Abstract
Nutritional competence is the ability of bacterial cells to utilize exogenous double-stranded DNA molecules as a nutrient source. We previously identified several genes in Escherichia coli that are important for this process and proposed a model, based on models of natural competence and transformation in bacteria, where it is assumed that single-stranded DNA (ssDNA) is degraded following entry into the cytoplasm. Since E. coli has several exonucleases, we determined whether they play a role in the long-term survival and the catabolism of DNA as a nutrient. We show here that mutants lacking either ExoI, ExoVII, ExoX, or RecJ are viable during all phases of the bacterial life cycle yet cannot compete with wild-type cells during long-term stationary-phase incubation. We also show that nuclease mutants, alone or in combination, are defective in DNA catabolism, with the exception of the ExoX− single mutant. The ExoX− mutant consumes double-stranded DNA better than wild-type cells, possibly implying the presence of two pathways in E. coli for the processing of ssDNA as it enters the cytoplasm.
Bacteria occupy virtually all niches capable of supporting life, usually in environments where nutrient availability is poor and competition for resources is intense (12). Therefore, starvation is a common state for most bacterial populations for the majority of their existence. Such conditions can be approximated by long-term incubation in the stationary phase of the bacterial life cycle (12).
We showed previously that “nutritional” competence is beneficial for bacterial cells, specifically Escherichia coli cells, where extracellular double-stranded DNA molecules (dsDNA) can serve as a nutrient source, and it is necessary for competitive survival (13, 25). A model for DNA uptake and catabolism was proposed in which dsDNA binds to the cell, enters the periplasm, and then is transported across the inner membrane where it is processed into single-stranded DNA (ssDNA) as it enters the cytoplasm (6, 7, 9, 25, 28, 29). In the final step, intracellular exonucleases degrade the ssDNA inside the cytoplasm, where degradation products can serve as the sole source of carbon and energy for the cell (13, 25). E. coli contains several nucleases that are able to cleave ssDNA and/or dsDNA molecules. For example, exonuclease I, exonuclease VII, and RecJ digest ssDNA (4, 20, 22), and exonuclease X digests both dsDNA and ssDNA (31). It is possible that one or more of these enzymes participate in the digestion of imported DNA during DNA uptake.
We wanted to determine whether known intracellular ssDNA nucleases play a role in the catabolism of DNA as a nutrient. Exonucleases I, VII, and X and RecJ were chosen because they specifically target ssDNA in the cytoplasm (Table 1). Single and multiple null mutants were constructed since these exonucleases can perform redundant functions in DNA repair and recombination (2, 16, 32). Exonuclease mutants showed the stationary-phase-specific competition-defective (SPCD) phenotype (13, 25) during long-term stationary-phase incubation and are defective in the use of dsDNA as the sole source of carbon and energy, with the exception of the ExoX− single mutant.
TABLE 1.
Characteristics of E. coli ssDNA exonucleases
| Genea | Protein | Polarity | Target | Location (min) |
|---|---|---|---|---|
| xonA, sbcA | ExoI | 3′→5′ | ssDNA | 44.85 |
| recJ | RecJ | 5′→3′ | ssDNA | 65.41 |
| xseA | ExoVII | 3′⇆5′ | ssDNA | 56.74 |
| exoX, yobC | ExoX | 3′→5′ | ssDNA/dsDNA | 41.48 |
Other commonly used gene names are indicated as appropriate.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All experiments were performed with strains derived from the E. coli K-12 strain ZK126 and are listed in Table 2.
TABLE 2.
Bacterial strains
| Strain | Relevant genotype/phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| ZK126 | W3110 ΔlacU169 tna-2 | 34 |
| ZK1142 | ZK126 Nalr | 34 |
| ZK1143 | ZK126 Strr | 34 |
| SF2526 | ZK126 xonA::Camr | This work |
| SF2527 | ZK126 recJ::Camr | This work |
| SF2528 | ZK126 xseA::Camr | This work |
| SF2529 | ZK126 exoX::Camr | This work |
| SF2530 | ZK126 xonA::Kanr | This work |
| SF2531 | ZK126 recJ::Kanr | This work |
| SF2532 | ZK126 xseA::Kanr | This work |
| SF2533 | ZK126 exoX::Kanr | This work |
| SF2534 | ZK126 xonA::FRT | This work |
| SF2535 | ZK126 recJ::FRT | This work |
| SF2536 | ZK126 xseA::FRT | This work |
| SF2537 | ZK126 exoX::FRT | This work |
| SF2538 | ZK126 exoX::FRT xonA::FRT | This work |
| SF2539 | ZK126 xseA::FRT xonA::FRT | This work |
| SF2540 | ZK126 xseA::FRT recJ::FRT | This work |
| SF2541 | ZK126 exoX::FRT xonA::FRT recJ::FRT | This work |
| SF2542 | ZK126 exoX::FRT xonA::FRT xseA::FRT | This work |
| SF2543 | ZK126 xseA::FRT recJ::FRT xonA::FRT exoX::FRT | This work |
| SF2544 | ZK126 xseA::FRT exoX::FRT xonA::FRT recJ::Camr | This work |
FRT refers to an eliminated drug resistance cassette using a helper plasmid encoding the FLP recombinase, leaving a single FLP recombination (FRT) site (see reference 8).
Gene disruptions.
Each gene of interest was replaced either with a cassette expressing chloramphenicol (Camr) or kanamycin (Kanr) resistance using a Red recombinase-mediated system (8) with the following modifications: template DNA for PCRs was not digested before electroporation, and 15 ml of cells (OD600, ∼0.6) per knockout were made electrocompetent immediately prior to electroporation (25). Also, a greater number of transformants was obtained when the Red recombinase was induced for 1 h before preparing cells for electroporation. Table 3 lists the primers used to generate PCR fragments containing the Camr or Kanr cassette flanked by regions of homology for each particular gene.
TABLE 3.
Primers used to generate null mutations in different exonuclease genes
| Gene | Primera | Sequence |
|---|---|---|
| xonA | H1P1 | ATGATGAATGACGGTAAGCAACAATCTACCTTTTTGTTTGTGTAGGCTGGAGCTGCTTC |
| H2P2 | GACAATCTCTTCCGCGTACTGCCAAAGTGCTTTTAACAGCATATGAATATCCTCCTTAG | |
| CHECK | CCAGGACCCGGCATGAATCTCTG | |
| recJ | H1P1 | AAACAACAGATACAACTTCGTCGCCGTGAAGTCGATGAAAGTGTAGGCTGGAGCTGCTTC |
| H2P2 | TGGCCAGATATTGTCGATGATAATTTGCAGGCTGCGGTTGCATATGAATATCCTCCTTAG | |
| CHECK | GGGATCACCGTGCGTTATCTTGC | |
| xseA | H1P1 | TTACCTTCTCAATCCCCTGCAATTTTTACCGTTAGTCGCCGTGTAGGCTGGAGCTGCTTC |
| H2P2 | CTTTCTATCCAGCCGTCTTCCAGACGTGTGGTTAGCATTTCATATGAATATCCTCCTTAG | |
| CHECK | CGTTACGCTCGGTCAGTTCTTTCAC | |
| exoX | H1P1 | ATGTTGCGCATTATCGATACAGAAACCTGCGGTTTGCAGGTGTAGGCTGGAGCTGCTTC |
| H2P2 | AGTATTTTCCAGATAATGTTTCAGTGTTAAACGCAGCTCCATATGAATATCCTCCTTAG | |
| CHECK | GGATCTACCATCAGAGTCATGGTGC |
H1 corresponds to approximately the first 12 codons and H2 corresponds to the last 12 codons of each particular gene. P1 and P2 are underlined and correspond to the regions flanking the antibiotic resistance cassette located on the pKD3 and pKD4 plasmids (8).
The PCR products were purified prior to electroporation using a QIAquick PCR purification kit (Qiagen). All mutants were colony purified and tested for the absence of the Red recombinase-expressing plasmid pKD46 by plating for antibiotic sensitivity on agar plates containing ampicillin (150 μg/ml). To verify each mutation, PCRs were performed using one primer specific to a region upstream of the insertion point (called “check” primers [Table 3]) and another primer complementary to the sequence of P1 (P1-C, CGAAGCAGCTCCAGCCTACAC) to amplify a band of specific size. Once confirmed, the drug resistance cassettes flanked by the FLP recombination target sites were removed by introducing the FLP recombinase-expressing plasmid pCP20 as specified by the protocol (8), with the minor modification that colonies were additionally streaked on Luria-Bertani (LB) plates to ensure the complete loss of the pCP20 plasmid. Mutants with more than one exonuclease gene eliminated were constructed by the bacteriophage P1-mediated transduction of the Camr or Kanr allele, followed by removal of the Camr or Kanr cassette by FLP recombination as appropriate. Each mutant strain with multiple gene knockouts was verified by PCR.
Long-term survival assays in LB and defined minimal media.
E. coli wild-type (ZK126) and mutant strains were separately incubated overnight in 5 ml of LB broth (Difco) or in M63 minimal medium containing 0.2% glucose at 37°C with aeration in a humidified warm room (65% relative humidity). Titers were determined by the periodic sampling of the cultures and serial dilution and plating on LB plates and are accurate to within threefold (19). The limit of detection was <1,000 CFU/ml.
Batch culture competition assays.
The E. coli wild-type, ZK1142 (Nalr) or ZK1143 (Strr), and mutant strains were separately incubated overnight in 5 ml of LB broth (Difco) at 37°C with aeration. Cultures were then diluted 1:1,000 (vol/vol) in fresh LB broth, either individually or in coculture, by inoculating 5 μl of LB broth with each bacterial strain (13). Subpopulation titers were determined by periodic sampling of the cultures, serial dilution, and plating on medium containing appropriate antibiotics (nalidixic acid at 20 μg/ml, streptomycin at 50 μg/ml, Kan at 50 μg/ml, or Cam at 20 μg/ml) to determine the relative fitness (10, 14). All antibiotics were purchased from Sigma-Aldrich. Experiments were performed in triplicate for each mutant strain. The limit of detection in all experiments was <1,000 CFU/ml.
Preparation of minimal medium supplemented with purified DNA.
M63 medium was prepared as described previously (24, 26). Sonicated salmon sperm DNA (Sigma-Aldrich) was prepared as previously described (13, 25), including multiple rounds of organic extraction (phenol, phenol-chloroform, chloroform, and ether) to remove the contaminating proteins. For each “DNA eating” experiment, purified salmon sperm DNA was freshly precipitated with ethanol-acetate and resuspended to a final concentration of 0.1% (wt/vol) in 1-ml M63 cultures. Media were prepared and all experiments were performed in new acid-treated glassware to eliminate the possibility of contamination by other nutrients. Cultures were inoculated at a dilution of ∼1:10,000,000 (vol/vol; ∼102 CFU/ml), and viable cell counts were determined periodically for 48 h by plating on LB plates. Experiments were performed several times for each mutant (n = 2 to 4).
RESULTS
Stationary phase survival in LB and M63 media.
Based on the DNA eating model, the prediction was made that one or more exonucleases might be involved in the degradation of ssDNA inside the cytoplasm. E. coli has four potent exonucleases targeting ssDNA: ExoI, RecJ, ExoVII, and ExoX. To evaluate the role of these enzymes in the catabolism of internalized ssDNA, isogenic null mutants were constructed using the method of Datsenko and Wanner (8). Due to the fact that there is redundancy in the functions of these nucleases, we also created double, triple, and quadruple mutants to observe any deficiency in the DNA consumption process.
It was previously reported that multiple exonuclease mutants are viable, although quadruple mutants show a reduction in viability after incubation at low temperature in rich medium (2, 32). Here we created new mutant strains in order to (i) maintain the isogenicity of the markers used, (ii) remove, as much as was practical, the possible effects of the presence of multiple antibiotic resistance markers which might have differential effects on fitness in competition experiments, and (iii) create prototrophic mutant strains appropriate for use in the DNA eating assays.
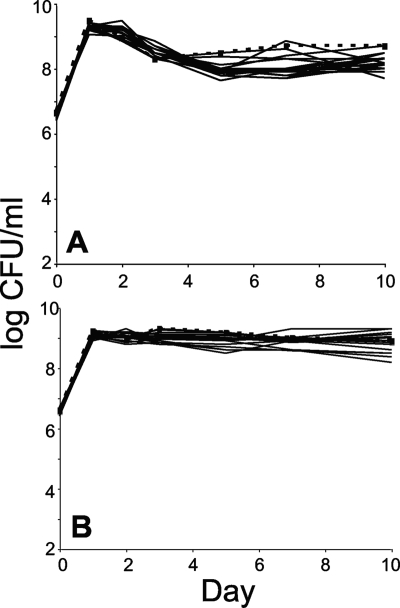
Once the mutants were constructed (Table 2), it was crucial to determine the ability of the null mutants to grow in LB broth and minimal medium supplemented with glucose to eliminate the possibility of gross fitness defects as shown previously (2, 32). Figure 1 shows the monoculture growth patterns of the wild type and the null mutants in different media. No significant defects were observed for any mutant strains, including the quadruple mutant, indicating that the exonucleases are not essential for survival under these conditions during the exponential, stationary, or long-term stationary phases.
FIG. 1.
Exonuclease null mutants show no growth defect during long-term stationary phase incubation. Cells were continuously incubated for 10 days in LB medium (A) or M63 minimal medium supplemented with 0.2% glucose (B). Wild-type cells are represented by filled squares and a dashed line; mutants are represented by solid lines. Each mutant listed in Table 4 is presented here. However, strains are not distinguished since all values agree within the titering error (see reference 19). The limit of detection is <1,000 CFU/ml. Representative data are shown.
Exonuclease mutants show a SPCD phenotype when coincubated with wild-type cells during the long-term stationary phase.
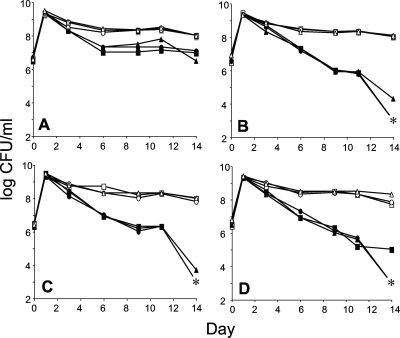
Since the exonuclease mutants show similar patterns of long-term survival when incubated in monoculture, we next assayed for the SPCD phenotype, which reflects a deficit in some activity that is only revealed in competition with otherwise wild-type strains during the long-term stationary phase (14, 25). The mutants exhibit no fitness loss during the exponential phase of growth or upon entry into stationary phase when cocultured with the wild-type parent. However, after several days of coincubation in stationary phase with the wild-type cells, all mutants are outcompeted and most of them are completely lost after 2 weeks of continuous batch coculture (Fig. 2). Such a phenotype is indicative of SPCD and shows that exonucleases are important during long-term survival in stationary phase. While the exoI mutant shows a reduction in competitive fitness, unlike the other three single mutant strains that are outcompeted, its final population density stabilizes at 10% of that of the wild type. This suggests that the loss of ExoI function is being compensated for by other proteins, perhaps due to functional redundancy or because the role of ExoI is less important under these conditions.
FIG. 2.
Survival patterns of different exonuclease mutants competing with wild-type parental strains when cocultured in LB broth. Cultures were continuously incubated for 14 days at 37°C with aeration. Each competition was performed in triplicate; different symbols represent different competition pairs. Strains contain a null mutation for exonuclease I (A), RecJ exonuclease (B), exonuclease VII (C), or exonuclease X (D). Mutant and wild-type strains are represented by filled and open symbols, respectively. Asterisks indicate no detectable counts (the limit of detection is <1,000 CFU/ml).
Ability of exonuclease mutants to utilize dsDNA as a sole source of carbon and energy.
To address the ability of exonuclease null mutants to utilize dsDNA, a DNA eating assay was performed in which the only source of carbon and energy supplied was highly purified sonicated salmon sperm DNA at a concentration of 0.1% (wt/vol) (13, 25). Wild-type cells and different mutants were inoculated at ∼100 CFU/ml into minimal medium supplemented with dsDNA and incubated at 37°C with aeration for 2 days. Cell titers were determined over 48 h, and final growth yields were calculated (Table 4). The wild-type cells showed an increase in cell density of more than 200-fold compared to the mutant strains, which showed little or no growth with one exception, the ExoX− strain. The DNA eating phenotype of the exoX single mutant was unexpectedly high, and the mutant showed a growth yield of ∼1,450-fold, which is, on average, seven times greater than that of the wild-type cells.
TABLE 4.
Average growth yields of wild-type and exonuclease null mutant cells
| Strain | Presence or mutation ofa:
|
Average yield (fold) (±SD)b | |||
|---|---|---|---|---|---|
| xonA (ExoI) | recJ (RecJ) | xseA (ExoVII) | exoX (ExoX) | ||
| Wild type | + | + | + | + | 223 ± 103 |
| xonA strain | − | + | + | + | 3.5 ± 1.1 |
| recJ strain | + | − | + | + | 4.0 ± 3.2 |
| xseA strain | + | + | − | + | 2.6 ± 0.5 |
| yobC strain | + | + | + | − | 1,458 ± 710 |
| xonA yobC strain | − | + | + | − | 2.3 ± 1.4 |
| xonA xseA strain | − | + | − | + | 2.0 ± 0.2 |
| recJ xseA strain | + | − | − | + | 1.4 ± 1.1 |
| xonA recJ yobC strain | − | − | + | − | 2.9 ± 0.6 |
| xonA xseA yobC strain | − | + | − | − | 2.3 ± 1.4 |
| xonA recJ xseA yobC strain | − | − | − | − | 3.0 ± 2.7 |
+, presence of a gene; −, mutation of a gene. Experiments were performed several times for each mutant (n = 2 to 4).
Cells were grown in M63 minimal medium supplemented with 0.1% ultra-pure sonicated salmon sperm DNA as the sole source of carbon and energy. Growth yields were determined by dividing the number of cells after 48 h of incubation by the number of cells at inoculation. SD, standard deviation.
DISCUSSION
The working model for the use of DNA as a nutrient predicts that when extracellular dsDNA is taken up by cells, only ssDNA is ultimately transported into the cytoplasm, where it is degraded by different single-stranded exonucleases; the second strand may be unwound and degraded in the periplasm. We show here that intracellular exonucleases (ExoI, RecJ, ExoVII, and ExoX) play a significant role in the catabolism of DNA as a nutrient, since all strains deficient in any one of them exhibit the SPCD phenotype during long-term coculture with wild-type cells. In addition, cells lacking any individual nuclease are altered in their ability to grow in minimal medium containing dsDNA as a carbon and energy source.
Most of these exonucleases are evolutionarily conserved, except for ExoX which is found primarily in the gammaproteobacteria (31; V. Palchevskiy, unpublished observation), and are involved in a variety of cellular functions. ExoI is specific for ssDNA, hydrolyzes DNA through its 3′→5′ activity, and is involved in DNA repair and DNA restriction to control undesirable levels of DNA recombination by suppressing illegitimate recombination (1, 27, 33). RecJ has an essential role in recBCD-independent recombination pathways (21). ExoVII is specific for ssDNA and can initiate hydrolysis at either the 3′ or 5′ end of DNA, generating oligonucleotides as products (3-5). In addition, xseA mutants have an increased frequency of recombination and are sensitive to nalidixic acid. ExoX is a DNA repair enzyme able to degrade both ssDNA and dsDNA, exhibiting 3′→5′ polarity with high affinity for ssDNA (32). An exoX mutant has increased sensitivity to UV irradiation in a background strain lacking several other DNA repair enzymes. ExoX has also been shown to be involved in the stabilization of tandem repeats preventing deletion events (11). All four of these exonucleases are also involved in mismatch-repair where incorrect nucleotides are removed by a strand-specific excision reaction after replication (11, 18, 30).
The DNA eating assay shows that single mutants lacking either ExoI, ExoVII, or RecJ are unable to utilize dsDNA as a sole nutrient source, while exoX mutants can consume dsDNA much better than wild-type cells. These findings were unexpected, as it was predicted that due to the redundancy of functions of these exonucleases, a deficiency phenotype for the DNA consumption process might only appear in multiple mutants. The differences observed are not due to altered transcription levels or the presence of extracellular nucleases released into the medium in the different mutant strains (data not shown). Therefore, it is possible that these exonucleases are working as part of a complex, and the removal of any of them disrupts the normal function of a particular complex. For example, perhaps ExoI, ExoVII, and RecJ work together as part of a DNA catabolism system, while ExoX might also play a role in some other process, such as DNA repair, which directs DNA away from the catabolic system. Therefore, when we created the exoX mutant, its complex or pathway was disrupted, leaving more DNA available for the catabolic system, resulting in greater growth yields of the exoX mutant strain. However, when the exoX mutation is combined with any other nuclease mutation, growth yields return to the low values because the catabolic complex is now also disrupted. The interactions of exonucleases with ssDNA-binding protein might be of interest here, since several exonucleases have been shown to interact with ssDNA-binding protein-bound ssDNA (15, 17, 23).
Taken together, the data presented here support a model in which only ssDNA enters the cytoplasm during the DNA eating process. However, apparently, the polarity of degradation appears to be unimportant since the deletion of either 3′→5′ or 5′→3′ exonucleases lacked the ability to catabolize DNA. The fact that the deletion of three of the nucleases studied impairs the cell's ability to “eat” DNA, while the mutation of a fourth nuclease increases DNA catabolism, suggests that these nucleases may function in concert with each other and/or other proteins in heretofore uncharacterized complexes.
Acknowledgments
We thank Christopher Corzett, Kavita Chavan, Alison Kraigsley, Evan Pepper, Meghann Ribbens, and Miriam Susskind for comments on the manuscript.
This study was supported by an NSF career award to S.E.F. (MCB-0237975). V.P. was a recipient of a National Institute of Aging predoctoral training grant (AG000093).
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Allgood, N. D., and T. J. Silhavy. 1991. Escherichia coli xonA (sbcB) mutants enhance illegitimate recombination. Genetics 127671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burdett, V., C. Baitinger, M. Viswanathan, S. T. Lovett, and P. Modrich. 2001. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. USA 986765-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chase, J. W., and C. C. Richardson. 1977. Escherichia coli mutants deficient in exonuclease VII. J. Bacteriol. 129934-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chase, J. W., and C. C. Richardson. 1974. Exonuclease VII of Escherichia coli. Mechanism of action. J. Biol. Chem. 2494553-4561. [PubMed] [Google Scholar]
- 5.Chase, J. W., and C. C. Richardson. 1974. Exonuclease VII of Escherichia coli. Purification and properties. J. Biol. Chem. 2494545-4552. [PubMed] [Google Scholar]
- 6.Chen, I., and D. Dubnau. 2003. DNA transport during transformation. Front. Biosci. 8s544-s556. [DOI] [PubMed] [Google Scholar]
- 7.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2241-249. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53217-244. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 1857044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feschenko, V. V., L. A. Rajman, and S. T. Lovett. 2003. Stabilization of perfect and imperfect tandem repeats by single-strand DNA exonucleases. Proc. Natl. Acad. Sci. USA 1001134-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkel, S. E. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4113-120. [DOI] [PubMed] [Google Scholar]
- 13.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 1836288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 964023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, E. S., D. L. Cooper, N. S. Persky, V. A. Sutera, Jr., R. D. Whitaker, M. L. Montello, and S. T. Lovett. 2006. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 341084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, R. S., K. J. Ross, M. J. Lombardo, and S. M. Rosenberg. 1998. Mismatch repair in Escherichia coli cells lacking single-strand exonucleases ExoI, ExoVII, and RecJ. J. Bacteriol. 180989-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodskinson, M. R., L. M. Allen, D. P. Thomson, and J. R. Sayers. 2007. Molecular interactions of Escherichia coli ExoIX and identification of its associated 3′-5′ exonuclease activity. Nucleic Acids Res. 354094-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun, S. H., T. G. Kim, and C. Ban. 2006. DNA mismatch repair system. Classical and fresh roles. FEBS J. 2731609-1619. [DOI] [PubMed] [Google Scholar]
- 19.Kraigsley, A. M., and S. E. Finkel. 2009. Adaptive evolution in single species bacterial biofilms. FEMS Microbiol. Lett. 293135-140. [DOI] [PubMed] [Google Scholar]
- 20.Lehman, I. R., and A. L. Nussbaum. 1964. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase). J. Biol. Chem. 2392628-2636. [PubMed] [Google Scholar]
- 21.Lovett, S. T., and A. J. Clark. 1984. Genetic analysis of the recJ gene of Escherichia coli K-12. J. Bacteriol. 157190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovett, S. T., and R. Kolodner. 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 862627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, D., and J. L. Keck. 2008. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl. Acad. Sci. USA 1059169-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Palchevskiy, V., and S. E. Finkel. 2006. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J. Bacteriol. 1883902-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of beta-galactosidase in E. coli. J. Mol. Biol. 1165-178. [Google Scholar]
- 27.Sandigursky, M., and W. A. Franklin. 1992. DNA deoxyribophosphodiesterase of Escherichia coli is associated with exonuclease I. Nucleic Acids Res. 204699-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, H. O., D. B. Danner, and R. A. Deich. 1981. Genetic transformation. Annu. Rev. Biochem. 5041-68. [DOI] [PubMed] [Google Scholar]
- 29.Stewart, G. J., and C. A. Carlson. 1986. The biology of natural transformation. Annu. Rev. Microbiol. 40211-235. [DOI] [PubMed] [Google Scholar]
- 30.Viswanathan, M., and S. T. Lovett. 1998. Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance. Genetics 1497-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viswanathan, M., and S. T. Lovett. 1999. Exonuclease X of Escherichia coli. A novel 3′-5′ DNase and DnaQ superfamily member involved in DNA repair. J. Biol. Chem. 27430094-30100. [DOI] [PubMed] [Google Scholar]
- 32.Viswanathan, M., V. Burdett, C. Baitinger, P. Modrich, and S. T. Lovett. 2001. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 27631053-31058. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi, H., K. Hanada, Y. Asami, J. I. Kato, and H. Ikeda. 2000. Control of genetic stability in Escherichia coli: the SbcB 3′-5′ exonuclease suppresses illegitimate recombination promoted by the RecE 5′-3′ exonuclease. Genes Cells 5101-109. [DOI] [PubMed] [Google Scholar]
- 34.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 2591757-1760. [DOI] [PubMed] [Google Scholar]