One of the most widely used high-throughput technologies is the oligonucleotide microarray. From the initial development of microarrays, high expectations were held for their use to aid in answering biological questions, due to their ability to measure mRNA abundances on a genome scale. However, accumulating experience is revealing that even when questions of sample preparation, data processing, and dealing with the inherently noisy data (81) are set aside, the large amount of data generated has proven difficult to analyze and interpret (12). It is also often challenging to narrow down specific novel findings based solely on expression profiling data.
Here, we present a downloadable compendium of gene expression profiles for Escherichia coli and discuss the experience from one lab in which expression profiling data have been employed in a myriad of studies of E. coli. We will try to address two classes of expression profiling data usage: (i) how expression profiling can be analyzed using more traditional statistical methods to provide biological understanding and (ii) how genome-scale models form a context within which expression profiling data content increases in value.
THE DATA
We present a database of 213 expression profiles produced in our laboratory and used in many of the case studies reviewed here. These profiles represent measurements for about 70 combinations of variations in experimental conditions and genetic variations in E. coli K-12 MG1655. In this set of experiments, the following parameters were varied: (i) the carbon source, (ii) the terminal electron acceptor, (iii) the temperature, (iv) the number of days the bacteria were grown at mid-log phase, and (v) the genotype (wild-type and gene deletion strains were used). These data and the corresponding minimum information about a microarray experiment (9) may be freely downloaded at http://systemsbiology.ucsd.edu/In_Silico_Organisms/E_coli/E_coli_expression2.
LEVELS OF ANALYSIS
Much insight into the function of E. coli has been gained over the past decade through the statistical analysis of cDNA and oligonucleotide arrays. Methods such as the genome-scale analysis of differential gene expression patterns (21), clustering and classification (17), and gene set enrichment (76) have been successfully deployed. Overall, our experience shows that informative results can also be obtained if data are analyzed in the context of a genome-scale network reconstruction and with the use of an in silico model. Furthermore, greater success in data analysis has been achieved on a fine-grain level for more specific hypothesis generation and assessment than on a coarse-grain genome-scale level. The lessons learned from the analysis of this data set can be classified by the granularity of the analysis and the level of use of the in silico model.
(i) Genome-scale analysis without using a modeling framework.
E. coli grown to mid-log phase on various carbon sources and/or with various gene knockouts has been shown to usually evolve a growth phenotype as predicted computationally. However, only a modest level of understanding of the optimal growth phenotype was obtained through the analysis of gene expression data alone.
(ii) Regulon- and network-level analysis in the modeling framework.
Several studies in which computational models have been tested to predict global properties of E. coli metabolism and transcriptional regulation, to suggest pathway usage in strains that do not grow at the optimal rate, and to predict gene expression changes under growth condition shifts have been conducted. Here, expression profiling data have played a beneficial role in both validating computational predictions and providing necessary information to predict novel regulatory interactions.
(iii) Gene-level analysis using in silico models.
The most informative use of gene expression profiling data was found when simulations predicted growth phenotypes that conflicted with experimental observations (growth/no growth), suggesting that undiscovered pathways still existed. Microarrays were used in this setting to identify individual genes that may contribute to computationally predicted pathways.
Each of these levels of analysis will be discussed and examples will be provided below.
GENOME-LEVEL ANALYSIS WITHOUT USING IN SILICO MODELS
Characterizing disparate paths to common end points in adaptive evolution.
Wild-type E. coli K-12 has been evolved to improve the growth rate by using serial passaging in the mid- to late log phase of growth (45, 47). Interestingly, when parallel cultures are evolved in this way, their evolutionary paths are not the same, though they usually evolve to have similar growth rates, substrate uptake rates, and oxygen uptake rates at the group level (31, 32). In an effort to understand these different evolutionary pathways toward similar end points, gene expression profiles at different time points in the evolution of strains grown on lactate or glycerol were obtained and analyzed (31).
Cultures grown on glycerol and lactate experienced significant changes in the expression of 39 and 18% of genes, respectively (compared to expression during growth on glucose), on the first day. At the evolutionary end points (44 and 60 days, respectively), the strains evolved on glycerol and lactate had only 11 and 7% of the genes differentially expressed relative to the expression state prior to adaption to the new carbon substrate. Thus, it is apparent that most genes with altered expression on the first day will return to preevolution transcription rates by the evolutionary end point. These changes were likely due to some response to the initial change in the substrate and thus were compensated for over the course of adaptation, since regulatory mechanisms that were active on day 1 (phoB, cre, crp, and rpoN) were not active thereafter. Even more interesting is that few genes that evolved differential expression patterns over time were differentially expressed in most of the replicates run in parallel. For example, in strains evolved on glycerol, 23 genes involved in cell motility were consistently downregulated in almost all replicates, leading to decreased cell motility. The strains evolved on lactate had only two genes which were consistently differentially expressed, one of which is necessary for the phosphorylation of pyruvate following lactate uptake. However, when gene expression changes were compared with fluxomic data for the same strains, little correlation was found (41), suggesting that more complex regulation was occurring, possibly at several levels that are hard to elucidate from gene expression profiling alone.
Despite the fact that the growth phenotypes converged to similar phenotypic end points, the gene expression profiles of the evolved strains varied widely, allowing many pathways to similar phenotypes in the fitness landscape. In addition, the gene expression profiles showed highly consistent regulatory responses upon the initial introduction of E. coli to a new growth substrate, followed by the consistent relaxation of these changes. However, a purely statistical genome-scale analysis of the gene expression data suggested only some general principles in adaptive evolution but failed to elucidate any specifics in the mechanisms underlying them. The profiles did not even suggest the genetic basis that was subsequently established using whole-genome resequencing (40). The remaining examples will demonstrate how the contents of gene expression profiles were more effectively analyzed in the context of genome-scale models for network-level questions and gene-level discoveries.
USING IN SILICO MODELS: PROVIDING CONTEXT FOR CONTENT
Genome-scale models and gene expression profiling to study the genotype-phenotype relationship.
With the availability of annotated genome sequences, constraint-based models of microbial metabolism on the genome scale have been built (24, 25). The genome-scale E. coli network reconstructions that have appeared (24, 29, 67) are highly curated knowledge bases of all known biochemical transformations for the organism of interest (66). Network reconstructions can be converted into a mathematical format and used to produce in silico models (28, 55). Since these advancements, scores of studies have attempted to mechanistically describe genotype-phenotype relationships using genome-scale metabolic network reconstructions and in silico models. Computational models of E. coli have been employed for metabolic engineering (3, 34, 84), exploratory analyses of network properties (2, 11, 65), gene function discovery (15, 68, 70), and studies of phenotypic behavior and component essentiality (36, 46, 58) and properties of evolution (30, 45, 61, 62) described in more than a hundred publications and patents. Applications of constraint-based modeling for E. coli have been reviewed recently (28). In these applications, constraint-based modeling has demonstrated some of its strengths, including its abilities (i) to scale to genome-size networks, (ii) to be easily integrated with a wide range of analytical methods (55, 64), and (iii) to provide useful predictions despite incomplete knowledge (20, 53).
The E. coli network reconstructions have proven useful in biological discovery when coupled with high-throughput data. Gene expression profiling data have been used in conjunction with E. coli genome-scale metabolic models to discover functions of uncharacterized open reading frames (ORFs) (68), gain insight into the growth phenotypes of mutant strains (30, 38), validate computational predictions of metabolic network functional modes (5), and predict transcriptional regulatory rules (20). Table 1 provides a more comprehensive list of studies in which gene expression data have been analyzed in the context of genome-scale reconstructions and models of E. coli metabolism and transcriptional regulation.
TABLE 1.
Studies in which gene expression profiling was used in conjunction with constraint-based modeling for E. coli
| Purpose(s) of study(ies) | Reference(s) |
|---|---|
| Discovery of novel gene function | 35, 68 |
| Prediction of gene expression levels/changes | 19, 20, 73, 75 |
| Prediction of transcriptional regulatory | |
| rules/gene coregulation | 20, 60 |
| Characterization of adaptive evolution on | |
| alternative carbon sources/in mutant strains | 6, 30, 33, 38 |
| Validation of regulatory/metabolic | |
| network properties | 5, 65 |
| Modeling and characterization of gene expression changes under | |
| mixed-substrate conditions | 7, 80 |
| Metabolic engineering and strain | |
| design/characterization | 54 |
| Validation of gene annotation/ | |
| gene-finding algorithm | 51, 60 |
| Construction of context-specific models | 6 |
Gene expression profiling aiding in the analysis of in silico results.
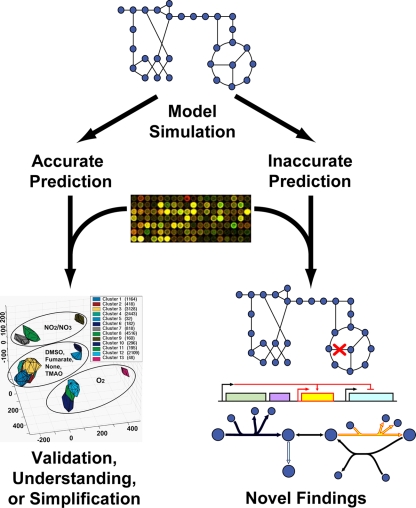
Gene expression profiling may be used with constraint-based modeling to validate computational methods/outcomes, verify model completeness, or predict missing pathways. As is common with any modeling framework, much may be learned from integrating data with computational outcomes (Fig. 1). Accurate predictions may allow for a more detailed understanding of the biology (26, 30, 45) or help simplify complex biological systems by suggesting essential variables (5). Incorrect predictions, on the other hand, can be used to guide discovery. Reconciling incorrect predictions with measured data can help elucidate novel pathways (68), bottleneck reactions (38), or regulatory interactions (20, 68). Gene expression data have been used in several cases to derive novel knowledge from both accurate and inaccurate in silico predictions for E. coli, as will be demonstrated here.
FIG. 1.
Potential outcomes from simulation and analysis of genome-scale models (5). Like any modeling framework, constraint-based models can lead to either accurate or inaccurate predictions. In cases in which an accurate result is found, microarray data have been used to validate the model and methods of analysis, provide data for potential insight into the biology reflected by the simulations, or suggest simplifications for an otherwise complex biological system. For example, as depicted in the graph, E. coli metabolism and its associated TRN exhibit few functional modes, affected mostly by the electron acceptor used. Activity profiles formed three clusters, separated as a function of the electron acceptor (represented by the ellipses), the carbon source (represented by separation inside each ellipse), and to a lesser degree, the nitrogen source. If, however, the simulations yield inaccurate predictions, novel findings can be obtained through alternative analytical methods. Predictions of inactive pathways, novel pathways, or new regulatory rules are just examples of new knowledge that may be gained this way. DMSO, dimethyl sulfoxide; TMAO, trimethylamine N- oxide.
USING IN SILICO MODELS: VALIDATING ACCURATE PREDICTIONS
Computational predictions suggest that the E. coli transcriptional regulatory network (TRN) for metabolism demonstrates few functional states.
Utilizing a metabolic reconstruction with all known transcriptional regulatory rules (20), E. coli was grown in silico under 15,580 different medium conditions. The functional state of the TRN that governs metabolism was described with activity profiles that were generated by combining the gene expression predictions and on/off regulatory logic. In this process, it was demonstrated that the regulatory network, as it is currently known, that governs metabolism in E. coli exhibits few functional states.
These activity profiles were projected into a transcriptional regulatory space and then subjected to clustering and dimensionality reduction. As shown in Fig. 1, these profiles clustered into effectively three dimensions. Clusters were separated as a function of the electron acceptor, the carbon source, and to a lesser degree, the nitrogen source. To test these computational predictions, the activity profiles for nine growth conditions used to produce a number of gene expression profiles were computed. The spatial organization of the gene expression profiling data was then shown to correlate well with the organization of the computational predictions shown in Fig. 1, despite the intrinsic noise of the microarray data. Validation from the gene expression profiling data not only provided support for the algorithm used, but also supported the hypothesis that while complex cellular networks have the potential to generate many different behaviors, the number that cells utilize is relatively small. Also, in the case of E. coli, this behavior is dependent mostly on the type of terminal electron acceptor which is available to the organism and the presence or absence of glucose or gluconate.
Beyond insights into E. coli biology, two central lessons that came out of this study were as follows. First, the summary of these functional states described through simulation is not possible with only a logical connectivity diagram of the TRN; thus, it is apparent that genome-scale computational models will continue to play an important role for the understanding of biological systems. Second, this study demonstrated that rational analysis of gene expression profiling data in the context of mechanistic models can provide insights into biology that otherwise may be missed.
Analyzing gene expression profiles through context-specific models.
Since a reconstruction is a knowledge base that contains details about all known biochemical transformations, it serves as a superset of all reactions that are actually functioning at any one time. For that reason, model accuracy can be improved if context-specific models are constructed by constraining the flux through reactions (1, 7) and/or removing reactions associated with genes that would not be transcribed or active under the given conditions (18-20, 39). When regulatory mechanisms are poorly understood, a few approaches can be employed that use high-throughput data to build context-specific models or analyze such models on a gene-by-gene basis (1, 74, 83) or on the basis of functional modules (71).
One method, initially tested on E. coli, accurately predicts context-specific models based on measured gene expression levels. This algorithm, called GIMME (6), utilizes an optimization framework and scoring algorithm to determine the best model structure based on gene expression levels and known functionalities of the cell under the given conditions. This algorithm was employed with gene expression profiling data for strains from three different studies: (i) strains of E. coli that were evolved on glycerol or lactate (31); (ii) E. coli mutant strains that were designed using computational predictions (34) to produce lactate (42); and (iii) wild-type and mutant strains that were grown under conditions that included different terminal electron acceptors (20).
Each data set was used to generate a context-specific model. A scoring algorithm was used to test if the microarray data were consistent with the expected cellular objectives.
First, models of wild-type strains of E. coli and strains evolved on glycerol and lactate were generated. It has been shown previously that wild-type E. coli does not grow optimally when immediately grown under an alternative growth condition; however, strains grow faster and more efficiently when grown at mid-log phase for extended periods of time (45). In addition, strains evolved on lactate or glycerol usually grow more rapidly than wild-type E. coli on many different substrates (31). In this study, when models from the evolved-strain data were compared with models from wild-type gene expression data, the evolved-strain models were found to be more consistent with the required network topology for optimal growth under all tested growth conditions. This finding supports the hypothesis that E. coli alters its gene expression profile in adaptive evolution, thereby allowing for more efficient network topology and for improved growth on a new substrate.
In the second case, a computational algorithm was used to predict gene deletion mutant strains of E. coli that would produce and excrete lactate in order to grow optimally (42). The gene expression data from a strain predicted with the OptKnock algorithm were much more consistent with lactate production than the gene expression data from wild-type E. coli (P = 0.0014). This result provides added support for the notion that E. coli can rearrange its gene expression pattern to cope with a gene deletion and that this gene expression pattern is consistent with the computationally predicted phenotype.
The third case employed data gathered from 21 different unevolved-strain and electron acceptor combinations. Data from each comparison were employed to determine how consistent the gene expression profiles were with the growth characteristics under the three electron acceptor conditions (aerobic, anaerobic, and anaerobic with nitrate). When pairwise comparisons of all 21 combinations were made, it was clear that groups of arrays from the three electron acceptor conditions were more consistent with the required network topology for growth under the corresponding conditions than data from strains grown under other conditions. In fact, all statistically significant comparisons demonstrated that data from aerobic growth were more consistent with the required network topology for growth on oxygen than the results from microarray data for strains grown anaerobically with or without nitrate. This comparison held true also for anaerobic growth in 99% of the cases and for growth under anaerobic conditions with nitrate supplementation in 90% of the cases. Since the data were gathered within a day after strains were introduced to these growth conditions, these results add yet more support for the idea that the E. coli TRN is wired for rapid response to changes in terminal electron acceptors, thus allowing near optimal growth.
In this study, only a brief look at the results of mapping the gene expression profiling data to the E. coli metabolic network was taken, since the goal was primarily to validate the method. This study did, however, provide additional evidence for hypotheses concerning gene expression states in evolved strains and engineered strains and under various growth conditions.
In addition, recently developed alternative methods allow the construction of context-specific models based on previously established network reconstructions by the direct mapping of gene expression data to a reconstruction (82) or the use of mixed-integer linear programming to determine an optimal network based on gene expression and proteomic data (74). Since neither of these approaches has been applied to E. coli in studies reported in the literature, it is expected that a more in-depth analysis of the gene expression data mapped onto the E. coli metabolic network by these novel methods will lead to additional insights into E. coli metabolism and transcriptional regulation.
USING IN SILICO MODELS: FALSE PREDICTIONS DRIVE DISCOVERY
Experimental validation of simulation results is necessary when testing novel hypotheses; however, it is not uncommon that the model fails to predict the experimental results accurately. Fortunately, such inconsistencies can drive biological discovery and elucidate novel understanding of biological mechanisms.
Refining regulatory network reconstruction.
Gene expression profiling data have been used to iteratively refine genome-scale models of metabolism and transcriptional regulation (20, 39). A computational model of metabolism was used to predict growth phenotypes for 13,750 different combinations of nitrogen sources, carbon substrates, and nonessential-gene knockout mutants of E. coli (8, 37). When the model of metabolism was coupled with all known transcriptional regulatory rules, it accurately predicted the growth phenotype in 78.7% of the simulations. To improve the predictive power of this model, a dual-perturbation study was conducted in which the aerobic/anaerobic conditions were tested using six knockout strains in which key transcriptional regulators in the oxygen response system were removed (ΔarcA, ΔappY, Δfnr, ΔoxyR, and ΔsoxS strains and the dual-knockout ΔarcA Δfnr strain). Gene expression profiles were obtained for wild-type E. coli and each of the knockout strains. Computational predictions of differential gene expression patterns were also made by using the models; however, the degree of overlap between measured gene expression changes and computationally predicted changes was low (only ∼15% of the differentially expressed genes were predicted by the model).
Novel regulatory rules were predicted by conducting a two-way analysis of variation comparing the microarray data set for each knockout strain in this study against the wild-type array data. The results allowed many regulatory rules to be rewritten, relaxed, or removed. These updated regulatory rules were then again tested by allowing the model to predict gene expression changes. The updated model accurately predicted 100 of the 151 measured gene expression changes and gave 0 false positives.
In this study, the iterative modification of gene regulatory rules led to three conclusions that suggest the need for the analysis of microarray data in the modeling context. First, the Boolean rule prediction can be difficult under some knockout conditions, due to complex interactions with other transcription factors. Second, when a transcription factor is activated, the levels of mRNA for the regulatory protein can either increase or decrease. Therefore, the identification of gene regulatory networks will not occur solely by the identification of correlated gene sets through statistical analysis. Third, differential gene expression patterns are the effects of complex interactions and indirect effects. Because transcription factors can be affected by many factors such as internal metabolite concentrations and metabolic by-products, the elucidation of gene regulatory networks will not likely occur without the aid of computational models and follow-up experiments (4).
Characterizing metabolic network mutants in adaptive evolution by introducing network information into analyses.
Growth profiles for in silico E. coli gene knockout mutants on different growth substrates were computed. When the genes were knocked out in vitro and the strains were evolved to the end point (600 to 800 generations), it was demonstrated that 78% of these strains evolved to computationally predicted growth rates (30). Many of the failed predictions involved Δppc and glucose-grown Δtpi strains. These mutants were later subjected to isotopomer metabolic flux analysis and gene expression profiling (33).
Both mutants initially suffered impaired growth phenotypes but rapidly evolved reproducibly improved phenotypes. Many recovered wild-type-like phenotypes at the end of the evolution time course. The metabolic flux analysis revealed that, initially, the mutants diverted flux around the genetic lesions through normally latent pathways like the glucose-inhibited glyoxylate shuttle or the methylglyoxyal bypass. The fluxomic data suggested that mutants evolving in parallel relied on the same adaptation principle of the redirection of local flux around the lesion to cope with the altered metabolomic balance.
Since fluxomic data only suggested sites of altered activity, gene expression profiling was employed to find a genetic basis for the altered pathway utilization. The Δtpi strains demonstrated changes in gene expression among the reactions involved in the methylglyoxyal bypass. Analyses of changes in gene expression between the wild type and the evolved Δppc strains showed the upregulation of the glyoxylate shunt and the downregulation of the lower part of the tricarboxylic acid cycle in the mutant strains. In addition, adaptive mechanisms in the TRN that would help the cells meet energy demands were seen; nevertheless, it was apparent that the mechanisms underlying adaptive evolution include differential gene expression patterns, which aid in the rerouting of metabolic fluxes around genetic lesions and the adjustment of downstream fluxes to achieve one of potentially many optimal states.
Reconciling suboptimal growth with pathway inactivation predictions.
As discussed above, in a few cases, mutant strains of E. coli failed to evolve completely to achieve the computationally predicted growth rates (30). It is believed that computational overpredictions such as these are the results of reactions that are unable to operate at full capacity. Based on this assumption, a computational method was developed to find the reactions that acted as bottlenecks in the metabolic network when growth rates were incorrectly predicted (38).
The method, called optimal metabolic network identification, was applied to five E. coli mutant strains that all grew suboptimally, even after adaptive evolution (30). For each case, data from the 13C-labeled-tracer experiments just discussed (33) were used to predict sets of bottleneck reactions that, if removed from the model, would improve model predictions for the intracellular flux and growth rate. To validate these sets of bottleneck reactions, gene expression data were analyzed to see if the genes associated with the bottleneck reactions were consistently downregulated. In this analysis, it was found that most genes associated with bottleneck reactions were, in fact, downregulated.
The utility of this method was further validated with a double-gene-deletion strain (ΔpfkA Δpta) (34, 42) for which there were no flux data. By using only uptake and secretion rates for a few metabolites, bottleneck reactions (e.g., those of pyruvate dehydrogenase and ATP synthase) were predicted, and the genes associated with these reactions were evaluated in relation to the gene expression data. Once again, there was strong concordance between the predicted sets of genes and the measured expression levels of those genes.
The comparison of gene expression data with the bottleneck predictions demonstrated the downregulation of many genes associated with the bottleneck reactions, thus providing support for these predictions and suggesting that transcriptional regulatory mechanisms likely play a part in inhibiting these strains from reaching an optimal growth rate.
GENE-LEVEL ANALYSIS WITHIN THE GENOME-SCALE FRAMEWORK
Model-driven discovery of metabolic pathways and gene function.
In terms of metabolism and transcriptional regulation, E. coli is arguably one of the best-characterized model organisms (77). However, despite this depth of knowledge, it is apparent that there are still unknown metabolic pathways (56, 63, 68), many uncharacterized regulatory interactions (20, 73, 77), and hundreds of ORFs with unknown functions (69).
Recently, a model-driven approach was undertaken to reconcile cases in which a genome-scale model of E. coli failed to predict the experimental growth phenotype data (68). There were 50 cases in which the failure was due to incomplete knowledge of E. coli metabolism; therefore, a computational algorithm was used to predict potential reactions or transporters that could reconcile the model predictions and experimental results. The algorithm queried a database of all known metabolic reactions in living organisms (49) and computed the minimum number of reactions needed to restore in silico growth in the model. A subset of the predicted solutions was chosen for experimental verification, leading to the annotation of eight ORFs.
The annotation of three ORFs in this study was facilitated with the use of Affymetrix gene expression data. As one example, the in silico model predicted that E. coli was not able to grow on l-galactonate, as there were no known genes for such a pathway. Candidate genes for l-galactonate oxidation and transport were elucidated from Affymetrix gene expression profiles for E. coli grown on l-galactonate. When compared to an assortment of arrays from other growth conditions, these arrays showed the strong upregulation of two ORFs: yjjL (44-fold increase) and yjjN (23-fold increase). After additional experiments (gene knockout screens and reverse transcriptase PCR analyses), these genes were annotated as follows: yjjL encodes a product that transports l-galactonate, yjjN is responsible for the l-galactonate oxidoreductase activity, and yjjM regulates the gene expression.
Thus, it is apparent that gene expression profiling may be used in tandem with in silico modeling to demonstrate where knowledge is incomplete. While successful in silico predictions can help to validate a model, this study showed (i) that failed predictions may be used to algorithmically generate experimentally testable hypotheses and lead to the refinement of the genome annotation on a gene-by-gene basis and (ii) that microarray data are more informative when used to find answers to fine-grain hypotheses.
CONCLUSIONS AND FUTURE DIRECTIONS
Several forms of statistical analyses of genome-scale gene expression data have been practiced extensively over the past decade. Numerous studies of E. coli physiological functions and responses have benefitted from these approaches (13, 23, 48, 52, 79, 85). However, while purely statistical data analysis approaches play an important role, our experience and evidence from recent studies have shown that the analysis of gene expression data in the context of an in silico model offers an alternative approach that allows better extraction of mechanistic, functional, and fine-grain knowledge from data (14, 22, 50, 51, 74). The modeling framework has been used in conjunction with gene expression profiling to discover properties of E. coli metabolism and transcriptional regulation. In addition, these approaches have been used together to identify inaccurate predictions and help identify novel pathways and annotate uncharacterized genes. This modeling context for gene expression profiling content will likely open up new possibilities for biological discovery in E. coli and other organisms.
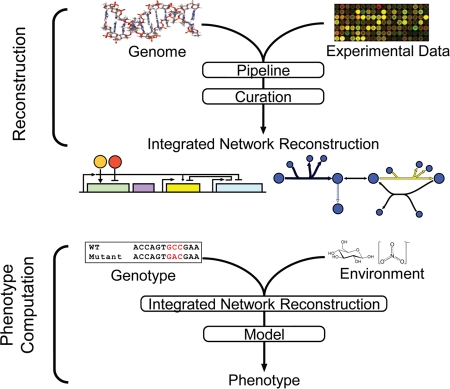
The microarray platform has been in successful use for over a decade and has helped pave the way for the improved analysis of novel sequencing technologies (57, 59, 72) which will likely replace much of the work currently done on a microarray, as these technologies offer an improved dynamic range and the identification of all RNAs, not just those which are able to bind to a complementary sequence. However, the computational framework in constraint-based modeling will remain a useful platform for the analysis of these and other high-throughput technologies. In fact, methods to create a data integration pipeline that will incorporate gene expression profiling data and transcription factor binding information, etc., to help build more complete genome-scale networks are being developed (4, 10, 16, 27, 43, 44). These methods will effectively streamline constraint-based modeling into a two-step process (Fig. 2). This process will lead to the rapid reconstruction of networks (including those of metabolism, transcriptional regulation, and the machinery used for transcription and translation) (78). Such networks can, in turn, aid in improving data analysis. Gene expression profiling was one of the first high-throughput methods that has allowed for prediction with and validation of genome-scale models. Genome-scale models, in turn, can now utilize this and other high-throughput data sources for further discovery.
FIG. 2.
Pipeline of data to generate genome-scale in silico models that are predictive. Constraint-based modeling is evolving into a process with two steps in computation: (i) reconstruction and (ii) simulation. Novel computational methods are able to integrate genome sequences and various high-throughput data types (gene expression data and chromatin immunoprecipitation-on-chip/chromatin immunoprecipitation-sequencing data, etc.) to aid in the reconstruction of metabolic and transcription/translation networks and TRNs. After curation, an integrated network reconstruction can be converted into a model based on the organism's genotype and environmental conditions. From that point, hypotheses can be tested in silico and exploratory analysis can be conducted. WT, wild-type.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Akesson, M., J. Forster, and J. Nielsen. 2004. Integration of gene expression data into genome-scale metabolic models. Metab. Eng. 6285-293. [DOI] [PubMed] [Google Scholar]
- 2.Almaas, E., B. Kovacs, T. Vicsek, Z. N. Oltvai, and A. L. Barabasi. 2004. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427839-843. [DOI] [PubMed] [Google Scholar]
- 3.Alper, H., Y. S. Jin, J. F. Moxley, and G. Stephanopoulos. 2005. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 7155-164. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, C. L., and B. O. Palsson. 2006. Iterative reconstruction of transcriptional regulatory networks: an algorithmic approach. PLoS Comput. Biol. 2e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett, C. L., C. D. Herring, J. L. Reed, and B. O. Palsson. 2005. The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states. Proc. Natl. Acad. Sci. USA 10219103-19108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, S. A., and B. O. Palsson. 2008. Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 4e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beg, Q. K., A. Vazquez, J. Ernst, M. A. de Menezes, Z. Bar-Joseph, A. L. Barabasi, and Z. N. Oltvai. 2007. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc. Natl. Acad. Sci. USA 10412663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4309-314. [DOI] [PubMed] [Google Scholar]
- 9.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29365-371. [DOI] [PubMed] [Google Scholar]
- 10.Breitling, R., D. Vitkup, and M. P. Barrett. 2008. New surveyor tools for charting microbial metabolic maps. Nat. Rev. Microbiol. 6156-161. [DOI] [PubMed] [Google Scholar]
- 11.Burgard, A. P., E. V. Nikolaev, C. H. Schilling, and C. D. Maranas. 2004. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 14301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butte, A. 2002. The use and analysis of microarray data. Nat. Rev. Drug Discov. 1951-960. [DOI] [PubMed] [Google Scholar]
- 13.Cardinale, C. J., R. S. Washburn, V. R. Tadigotla, L. M. Brown, M. E. Gottesman, and E. Nudler. 2008. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chechik, G., E. Oh, O. Rando, J. Weissman, A. Regev, and D. Koller. 2008. Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat. Biotechnol. 261251-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, L., and D. Vitkup. 2006. Predicting genes for orphan metabolic activities using phylogenetic profiles. Genome Biol. 7R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho, B. K., C. L. Barrett, E. M. Knight, Y. S. Park, and B. O. Palsson. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. USA 10519462-109467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clare, A., and R. D. King. 2002. How well do we understand the clusters found in microarray data? In Silico Biol. 2511-522. [PubMed] [Google Scholar]
- 18.Covert, M. W., and B. O. Palsson. 2003. Constraints-based models: regulation of gene expression reduces the steady-state solution space. J. Theor. Biol. 221309-325. [DOI] [PubMed] [Google Scholar]
- 19.Covert, M. W., and B. O. Palsson. 2002. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J. Biol. Chem. 27728058-28064. [DOI] [PubMed] [Google Scholar]
- 20.Covert, M. W., E. M. Knight, J. L. Reed, M. J. Herrgard, and B. O. Palsson. 2004. Integrating high-throughput and computational data elucidates bacterial networks. Nature 42992-96. [DOI] [PubMed] [Google Scholar]
- 21.Cui, X., and G. A. Churchill. 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David, H., G. Hofmann, A. P. Oliveira, H. Jarmer, and J. Nielsen. 2006. Metabolic network driven analysis of genome-wide transcription data from Aspergillus nidulans. Genome Biol. 7R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 1835239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards, J. S., and B. O. Palsson. 2000. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 975528-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards, J. S., and B. O. Palsson. 1999. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 27417410-17416. [DOI] [PubMed] [Google Scholar]
- 26.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19125-130. [DOI] [PubMed] [Google Scholar]
- 27.Faith, J. J., B. Hayete, J. T. Thaden, I. Mogno, J. Wierzbowski, G. Cottarel, S. Kasif, J. J. Collins, and T. S. Gardner. 2007. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feist, A. M., and B. O. Palsson. 2008. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 26659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feist, A. M., C. S. Henry, J. L. Reed, M. Krummenacker, A. R. Joyce, P. D. Karp, L. J. Broadbelt, V. Hatzimanikatis, and B. O. Palsson. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fong, S. S., and B. O. Palsson. 2004. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat. Genet. 361056-1058. [DOI] [PubMed] [Google Scholar]
- 31.Fong, S. S., A. R. Joyce, and B. O. Palsson. 2005. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 151365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fong, S. S., J. Y. Marciniak, and B. O. Palsson. 2003. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J. Bacteriol. 1856400-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong, S. S., A. Nanchen, B. O. Palsson, and U. Sauer. 2006. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J. Biol. Chem. 2818024-8033. [DOI] [PubMed] [Google Scholar]
- 34.Fong, S. S., A. P. Burgard, C. D. Herring, E. M. Knight, F. R. Blattner, C. D. Maranas, and B. O. Palsson. 2005. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 91643-648. [DOI] [PubMed] [Google Scholar]
- 35.Fuhrer, T., L. Chen, U. Sauer, and D. Vitkup. 2007. Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J. Bacteriol. 1898073-8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghim, C. M., K. I. Goh, and B. Kahng. 2005. Lethality and synthetic lethality in the genome-wide metabolic network of Escherichia coli. J. Theor. Biol. 237401-411. [DOI] [PubMed] [Google Scholar]
- 37.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrgard, M. J., S. S. Fong, and B. O. Palsson. 2006. Identification of genome-scale metabolic network models using experimentally measured flux profiles. PLoS Comput. Biol. 2e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrgard, M. J., B. S. Lee, V. Portnoy, and B. O. Palsson. 2006. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 16627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herring, C. D., A. Raghunathan, C. Honisch, T. Patel, M. K. Applebee, A. R. Joyce, T. J. Albert, F. R. Blattner, D. van den Boom, C. R. Cantor, and B. O. Palsson. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 381406-1412. [DOI] [PubMed] [Google Scholar]
- 41.Hua, Q., A. R. Joyce, B. O. Palsson, and S. S. Fong. 2007. Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Appl. Environ. Microbiol. 734639-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hua, Q., A. R. Joyce, S. S. Fong, and B. O. Palsson. 2006. Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains. Biotechnol. Bioeng. 95992-1002. [DOI] [PubMed] [Google Scholar]
- 43.Hwang, D., A. G. Rust, S. Ramsey, J. J. Smith, D. M. Leslie, A. D. Weston, P. de Atauri, J. D. Aitchison, L. Hood, A. F. Siegel, and H. Bolouri. 2005. A data integration methodology for systems biology. Proc. Natl. Acad. Sci. USA 10217296-17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang, D., J. J. Smith, D. M. Leslie, A. D. Weston, A. G. Rust, S. Ramsey, P. de Atauri, A. F. Siegel, H. Bolouri, J. D. Aitchison, and L. Hood. 2005. A data integration methodology for systems biology: experimental verification. Proc. Natl. Acad. Sci. USA 10217302-17307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420186-189. [DOI] [PubMed] [Google Scholar]
- 46.Imielinski, M., C. Belta, A. Halasz, and H. Rubin. 2005. Investigating metabolite essentiality through genome-scale analysis of Escherichia coli production capabilities. Bioinformatics 212008-2016. [DOI] [PubMed] [Google Scholar]
- 47.Isalan, M., C. Lemerle, K. Michalodimitrakis, C. Horn, P. Beltrao, E. Raineri, M. Garriga-Canut, and L. Serrano. 2008. Evolvability and hierarchy in rewired bacterial gene networks. Nature 452840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Justino, M. C., J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 2802636-2643. [DOI] [PubMed] [Google Scholar]
- 49.Kanehisa, M., S. Goto, S. Kawashima, Y. Okuno, and M. Hattori. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32D277-D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kharchenko, P., D. Vitkup, and G. M. Church. 2004. Filling gaps in a metabolic network using expression information. Bioinformatics 20(Suppl. 1)i178-i185. [DOI] [PubMed] [Google Scholar]
- 51.Kharchenko, P., L. Chen, Y. Freund, D. Vitkup, and G. M. Church. 2006. Identifying metabolic enzymes with multiple types of association evidence. BMC Bioinformatics 7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 9712170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kummel, A., S. Panke, and M. Heinemann. 2006. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 22006.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee, K. H., J. H. Park, T. Y. Kim, H. U. Kim, and S. Y. Lee. 2007. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol. Syst. Biol. 3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis, N. E., N. Jamshidi, I. Thiele, and B. Ø. Palsson. Metabolic systems biology: a constraint-based approach. In Robert A. Meyers (ed.), Encyclopedia of complexity and systems science, in press. Springer, New York, NY.
- 56.Loh, K. D., P. Gyaneshwar, E. Markenscoff Papadimitriou, R. Fong, K. S. Kim, R. Parales, Z. Zhou, W. Inwood, and S. Kustu. 2006. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. USA 1035114-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mardis, E. R. 2008. The impact of next-generation sequencing technology on genetics. Trends Genet. 24133-141. [DOI] [PubMed] [Google Scholar]
- 58.Motter, A. E., N. Gulbahce, E. Almaas, and A. L. Barabasi. 2008. Predicting synthetic rescues in metabolic networks. Mol. Syst. Biol. 4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagalakshmi, U., Z. Wang, K. Waern, C. Shou, D. Raha, M. Gerstein, and M. Snyder. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 3201344-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Notebaart, R. A., B. Teusink, R. J. Siezen, and B. Papp. 2008. Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Comput. Biol. 4e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pal, C., B. Papp, and M. J. Lercher. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 371372-1375. [DOI] [PubMed] [Google Scholar]
- 62.Pal, C., B. Papp, M. J. Lercher, P. Csermely, S. G. Oliver, and L. D. Hurst. 2006. Chance and necessity in the evolution of minimal metabolic networks. Nature 440667-670. [DOI] [PubMed] [Google Scholar]
- 63.Piskur, J., K. D. Schnackerz, G. Andersen, and O. Bjornberg. 2007. Comparative genomics reveals novel biochemical pathways. Trends Genet. 23369-372. [DOI] [PubMed] [Google Scholar]
- 64.Price, N. D., J. L. Reed, and B. O. Palsson. 2004. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat. Rev. Microbiol. 2886-897. [DOI] [PubMed] [Google Scholar]
- 65.Reed, J. L., and B. O. Palsson. 2004. Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: assessment of correlated reaction subsets that comprise network states. Genome Res. 141797-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed, J. L., I. Famili, I. Thiele, and B. O. Palsson. 2006. Towards multidimensional genome annotation. Nat. Rev. Genet. 7130-141. [DOI] [PubMed] [Google Scholar]
- 67.Reed, J. L., T. D. Vo, C. H. Schilling, and B. O. Palsson. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reed, J. L., T. R. Patel, K. H. Chen, A. R. Joyce, M. K. Applebee, C. D. Herring, O. T. Bui, E. M. Knight, S. S. Fong, and B. O. Palsson. 2006. Systems approach to refining genome annotation. Proc. Natl. Acad. Sci. USA 10317480-17484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riley, M., T. Abe, M. B. Arnaud, M. K. Berlyn, F. R. Blattner, R. R. Chaudhuri, J. D. Glasner, T. Horiuchi, I. M. Keseler, T. Kosuge, H. Mori, N. T. Perna, G. Plunkett III, K. E. Rudd, M. H. Serres, G. H. Thomas, N. R. Thomson, D. Wishart, and B. L. Wanner. 2006. Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satish Kumar, V., M. S. Dasika, and C. D. Maranas. 2007. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics 8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwartz, J. M., C. Gaugain, J. C. Nacher, A. de Daruvar, and M. Kanehisa. 2007. Observing metabolic functions at the genome scale. Genome Biol. 8R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shendure, J., and H. Ji. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 261135-1145. [DOI] [PubMed] [Google Scholar]
- 73.Shlomi, T., Y. Eisenberg, R. Sharan, and E. Ruppin. 2007. A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol. Syst. Biol. 3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shlomi, T., M. N. Cabili, M. J. Herrgard, B. O. Palsson, and E. Ruppin. 2008. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 261003-1010. [DOI] [PubMed] [Google Scholar]
- 75.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420190-193. [DOI] [PubMed] [Google Scholar]
- 76.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 10215545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thieffry, D., A. M. Huerta, E. Perez-Rueda, and J. Collado-Vides. 1998. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays 20433-440. [DOI] [PubMed] [Google Scholar]
- 78.Thiele, I., N. Jamshidi, R. M. T. Fleming, and B. Ø. Palsson. 2009. Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput. Biol. 5e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Traxler, M. F., D. E. Chang, and T. Conway. 2006. Guanosine 3′,5′-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. USA 1032374-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vazquez, A., Q. K. Beg, M. A. Demenezes, J. Ernst, Z. Bar-Joseph, A. L. Barabasi, L. G. Boros, and Z. N. Oltvai. 2008. Impact of the solvent capacity constraint on E. coli metabolism. BMC Syst. Biol. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verducci, J. S., V. F. Melfi, S. Lin, Z. Wang, S. Roy, and C. K. Sen. 2006. Microarray analysis of gene expression: considerations in data mining and statistical treatment. Physiol. Genomics 25355-363. [DOI] [PubMed] [Google Scholar]
- 82.Vo, T. D., W. N. Paul Lee, and B. O. Palsson. 2007. Systems analysis of energy metabolism elucidates the affected respiratory chain complex in Leigh's syndrome. Mol. Genet. Metab. 9115-22. [DOI] [PubMed] [Google Scholar]
- 83.Vo, T. D., H. J. Greenberg, and B. O. Palsson. 2004. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J. Biol. Chem. 27939532-39540. [DOI] [PubMed] [Google Scholar]
- 84.Wang, Q., X. Chen, Y. Yang, and X. Zhao. 2006. Genome-scale in silico aided metabolic analysis and flux comparisons of Escherichia coli to improve succinate production. Appl. Microbiol. Biotechnol. 73887-894. [DOI] [PubMed] [Google Scholar]
- 85.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 1834562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]