagC, N-acetylmannosamine-6-phosphate epimerase gene nanE, N-acetylglucosamine-6-phosphate deacetylase gene nagA, and glucosamine-6-phosphate deaminase gene nagB) from the eight M. synoviae strains we sequenced plus strain 53 (42). For context, the orthologous sialidase gene and genes of the N-acetylneuraminate catabolism pathway from 11 strains of C. perfringens and 15 strains of Streptococcus pneumoniae (see Table S1 in the supplemental material) were analyzed in a similar manner.
Statistical procedures.
The statistical significance of global selection on each protein was determined by using the Selecton software to perform a likelihood ratio test for the M8 model and a null model (M8a) that infers neutral or stabilizing selection based on deviations from a fixed ωs value of 1. In the absence of diversifying selection, the difference between the likelihood scores generated by the M8 and M8a models follows a χ2 distribution (39). Significant deviations from that distribution thus indicate diversifying selection. Within each gene, confidence intervals from 5% to 95% were determined from the observed distribution of ω for each codon. For sites with an ω of >1, if the lower bound of the confidence interval was also >1, then the inference of diversifying selection at the site was considered to be meaningful (38).
Topological projections.
The 16 primary amino acid sequences for M. synoviae sialidase were aligned with the primary sequence of the orthologous C. perfringens sialidase catalytic domain (GenBank accession no. 2BF6_A [8]) using CLUSTALW v1.82 and then projected onto the three-dimensional crystal structure of the domain (Protein Data Bank no. 2BF6 [29]) using FirstGlance v1.42 (http://firstglance.jmol.org) and ConSurf v3.0 (http://consurf.tau.ac.il) software (21). The ω value for each aligned M. synoviae residue was depicted with the ratios color coded on a seven-step scale, using the Java chemical structure viewer Jmol (http://www.jmol.org).
RESULTS
Global selection.
Diversifying selection on average over all codons in the sialidase gene from M. synoviae was inferred using both the M8 (P < 0.001) and MEC models (Akaike information criterion score < M8a [7]). Across 16 strains of M. synoviae, 59 sialidase residues (≈6% of the protein) had an ω of >3 in the M8 model (Fig. 1A). In striking contrast, there was no evidence of diversifying selection acting on the homologous sialidase gene from M. gallisepticum (P > 0.05), which had only three codons with ω values as high as 1.5 in the M8 model and the majority being far less than 0.5 in both M8 and MEC models. Each of the enzymes constituting the pathway for N-acetylneuraminate catabolism by M. synoviae was also inferred to be under neutral or stabilizing selection (P > 0.05 in the M8 model) (Fig. 1B). None of the orthologous sequences from 26 strains of C. perfringens and S. pneumoniae showed any evidence of diversifying selection (P > 0.05 in the M8 model), and most were inferred to be under stabilizing selection with a global ω far less than 1 in both M8 and MEC models (Fig. 1).
FIG. 1.
Ka/Ks ratios of sialidase and the N-acetylneuraminate catabolism pathway. The Ka/Ks ratios (ω) from the M8 model are ordered from left to right along the x axis for each codon from the N terminus to the C terminus of sialidase (A) and enzymes constituting the N-acetylneuraminate catabolism pathway (B). An ω value of ≈1 implies no strong selection of any type; ω of <1 indicates stabilizing selection, while an ω of >1 indicates diversifying selection. The M. gallisepticum genome encodes sialidase but not the N-acetylneuraminate catabolism pathway.
Selection on individual residues.
The ω values for individual codons within certain protein-coding sequences suggested that specific residues may be selected individually regardless of the form of selection governing the sequence globally. For example, the M8 model estimated that M. synoviae sialidase, inferred to be under diversifying selection globally, had only 22 residues with an ω confidence interval lower bound of >1. The S. pneumoniae sialidase, inferred to be under stabilizing selection globally, also had 17 such residues in the M8 model, although none of those were aligned with the selected residues in M. synoviae. The nanE genes of M. synoviae and C. perfringens each had 6 residues with an ω confidence interval lower bound of >1, although none were at equivalent aligned positions. No other gene had more than three such residues in the M8 model, and for all genes, the MEC model actually detected fewer residues with an ω confidence interval lower bound of >1 than the M8 model did. Conversely, two synonymous mutations were the only variations evident in the nucleotide sequences encoding the catalytic motifs of M. synoviae sialidase, and thus, those motifs were inferred to be under strong stabilizing selection when analyzed using either the M8 or MEC model (see Fig. S1 in the supplemental material).
Topological mapping of selected residues.
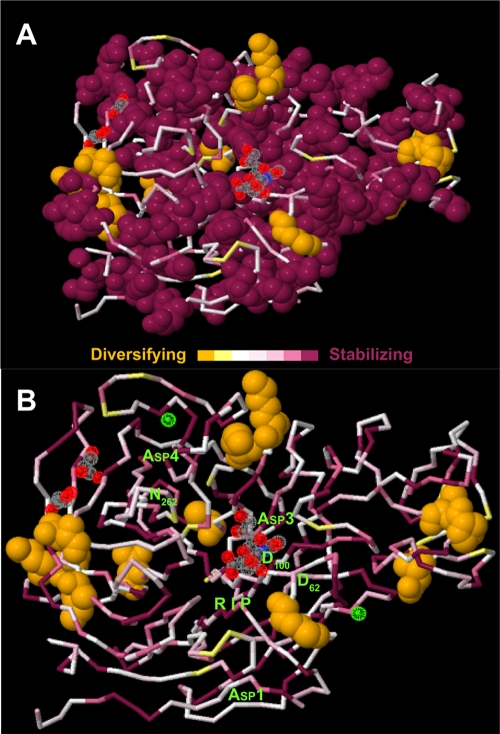
The 450 M. synoviae sialidase residues (≈48% of the protein) that were mapped onto the three-dimensional structure of the C. perfringens sialidase catalytic domain included 13 of the 22 individual residues inferred to be under diversifying selection. Those corresponded to residues 288, 289, 405, 422, 423, 481, 490, 492, 582, 594, 602, 603, and 622 of the C. perfringens sialidase (UniProtKB entry Q59310 [29]). They were all located on the predicted exterior surface of the protein, with the majority in regions of polar or cationic residues, and were excluded topologically from the catalytic sites or the sialic acid substrate binding groove (Fig. 2). The other 9 residues of the M. synoviae sialidase with an ω confidence interval lower bound of >1 were in unaligned regions of the catalytic domain or outside of the domain. In contrast, the catalytic sites and substrate binding groove were all populated with residues with low ω values. Trimming the unaligned regions had no effect on the mapped locations of the residues inferred to be under diversifying or stabilizing selection.
FIG. 2.
Topological projection of the M. synoviae sialidase catalytic domain. The primary amino acid sequences for M. synoviae sialidase were aligned with the orthologous C. perfringens sialidase catalytic domain and then projected onto the known three-dimensional structure of the domain. The Ka/Ks (ω) value for each aligned M. synoviae residue is color coded on a seven-step scale from violet (≪1) to orange (ω confidence interval lower bound of >1). A cocrystallized sialic acid molecule is shown in red, gray, and blue (center); cocrystallized glycerol molecules are also shown (left). (A) The residues of M. synoviae sialidase under significant diversifying selection (orange-colored van der Waals surfaces) map to regions of polar or cationic residues on the exterior of the protein, while those under stabilizing selection (violet-colored van der Waals surfaces) populate the catalytic sites and substrate binding groove. (B) Residues under diversifying selection (orange-colored van der Waals surfaces) are excluded from the catalytic sites and substrate binding groove. The second Asp box motif is obscured in this view. Cocrystallized calcium ions are shown (green).
DISCUSSION
Sialidase is a well-known bacterial virulence factor (9, 43) that was formerly considered to occur only rarely among mycoplasmas. Since the discovery of a sialidase gene in M. synoviae (42), we have explored the genetic basis for quantitative variation in sialidase-specific activity among strains and the observed trend for sialidase activity to correlate with M. synoviae strain virulence. Marked heterogeneity in the form of single-nucleotide polymorphisms exists within the locus of six contiguous M. synoviae genes associated with sialic acid scavenging and catabolism, particularly in the sialidase gene itself (25, 26). In the present study, we analyzed the polymorphisms in detail by estimating ω values globally for each of those enzymes and then for each amino acid residue to interpret the forms of natural selection acting at this locus. Because the time since sequence divergence among strains of M. synoviae is presumably shorter than the evolutionary time since the divergence of M. synoviae from an ancestor shared with M. gallisepticum or other Firmicutes, the polymorphisms reflect variation segregating within the population of conspecifics, but not fixed substitutions (20). The most deleterious changes are expected to be eliminated rapidly from the population of M. synoviae strains, but less-radical mutations may persist longer because selection is not instantaneous (36).
Global ω values for bacterial genes under weak to moderate stabilizing selection, whose corresponding proteins are subject to functional constraints such that nonsynonymous mutations are purged from a population, generally fall within the range of 0.04 to 0.2 (36). As expected for most enzymes, analysis using even the MEC model (10) indicated that the genes nanA, nagC, nanE, nagA, and nagB encoding the intracellular pathway for N-acetylneuraminate catabolism by M. synoviae experience this form of selection. This was supported by likelihood ratio tests using the M8 model, which weights all amino acid substitutions equally and is therefore less nuanced but globally more stringent (45). Examples such as nagC of M. synoviae and nagB of S. pneumoniae (Fig. 1) illustrate the need to interpret global ω values in the context of a null model especially when comparing sequences that have atypical baselines; for nagC of M. synoviae the baseline ω value in the M8a model was approximately 0.35. The unexpectedly high baseline for nagC of M. synoviae in both models may reflect its coding on the opposite strand from nanA, nanE, nagA, and nagB in the locus. There was no basis on which to suspect the theoretical alternative that extremely high selection coefficients promoting polymorphism in each of the enzymes in the pathway contributed to the ω values that are <1 (20).
Although earlier methodology was limited to global inferences, evidence of diversifying selection has been observed in viral, bacterial, protozoal, plant, and animal genes (5, 13, 18, 30, 37). Well-characterized examples of genes under diversifying selection include those encoding antigenic surface proteins (1, 41) and certain components of the adaptive immune system (14, 40). The highly significant likelihood ratio test of the M8 model for sialidase of M. synoviae, and its high proportion of individual residues with an ω confidence interval lower bound of >1, provided strong evidence that diversifying selection acts on the enzyme (6). Although some of the conserved (Ser, Gly, Thr) and all of the variable (X) residues in the Asp box motif Ser/Thr-X-Asp-X-Gly-X-Thr-Trp/Phe might tolerate minor codon variation, those sites were consistently invariant in the otherwise polymorphic protein. This was also expected, because Chien et al. (8) showed that the conserved motifs are critically important in maintaining the activity of the enzyme, but stabilizing selection can occur in some regions of a protein while diversifying selection operates in others (47). Projection onto the crystal structure of the C. perfringens sialidase catalytic domain showed that the selected residues with an ω of ≫1 mapped to the outer surface of the protein and were topologically distant from the conserved functional motifs. Thus, the more subtle differences in the topology of the enzyme that the diversifying mutations induce may be the basis for quantitative variation in sialidase activity of the magnitude observed among strains of M. synoviae, even while the core catalytic sites of the enzyme simultaneously experience strong stabilizing selection. These findings lead to the conclusion that the species M. synoviae is at its fittest when moderate diversity in specific sialidase activity is perpetuated among its strains.
Sialidase is believed to be a true homolog that was horizontally transferred between M. synoviae and M. gallisepticum, possibly during mixed infection of a common avian host (25, 42). Like the VlhA hemagglutinins, it is an immunodominant antigen during M. synoviae infection (3). If direct pressure to evade host antisialidase antibodies were the primary selective force, then the same inference of diversifying selection would be expected also for the sialidase homolog in M. gallisepticum. The evidence of neutral to moderate stabilizing selection obtained instead supports the conclusion that the sialidase in M. gallisepticum experiences different selective pressures than those most influential in the ecology of M. synoviae. Because ω can increase due to changing selection pressure that results from acquiring a new function (47), as would be possible following transfer into a different genomic context, the higher global ω obtained for the homolog in M. synoviae further corroborates the hypothesis that the direction of horizontal transfer was from M. gallisepticum to M. synoviae (25).
One directly relevant difference between the genomic contexts of the mycoplasmal sialidase homologs is the nutritive N-acetylneuraminate catabolism pathway present in M. synoviae, which is absent from M. gallisepticum. Selection might be predicted to favor sialidase variants that liberate the maximum amount of free sialic acid catabolizable for glycolysis by M. synoviae, but it is predicted to be neutral in this regard for M. gallisepticum. If linkage to this pathway were a primary force for variation, then the same inference of diversifying selection also might be expected for the sialidase orthologs in other Firmicutes that catabolize N-acetylneuraminate (24, 35). However, analyses of the orthologous sialidases in many strains of both comparatively closely related C. perfringens and the distantly related S. pneumoniae showed no evidence of diversifying selection in those genomic contexts, so the linkage seems unlikely to constitute the dominant influence on sialidase in M. synoviae.
Collectively, the findings lead to the conclusion that diversity in specific sialidase activity in M. synoviae is perpetuated by a nonnutritive function of the enzyme that is specific to the genomic context of the organism. The best-understood candidate is its potential role in modulating adherence to sialylated host cell surface receptors (25). If alleles of the primary M. synoviae cytadhesin VlhA have different receptor binding avidities, then the direct selection pressure of host anti-VlhA antibodies that drives allele switching might also constitute indirect pressure to perpetuate moderate variation in sialidase activity. Strains with higher-affinity adhesins would be expected to possess higher sialidase activity (44), resulting in the trend of the correlation of sialidase activity with M. synoviae strain virulence. Further study of the balance between variable sialidase activity and avidity of adherence to sialylated receptors can be expected to provide insight into the ecology of M. synoviae and illuminate key differences in the pathogeneses of M. synoviae and M. gallisepticum.
Supplementary Material
Acknowledgments
Sequencing was performed by the Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Laboratory at the University of Florida.
This work was supported by Public Health Service grant 1R01GM076584 from the National Institute of General Medical Sciences (D.R.B.).
Footnotes
Published ahead of print on 27 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrews, T. D., and T. Gojobori. 2004. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics 16625-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, L. 1989. The capacity of Ureaplasma urealyticum, Mycoplasma hominis and seven other Mycoplasma species for hemadsorption, sperm adsorption, hemolysis and peroxide formation. Arch. Exp. Veterinarmed. 43789-800. [PubMed] [Google Scholar]
- 3.Berčič, R. L., B. Slavec, M. Lavrič, M. Narat, A. Bidovec, P. Dovč, and D. Benčina. 2008. Identification of major immunogenic proteins of Mycoplasma synoviae isolates. Vet. Microbiol. 127147-154. [DOI] [PubMed] [Google Scholar]
- 4.Berčič, R. L., B. Slavec, M. Lavrič, M. Narat, O. Zorman-Rojs, P. Dovč, and D. Benčina. 2008. A survey of avian Mycoplasma species for neuraminidase enzymatic activity. Vet. Microbiol. 130391-397. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, J. G., A. M. Dean, and T. Mitchell-Olds. 2000. Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 975322-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, S., and J. M. Akey. 2006. Genomic insights into positive selection. Trends Genet. 22437-446. [DOI] [PubMed] [Google Scholar]
- 7.Burnham, K. P., and D. R. Anderson. 2002. Model selection and multimodel inference: a practical information-theoretic approach, 2nd ed. Springer-Verlag, New York, NY.
- 8.Chien, C. H., Y. J. Shann, and S. Y. Sheu. 1996. Site-directed mutations of the catalytic and conserved amino acids of the neuraminidase gene, nanH, of Clostridium perfringens ATCC 10543. Enzyme Microb. Technol. 19267-276. [DOI] [PubMed] [Google Scholar]
- 9.Corfield, T. 1992. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology 2509-521. [DOI] [PubMed] [Google Scholar]
- 10.Doron-Faigenboim, A., and T. Pupko. 2007. A combined empirical and mechanistic codon model. Mol. Biol. Evol. 24388-397. [DOI] [PubMed] [Google Scholar]
- 11.Goh, M. S., T. S. Gorton, M. H. Forsyth, K. E. Troy, and S. J. Geary. 1998. Molecular and biochemical analysis of a 105 kDa Mycoplasma gallisepticum cytadhesin (GapA). Microbiology 1442971-2978. [DOI] [PubMed] [Google Scholar]
- 12.Hudson, P., T. S. Gorton, L. Papazisi, K. Cecchini, S. Frasca, Jr., and S. J. Geary. 2006. Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect. Immun. 74931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, A. L. 1991. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics 127345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, A. L., and M. Nei. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335167-170. [DOI] [PubMed] [Google Scholar]
- 15.Hurst, L. D. 2002. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18486-487. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins, C., S. J. Geary, M. Gladd, and S. P. Djordjevic. 2007. The Mycoplasma gallisepticum OsmC-like protein MG1142 resides on the cell surface and binds heparin. Microbiology 1531455-1463. [DOI] [PubMed] [Google Scholar]
- 17.Jenkins, C., R. Samudral, S. J. Geary, and S. P. Djordjevic. 2008. Structural and functional characterization of an organic hydroperoxide resistance protein from Mycoplasma gallisepticum. J. Bacteriol. 1902206-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiggins, F. M., G. D. Hurst, and Z. Yang. 2002. Host-symbiont conflicts: positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae. Mol. Biol. Evol. 191341-1349. [DOI] [PubMed] [Google Scholar]
- 19.Kleineidam, R. G., S. Kruse, P. Roggentin, and R. Schauer. 2001. Elucidation of the role of functional amino acid residues of the small sialidase from Clostridium perfringens by site-directed mutagenesis. Biol. Chem. 382313-319. [DOI] [PubMed] [Google Scholar]
- 20.Kryazhimskiy, S., and J. B. Plotkin. 2008. The population genetics of dN/dS. PLoS Genet. 4e1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau, M., I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, and N. Ben-Tal. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33W299-W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockaby, S. B., F. J. Hoerr, L. H. Lauerman, B. F. Smith, A. M. Samoylov, M. A. Toivio-Kinnucan, and S. H. Kleven. 1999. Factors associated with virulence of Mycoplasma synoviae. Avian Dis. 43251-261. [PubMed] [Google Scholar]
- 23.Markham, P. F., M. D. Glew, K. G. Whithear, and I. D. Walker. 1993. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect. Immun. 61903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita, O., and A. Okabe. 2001. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon 391769-1780. [DOI] [PubMed] [Google Scholar]
- 25.May, M., and D. R. Brown. 2008. Genetic variation in sialidase and linkage to N-acetylneuraminate catabolism in Mycoplasma synoviae. Microb. Pathog. 4538-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May, M., S. H. Kleven, and D. R. Brown. 2007. Sialidase activity in Mycoplasma synoviae. Avian Dis. 51829-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May, M., L. Papazisi, T. S. Gorton, and S. J. Geary. 2006. Identification of fibronectin-binding proteins in Mycoplasma gallisepticum strain R. Infect. Immun. 741777-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyata, T., and T. Yasunaga. 1980. Molecular evolution of mRNA: a method for estimating evolutionary rates of synonymous and amino acid substitutions from homologous nucleotide sequences and its application. J. Mol. Evol. 1623-36. [DOI] [PubMed] [Google Scholar]
- 29.Newstead, S. L., J. A. Potter, J. C. Wilson, G. Xu, C. H. Chien, A. G. Watts, S. G. Withers, and G. L. Taylor. 2008. The structure of Clostridium perfringens NanI sialidase and its catalytic intermediates. J. Biol. Chem. 2839080-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, R., and Z. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noormohammadi, A. H., P. F. Markham, M. F. Duffy, K. G. Whithear, and G. F. Browning. 1998. Multigene families encoding the major hemagglutinins in phylogenetically distinct mycoplasmas. Infect. Immun. 663470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noormohammadi, A. H., P. F. Markham, A. Kanci, K. G. Whithear, and G. F. Browning. 2000. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae. Mol. Microbiol. 35911-923. [DOI] [PubMed] [Google Scholar]
- 33.Ohta, T. 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23263-286. [Google Scholar]
- 34.Papazisi, L., S. Frasca, Jr., M. Gladd, X. Liao, D. Yogev, and S. J. Geary. 2002. GapA and CrmA coexpression is essential for Mycoplasma gallisepticum cytadherence and virulence. Infect. Immun. 706839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paton, J. C., P. W. Andrew, G. J. Boulnois, and T. J. Mitchell. 1993. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. Annu. Rev. Microbiol. 4789-115. [DOI] [PubMed] [Google Scholar]
- 36.Rocha, E. P. C., J. M. Smith, L. D. Hurst, M. T. G. Holden, J. E. Cooper, N. H. Smith, and E. J. Feil. 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J. Theor. Biol. 239226-235. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 1022832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stern, A., A. Doron-Faigenboim, E. Erez, E. Martz, E. Bacharach, and T. Pupko. 2007. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 35W506-W511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson, W. J., R. Nielsen, and Q. Yang. 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2018-20. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka, T., and M. Nei. 1989. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol. Biol. Evol. 6447-459. [DOI] [PubMed] [Google Scholar]
- 41.Urwin, R., E. C. Holmes, A. J. Fox, J. P. Derrick, and M. C. Maiden. 2002. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 191686-1694. [DOI] [PubMed] [Google Scholar]
- 42.Vasconcelos, A. T. R., H. B. Ferreira, C. V. Bizarro, S. L. Bonatto, M. O. Carvalho, P. M. Pinto, D. F. Almeida, L. G. P. Almeida, R. Almeida, L. Alves-Filho, E. N. Assunção, V. A. C. Azevedo, M. R. Bogo, M. M. Brigido, M. Brocchi, H. A. Burity, A. A. Camargo, S. S. Camargo, M. S. Carepo, D. M. Carraro, J. C. de Mattos Cascardo, L. A. Castro, G. Cavalcanti, G. Chemale, R. G. Collevatti, C. W. Cunha, B. Dallagiovanna, B. P. Dambrós, O. A. Dellagostin, C. Falcão, F. Fantinatti-Garboggini, M. S. S. Felipe, L. Fiorentin, G. R. Franco, N. S. A. Freitas, D. Frías, T. B. Grangeiro, E. C. Grisard, C. T. Guimarães, M. Hungria, S. N. Jardim, M. A. Krieger, J. P. Laurino, L. F. A. Lima, M. I. Lopes, É. L. S. Loreto, H. M. F. Madeira, G. P. Manfio, A. Q. Maranhão, C. T. Martinkovics, S. R. B. Medeiros, M. A. M. Moreira, M. Neiva, C. E. Ramalho-Neto, M. F. Nicolás, S. C. Oliviera, R. F. C. Paixão, F. O. Pedrosa, S. D. J. Pena, M. Pereira, L. Pereira-Ferrari, I. Piffer, L. S. Pinto, D. P. Potrich, A. C. M. Salim, F. R. Santos, R. Schmitt, M. P. C. Schneider, A. Schrank, I. S. Schrank, A. F. Schuck, H. N. Seuanez, D. W. Silva, R. Silva, S. C. Silva, C. M. A. Soares, K. R. L. Souza, R. C. Souza, C. C. Staats, M. B. R. Steffens, S. M. R. Teixeira, T. P. Urmenyi, M. H. Vainstein, L. W. Zuccherato, A. J. G. Simpson, and A. Zaha. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 1875568-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vimr, E., and C. Lichtensteiger. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10254-257. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, R., M. Matrosovich, and H.-D. Klenk. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 12159-166. [DOI] [PubMed] [Google Scholar]
- 45.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Z., W. S. Wong, and R. Nielsen. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 221107-1118. [DOI] [PubMed] [Google Scholar]
- 47.Zelus, D., M. Robinson-Rechavi, M. Delacre, C. Auriault, and V. Laudet. 2000. Fast evolution of interleukin-2 in mammals and positive selection in ruminants. J. Mol. Evol. 51234-244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.