Abstract
Distinct features of the ribosomal peptide exit tunnel are known to be essential for recognition of specific amino acids of a nascent peptidyl-tRNA. Thus, a tryptophan residue at position 12 of the peptidyl-tRNA TnaC-tRNAPro leads to the creation of a free tryptophan binding site within the ribosome at which bound tryptophan inhibits normal ribosome functions. The ribosomal processes that are inhibited are hydrolysis of TnaC-tRNAPro by release factor 2 and peptidyl transfer of TnaC of TnaC-tRNAPro to puromycin. These events are normally performed in the ribosomal peptidyl transferase center. In the present study, changes of 23S rRNA nucleotides in the 2585 region of the peptidyl transferase center, G2583A and U2584C, were observed to reduce maximum induction of tna operon expression by tryptophan in vivo without affecting the concentration of tryptophan necessary to obtain 50% induction. The growth rate of strains with ribosomes with either of these changes was not altered appreciably. In vitro analyses with mutant ribosomes with these changes showed that tryptophan was not as efficient in protecting TnaC-tRNAPro from puromycin action as wild-type ribosomes. However, added tryptophan did prevent sparsomycin action as it normally does with wild-type ribosomes. These findings suggest that these two mutational changes act by reducing the ability of ribosome-bound tryptophan to inhibit peptidyl transferase activity rather than by reducing the ability of the ribosome to bind tryptophan. Thus, the present study identifies specific nucleotides within the ribosomal peptidyl transferase center that appear to be essential for effective tryptophan induction of tna operon expression.
The tryptophanase (tna) operon of Escherichia coli contains two major structural genes, tnaA, encoding tryptophanase, an enzyme responsible for the degradation of tryptophan (Trp), and tnaB, specifying a Trp-specific permease (6, 26). Degradation of Trp produces indole, pyruvate, and ammonia. The pyruvate and ammonia serve as sources of carbon and nitrogen, and the indole in some bacteria serves as a volatile quorum-sensing factor and in biofilm formation (29). In many bacterial species initiation of transcription of the tna operon is regulated by catabolite repression while transcription beyond the tna operon's leader region is regulated by Trp-mediated inhibition of Rho factor-dependent transcription termination (8). The transcript of the tna operon leader region contains the coding region for a 24-residue leader peptide, tnaC, followed by a Rho factor-binding site and then by a Rho factor termination site (26). In the presence of Trp, cleavage of the nascent TnaC-peptidyl-tRNA, TnaC-tRNAPro, at its UGA stop codon is inhibited (9). The resulting stalled translation complex blocks Rho factor's access to its binding site, preventing Rho-catalyzed transcription termination in the leader region of the operon (5, 10, 26). Several features of TnaC-tRNAPro required for inhibition of its cleavage have been identified. These include an essential Trp residue located at position 12 (W12), an aspartic residue located at position 16 (D16), the last amino acid in the TnaC peptide, proline 24 (P24), and the spacing between W12 and P24 of TnaC (3, 4) (Fig. 1). Interestingly, all of these residues and their positions are conserved among diverse eubacterial species (5). Several features of the ribosome required for Trp induction have also been identified. These include the lysine residue at position 90 of ribosomal protein L22, the 23S rRNA segment between nucleotides 749 to 752, and 23S rRNA nucleotide U2609 (4). These ribosomal residues are located in the ribosome peptide exit tunnel, near the putative position of W12 of TnaC-tRNAPro (4) (Fig. 1). Interactions between TnaC-tRNAPro and residues in the peptide exit tunnel contribute to the creation of a free Trp binding site, possibly at the ribosome active site, known as the peptidyl transferase center (PTC). At this site, the bound Trp presumably inhibits PTC activity, thereby preventing cleavage of the nascent TnaC-tRNAPro. Bound Trp has also been shown to inhibit the ribosomal action of some antibiotics, such as puromycin, an analog of aminoacyl-tRNA, and sparsomycin (2, 4, 8). Several other nascent peptides have been shown to act in cis, much like TnaC, inhibiting completion of their own synthesis (1, 4, 5, 7, 15, 17, 19, 27). With respect to TnaC-tRNA and other leader peptides, it has been suggested that interaction between features of these regulatory peptides and the peptide exit tunnel inhibits ribosomal PTC activity (1, 4, 5, 7, 15, 17, 19, 27). However, despite this apparent common feature, it is not known if or how the activity of the PTC is inhibited by these peptides, especially by TnaC-tRNAPro and bound Trp.
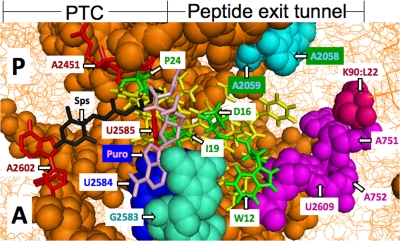
FIG. 1.
Model of the E. coli 50S ribosomal subunit containing the TnaC peptide in the peptide exit tunnel. This model is based on the ribosome structure of E. coli determined previously by Schuwirth et al. (23). Portions of the A and P sites of the peptidyl transferase center are displayed in this figure. Also displayed is the TnaC peptide, drawn as an α-helical sequence of amino acids located in the peptide exit tunnel. The TnaC residues Pro24 (P24), Asp16 (D16), Iso19 (I19), and Trp12 (W12) are in green. Each is highly conserved and has been shown to be essential for tna operon induction (5). The presumed locations of the antibiotics sparsomycin (Sps; black) (22) and puromycin (Puro; pink) (based on the ribosome structure of H. marismortui) (22) are shown in relation to the positions of the P site and A site (occupied by a puromycin molecule) in the ribosome. The positions of the ribosomal residues analyzed in this study, G2583 (pale green) and U2584 (blue), are also indicated. Some of the nucleotides involved in the peptidyl transferase reaction (red) and nucleotides affected by the sparsomycin interaction (cyan) are identified. The model was prepared using PyMOL software.
The PTC facilitates formation of peptide bonds and hydrolysis of peptidyl-tRNAs (25). The reactions involved in peptide bond formation and peptidyl-tRNA hydrolysis are chemically somewhat similar and are believed to involve nucleophilic attack of the substrate aminoacyl-tRNA or H2O attack by H2O positioned at the A site on the P-site substrate, peptidyl-tRNA (20). Hydrolysis of peptidyl-tRNA by H2O is assisted by a release factor (RF), either RF1 or RF2 in bacteria (20). High-resolution crystal structures of ribosomes revealed that the PTC center is composed predominantly of 23S rRNA (16, 23). The acceptor ends of A- and P-site tRNA substrates, as well as the functionally conserved GGQ sequence of RF1 and RF2 factors, are within the ribosome and are surrounded by PTC nucleotides such as C2063, A2451 and C2452, U2506, U2585, and A2602 of 23S rRNA (28, 33) (Fig. 1). These conserved nucleotides are at the core of the PTC center and are proposed to play roles in peptide bond formation and hydrolysis of peptidyl-tRNA (21, 22, 31).
In this study, we analyzed the effects of altering 23S rRNA nucleotides in and around the PTC on Trp induction of tna operon expression. We examined plasmids containing mutational changes at nucleotide positions 2583 to 2587, critical positions in the PTC (21). By introducing individual plasmids with each of these changes into a strain with all 23S rRNA chromosomal genes deleted (MG1655 Δ7) (4)—a strain containing a tnaA-lacZ reporter—we observed that changes of two nucleotides in the 2585 region, G2583 to A2583 and U2584 to C2584, reduced induction by Trp in vivo without reducing the affinity for Trp. It was also observed that these nucleotide replacements reduced the ability of Trp to inhibit puromycin action on TnaC-tRNA in vitro; however, these changes did not alter the ability of Trp to compete with sparsomycin. These findings identify specific nucleotides in the PTC that are essential for normal Trp induction of tna operon expression. The presumed positions of these nucleotides within the translating ribosome are shown in Fig. 1.
MATERIALS AND METHODS
Bacteria strains and plasmids.
The E. coli K12 strains used in this study were derived from strain SR-14, a strain lacking all chromosomal copies of rRNA operons (4). It contained the ampicillin resistance plasmid pNK, which has a wild-type rrnB operon (17). In this study pNK of this strain was replaced by other plasmids in which several nucleotides of the 2585 region (residues 2583 to 2587) of 23S rRNA in the PTC had been replaced previously (21). These altered plasmids were prepared from a kanamycin resistance plasmid containing a wild-type rrnB operon, pRB102 (21). SR-14 strains that were also examined contained the pK14-6 plasmid (a kanamycin resistance plasmid containing the wild-type rrnB operon) (S. Quan and C. L. Squires, unpublished data), pK14-6+A751 (rrnB operon with an insertion of an adenine at position 751 in the 23S rRNA gene) (17 and this study), and pK14-6 2609C (rrnB operon with a uridine-to-cytosine replacement at position 2609 in the 23S rRNA gene) (4 and this study).
Growth conditions and β-galactosidase assays.
In the experiments described in Fig. 2 and Table 1, plasmid-containing strains were grown with shaking at 37°C in minimal medium plus 0.2% glycerol, 0.05% acid casein hydrolysate, and 50 μg/ml kanamycin with different concentrations of the tna operon inducer, dl-1-methyl-tryptophan (dl-1MTrp). When each culture reached a density of 100 Klett units (660-nm filter), the cells were harvested and disrupted by sonication, and β-galactosidase activity was determined using standard procedures (4). The relationship between the β-galactosidase activity and the concentration of dl-1MTrp added resembled first-order enzyme kinetics (12). Accepting this assumption, the dl-1MTrp concentrations necessary to obtain 50% β-galactosidase activity (Km for an enzyme) were calculated using the Eadie-Hofstee procedure (12).
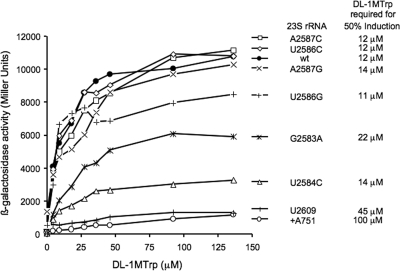
FIG. 2.
ß-Galactosidase levels of cultures of MG1655 Δ7 strains (4) with various 23S rRNA mutant plasmids grown with different concentrations of dl-1MTrp. Cell cultures grown in Vogel and Bonner minimal medium supplemented with 0.2% glycerol, 0.05% acid casein hydrolysate, and various concentrations of inducer were used to perform the β-galactosidase assays. The dl-1MTrp concentration necessary to reach 50% β-galactosidase activity expression was calculated using an Eadie-Hofstee diagram (see Materials and Methods).
TABLE 1.
tnaA-lacZ expression observed in various strains with the changes indicated, and their duplication times
| 23S rRNA gene mutation | tnaA-lacZ inductiona | Duplication time (min)b |
|---|---|---|
| None | + | 66 |
| +A751 | − | 78 |
| A2587C | + | 82 |
| A2587G | + | 72 |
| U2586C | + | 73 |
| U2586G | + | 75 |
| U2584C | − | 83 |
| G2583A | − | 85 |
| U2609C | − | 90 |
The SR-14 strain was transformed with plasmids containing each of the 23S rRNA genes indicated in the table and were grown on Vogel and Bonner (26) minimal agar plates supplemented with 0.2% glycerol, 0.05% ACH, 100 μg/ml l-Trp, and 40 μg/ml of the color-producing substrate X-Gal at 37°C for 16 h. The presence (+) or absence (−) of blue in the colonies indicated whether synthesis of β-galactosidase by the reporter gene fusion, tnaA-lacZ, was appreciably induced by l-Trp.
Cell growth of each strain in Vogel and Bonner (26) minimal medium supplemented with 0.2% glycerol and 0.05% ACH at 37°C was determined by measuring cell density using a Klett colorimeter equipped with a 660-nm filter, every hour. The duplication times shown were calculated using the log phase growth data obtained for each strain.
Isolation of biotinylated mRNA-ribosome complexes.
S-30 extracts were prepared from the cells of 3-liter cultures (optical density at 600 nm of 0.7) of every strain shown in Fig. 3 and 4 and grown in Luria broth plus 50 μg/ml kanamycin and 2% glucose. The harvested cells were resuspended in 20 ml of a solution containing 50 mM Tris-acetate (pH 8.0), 10 mM magnesium acetate, 175 mM potassium acetate, 10 mM ammonium acetate, and 2 mM dithiothreitol (DTT) and were then disrupted using a French press. The debris was separated by centrifugation at 30,000 × g for 30 min, and the supernatant was distributed as small samples, frozen, and stored at −70°C (4). Biotinylated mRNAs were prepared with DNA fragments obtained by PCR using plasmid pGF25-00 containing a wild-type tnaC gene (11). The in vitro translation reactions were performed in a reaction mixture (100 μl) containing 40 mM Tris-acetate (pH 8.0), 2 mM DTT, 0.5 mM GTP, 30 mM phosphoenolpyruvate, 0.3 U/ml pyruvate kinase, 2 mM ATP, 3.5% (wt/vol) polyethylene glycol 8000, 20 μg/ml folinic acid, 10 mM ammonium acetate, 175 mM potassium glutamate, 1 mM spermidine, 7.5 mM magnesium acetate, 20 μg/ml tRNA (E. coli mixture), 40 μg/ml mRNA, 0.37 mM of each amino acid, and 30 μl of the S-30 preparation previously treated with the antibody anti-RF2, as described previously (4, 8). In the experiment shown in Fig. 3, [35S]methionine (20 μCi; 3,000 Ci/mmol) was used to label the TnaC; the corresponding nonlabeled amino acid was not added. To purify the biotinylated mRNA complexes using streptavidin paramagnetic beads, 0.5 ml of a suspension of streptavidin beads previously washed and resuspended in a washing buffer containing 35 mM Tris-acetate (pH 8.0), 1 mM DTT, 10 mM ammonium acetate, 175 mM potassium glutamate, and 10 mM magnesium acetate was added to the total reaction mixture. The final mixture was incubated for 10 min at room temperature to allow biotin-streptavidin interactions. The streptavidin beads with bound biotinylated mRNA-ribosome complexes were then separated by using an electromagnetic field and were washed three times with washing buffer. Each of the final bead preparations was resuspended in 30 μl of washing buffer.
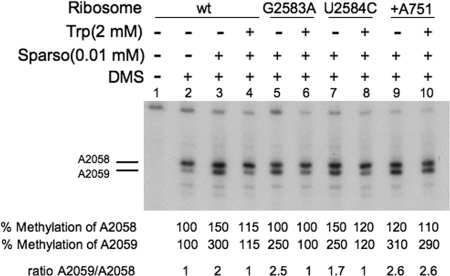
FIG. 3.
Action of the antibiotic sparsomycin on different isolated TnaC-tRNA-ribosome-mRNA complexes. Complexes isolated as indicated in the legend of Fig. 3 were prepared using the biotinylated tnaC mRNA designated for each panel. The isolated complexes were preincubated for 5 min with (+) or without (−) sparsomycin (Sparso) at 37°C. They were then incubated for five additional minutes at the same temperature in the presence (+) or absence (−) of Trp. Finally, the mixtures were exposed to the methylating reagent DMS to modify the A2058 and A2059 adenine nucleotides of 23S rRNA (positions indicated by horizontal lines). A sample was incubated in the absence of DMS as a methylation control. The rRNA isolated from the final mixtures was used to perform primer extension analyses. The percent methylation shown corresponds to the ratio of the cpm detected for each A2058 and A2059 band divided by the cpm for the corresponding band obtained with the control sample of ribosome-tnaC mRNA without tryptophan. wt, wild type.
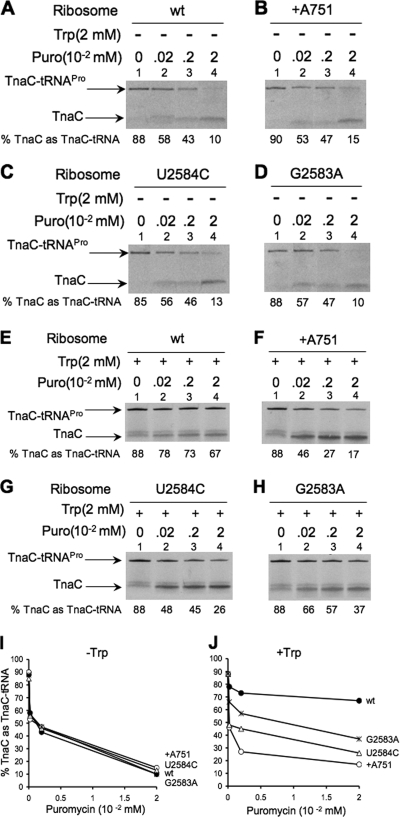
FIG. 4.
Competition assays between puromycin and Trp using different isolated TnaC-tRNA-ribosome complexes. In vitro translation reactions were performed with [35S]methionine to label the TnaC peptides. The cell extracts employed were pretreated with an anti-RF2 antiserum, and biotinylated tnaC mRNAs were used in each reaction mixture. The TnaC-tRNA-ribosome-mRNA complexes were isolated from the in vitro translation reactions by using streptavidin beads, and the isolated complexes were initially incubated with (+) or without (−) 2 mM Trp at 37°C for 5 min. Later, the complexes were incubated with different concentrations of puromycin (Puro) at the same temperature for 10 additional minutes. The final products of the reaction were resolved, and their presence was determined by measuring radioactivity. (A and E) Complexes containing wild-type 23S rRNA. (B and F) Complexes containing +A751 23S rRNA. (C and G) Complexes containing U2584C 23S rRNA. (D and H) Complexes containing G2583A 23S rRNA. (I) Plot of the data in panels A to D. (J) Plot of the data in panels E to H. The percentage of the TnaC as TnaC-tRNA was calculated by dividing the cpm in each TnaC-tRNA band by the combined cpm in the TnaC-tRNA and TnaC bands.
Puromycin protection assays.
The puromycin competition assays presented in Fig. 3 were performed with 10-μl bead complexes obtained from in vitro translation reactions carried out with [35S]methionine. Solutions were mixed with 1 μl of a 20 mM Trp solution. After incubation with Trp, the mixed solutions were incubated with different concentrations of puromycin. The puromycin was added to induce transfer of the TnaC peptide from TnaC-tRNAPro to this antibiotic molecule. After each reaction was stopped by the addition of 10 μl of a solution containing 10 mM Tris (pH 6.8), 10% sodium dodecyl sulfate, and 2 mM DTT, the components of the final reaction mixtures were resolved by electrophoresis on 10% Tricine-sodium dodecyl sulfate protein gels. The dried gels were exposed to X-ray films, and the sections of the dried gels that contained TnaC-tRNAPro and TnaC were excised. The radioactivity (counts per minute [cpm]) in these bands was measured using a scintillation counter. The amount of [35S]methionine-TnaC-tRNAPro remaining after the reaction with puromycin was determined by dividing the cpm in the band corresponding to TnaC-tRNAPro by the combined cpm in the bands corresponding to TnaC-tRNAPro and the TnaC peptide.
Footprinting assays.
In the experiments shown in Fig. 4, footprinting assays were performed as described previously (14). A total of 100 μl of isolated complexes resuspended in the appropriate buffer was modified by exposure to 4 μl of dimethyl sulfate (DMS) (1:6 dilution in ethanol) for 10 min at room temperature. DMS-methylation reactions were stopped by the addition of 50 μl of stop buffer (1 M Tris-HCl [pH 8.0], 1 M 2-mercaptoethanol, and 1 mM EDTA) to each reaction mixture. The final reaction mixtures were diluted with a 10 mM EDTA solution to reach a total volume of 500 μl, and the RNA was then extracted by treatment with phenol. Modification by methylation was monitored by primer extension analyses using avian myeloblastosis virus reverse transcriptase (Gibco) and 5′ 32P-labeled deoxyoligonucleotide primers (4). Primer 5′-CTATCCTACACTCAAGGCTC-3′, complementary to nucleotides 2102 to 2122 of 23S rRNA, was used to detect modified nucleotides in 23S rRNA. The positions of the modified nucleotides were identified by reference to dideoxy sequencing ladders obtained using a fmol DNA Cycle Sequencing System kit (Promega). DNA from plasmid pNK that contained the 23S rRNA gene (17) and the same primers were used for primer extension analyses of rRNA genes.
RESULTS AND DISCUSSION
Identification of mutants altered in the PTC which exhibit reduced tna operon induction.
Previously, we described studies with mutants bearing alterations in the 23S rRNA at nucleotides A751 to 752 and U2609 or changes of residue K90 of ribosomal protein L22, which reduced or prevented Trp-mediated induction of tna operon expression (Fig. 1) (3, 4). Our prior results suggested that interactions between these residues in the ribosome exit tunnel and W12 of TnaC help in creating a Trp binding site in or near the PTC (2). Once Trp is bound, the binding and action of several molecules, including RF2 and the antibiotics sparsomycin or puromycin, appeared to be affected (2). Like RF2, sparsomycin and puromycin usually interact with nucleotides in the PTC of the 50S ribosome subunit of E. coli (Fig. 1). In the PTC these molecules are surrounded by nucleotides believed to be involved in peptide bond synthesis and termination, including A2451, U2585, and A2602, among others (Fig. 1) (22, 24, 28, 31). It is conceivable that under induction conditions, the Trp residue bound within the ribosome affects the spatial distribution of nucleotides around these PTC positions.
Recently, a plasmid library was described in which there was complete randomization of nucleotides at critical regions of the PTC, i.e., in regions known to be essential for peptide bond formation (21). We obtained and examined 12 plasmids from this library to determine if changes in the nucleotides of the conserved 2585 region (residues 2583 to 2587) of the PTC affect Trp-mediated induction of tna operon expression. The mutations located in these plasmids were the following: A2587G, A2587T, A2587C, T2586G, T2586C, T2586A, T2584C, G2583A, G2583A T2586C, T2586A A2587G, and T2586A A2587G (21). The resident pNK ampicillin resistance plasmid (containing the wild-type rrnB gene) in a strain with a tnaA-lacZ reporter construct (4) that has deletions removing all seven chromosomal rrn gene genes was replaced by the kanamycin-resistant plasmids (21). Six of these 12 plasmid constructs were able to replace the resident plasmid, yielding viable progeny (Table 1). After successful exchanges, bacteria with the kanamycin resistance plasmids were plated on agar containing l-Trp and a blue-producing β-galactosidase substrate, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, as previously described (4). We observed that 2 out of the 12 transformed constructs affected tnaA-lacZ reporter gene induction by Trp (Table 1). These two constructs contained the nucleotide changes G2583A and U2584C in the 2585 region (their positions can be seen in Fig. 1). Other plasmids containing mutations in the same region, such as U2586 or A2587, did not affect tnaA-lacZ gene expression induced by Trp (Table 1). No major difference in the duplication time was observed for strains bearing plasmids with these mutations in the 2585 region (Table 1). However, all isolates with mutations in this region grew at a slightly slower rate than the wild-type strain (Table 1). These results indicate that some mutational changes to residues in the 2585 region do reduce Trp induction of tna operon expression in vivo without significantly affecting cell growth.
dl-1MTrp concentrations required to obtain 50% induction in strains bearing nucleotide changes in the 2585 region of the ribosomal PTC.
Mutational changes of the 23S rRNA nucleotides A751 to 752 and U2609 in the peptide exit tunnel have been shown to affect Trp binding within the ribosome (3). To determine if our newly identified PTC mutations affect Trp affinity for the ribosome, we determined the dl-1MTrp concentration required in vivo to obtain 50% induction of tnaA-lacZ expression (Fig. 2). dl-1MTrp, an analog of Trp, was used as an inducer instead of Trp because the strain employed in this study (SR-14) (see Materials and Methods) contains an intact tna operon including the structural gene for tryptophanase (4). The presence of tryptophanase would reduce the Trp concentration, thereby reducing expression of the tnaA-lacZ reporter gene, as has been shown previously (30). However, dl-1MTrp, an excellent inducer of tnaA-lacZ expression, is not cleaved by tryptophanase, and also it does not significantly affect Trp incorporation into proteins (30). β-Galactosidase levels were determined during growth in the presence of different dl-1MTrp concentrations using strains containing plasmids with mutations affecting nucleotides in the 2585 region and nucleotides around the peptide exit tunnel (Materials and Methods). The β-galactosidase activity values (Miller units) obtained for each strain were used to prepare the plot shown in Fig. 2. As can be seen in Fig. 2, the presence of low concentrations of dl-1-MTrp in wild-type cultures increased production of β-galactosidase substantially in vivo until a plateau was reached (at 45 μM dl-1MTrp), indicating maximum β-galactosidase production (Fig. 2). Induction was determined for all strains with plasmids bearing mutations in the 2585 region, G2583A, U2584C, U2586C/G, and A2586C/G. Of particular interest, strains with the changes G2583A and U2584C produced much lower levels of β-galactosidase activity, i.e., 50% and 25%, respectively, than the levels observed in the wild-type strain (Fig. 2). On the other hand, strains containing the adenine inserted at position 751 (+A751) and U2609C mutant plasmids produced even lower levels of ß-galactosidase than strains with the wild-type or other mutant plasmids (Fig. 2). These results indicate that, unlike the +A751 and U2609C mutations in the peptide exit tunnel, the G2583A and U2584C mutations in the PTC affect maximum induction by dl-1MTrp but apparently not inducer (dl-1MTrp) affinity. Thus, the dl-1MTrp concentration necessary to induce 50% expression in the mutant strains containing changes in the 2585 region did not appear to be significantly different from the concentration required for 50% induction in the wild-type strain (Fig. 2). These findings indicate that ribosomal affinity for dl-1MTrp is probably not reduced when the ribosome contains either of these mutations in the PTC. By contrast, the dl-1MTrp concentration necessary for 50% induction in strains bearing the mutations +A751 and U2609C were approximately 10 and 5 times higher than that required for 50% induction in the wild-type strain (Fig. 2), which agrees with previous findings indicating that these mutations in the peptide exit tunnel do affect Trp binding to the ribosome (2). In summary, the results presented in Fig. 2 suggest that the G2583A and U2584C mutations in the PTC of the ribosome do reduce tna operon induction by dl-1MTrp but without affecting dl-1MTrp affinity.
Competition between Trp and the antibiotic sparsomycin for binding to wild-type and mutant ribosomes.
In previous studies it was shown that a Trp concentration of ca. 2 mM was the most effective in inhibiting puromycin or RF2 action (2, 4). It was also shown that 2 mM Trp could effectively compete with 1/10 or less of the concentration of sparsomycin when sparsomycin binding at its binding site in the ribosome was examined (2). Sparsomycin is an antibiotic that also interacts with the ribosomal PTC (18). Trp was shown to reduce sparsomycin's ability to increase DMS methylation of nucleotide A2059 of 23S rRNA of wild-type ribosomes, suggesting that Trp and sparsomycin compete with one another (2). Thus, this type of analysis could also be used to determine if Trp binds within any mutant ribosome. Following pretreatment of wild-type and mutant ribosomal preparations with an anti-RF2 antiserum (see Materials and Methods), TnaC-tRNAPro-ribosome-mRNA complexes were isolated using streptavidin beads. Competition assays were then performed in vitro between added sparsomycin and Trp using these isolated complexes. As observed with wild-type complexes, sparsomycin-treated complexes with the G2583A and U2584C changes were protected from methylation by Trp (Fig. 3, compare lane 4 with lanes 6 and 8). By contrast, as observed before, Trp was unable to protect +A751 complexes from sparsomycin action (Fig. 3, compare lane 4 with lane 10) (3). These findings suggest that in complexes containing the G2583A and U2584C changes, Trp can bind as efficiently as it does in complexes containing wild-type ribosomes; however, it does not bind to ribosome complexes containing the +A751 change (Fig. 3).
Analysis, in vitro, of the ability of Trp to inhibit puromycin action on mutant and wild-type TnaC-tRNAPro ribosome complexes.
Trp has been observed to competitively inhibit hydrolysis of TnaC-tRNAPro by puromycin, an analog of an aminoacyl-tRNA that interacts at the A site of the PTC (Fig. 1) (9). In view of the results presented in Fig. 2 and 3, it was of interest to determine if Trp would inhibit puromycin action on PTC mutant ribosomes. TnaC-tRNAPro-ribosome-mRNA complexes obtained from anti-RF2 pretreated cell extracts were also used for this purpose. In the absence of Trp, the TnaC-tRNAPro within the mutant complexes was efficiently transferred to puromycin as well as the TnaC-tRNAPro within wild-type complexes (Fig. 4A to D). These results indicate that these mutations in 23S rRNA do not affect the action of puromycin in the ribosome, even with a limiting puromycin concentration; approximately 90% of the TnaC-tRNAPro was transferred in all complexes after the addition of 0.02 mM puromycin (Fig. 4A to D, compare the corresponding lanes 1 with 4). In the presence of Trp the results obtained were completely different. When 2 mM Trp was present, TnaC-tRNAPro within wild-type ribosome complexes was poorly cleaved despite an increase in the concentration of puromycin (Fig. 4E, compare lane 1 with lane 4). With mutant ribosome complexes, TnaC-tRNAPro was more sensitive to puromycin addition (Fig. 4F to H). We observed that the addition of 0.02 mM puromycin cleaved 80% of the TnaC-tRNAPro in the complexes with the +A751 mutant ribosomes (Fig. 4F, compare lane 1 with lane 4). However, at the same puromycin concentration, 70% and 58% of the TnaC-tRNAPro was cleaved in U2584C and G2583A mutant ribosomes, respectively (Fig. 4G and H, compare lane 1 with lane 4 for each). The results presented in Fig. 4J suggest that Trp protects G2583A and U2584C complexes from the action of puromycin more effectively than it protects +A751 complexes, but this protection is much less effective than is observed with wild-type complexes. These results are consistent with the findings obtained in vivo (Fig. 2), where induction of expression of the reporter gene in strains with G2583A and U2584C mutant 23S rRNA plasmids was lower than in strains with wild-type 23S rRNA plasmids but greater than in strains with the +A751 mutant 23S rRNA plasmid. These findings suggest that Trp may bind to G2583A and U2584C mutant ribosomes but that, when bound, Trp is incapable of protecting TnaC-tRNAPro from hydrolysis as efficiently as it does with wild-type ribosomes.
Conclusions.
The findings described in this paper demonstrate that two 23S rRNA nucleotides located in the PTC, G2583 and U2584, when replaced by other nucleotides, A and C, respectively, significantly reduce Trp induction of tna operon expression in vivo (Fig. 2). However, induction in vivo with these mutant ribosomes exhibits a response to changes in the concentration of the inducer, 1MTrp, similar to changes observed with wild-type ribosomes (Fig. 2). This suggests that these nucleotide changes may not reduce the affinity of the ribosome for the inducing Trp analog; rather, they may alter the sensitivity and responsiveness of the PTC. Positional changes in these two nucleotides could also affect the spatial location of other nearby PTC nucleotides, such as residue U2585 (Fig. 1), a nucleotide that is actively involved in peptide bond formation, and hydrolysis of peptidyl-tRNA induced by RF1 or RF2 (13, 22, 28, 31, 32). Therefore, structural changes that occur in the peptide exit tunnel upon interaction of its nucleotides with amino acid residues of the TnaC-tRNAPro molecule may normally be transmitted to nucleotides G2583 and U2584, altering their position in the ribosome and disturbing the PTC-RF2 interactions and thereby contributing to inhibition of RF2 action by Trp. Additional experiments must be performed to verify these assumptions.
Acknowledgments
We are grateful to Anastasia Levitin, Selwyn Quan, and Catherine Squires for helpful discussions concerning the experiments described in this paper. We also thank Selwyn Quan and Catherine Squires for the pK4-16 plasmid, and we thank Tsutomu Suzuki and collaborators for plasmids from their randomized rRNA plasmid library.
This work was supported by National Science Foundation grant MCB-0615390 to C.Y. and University of Alabama-Huntsville start-up funds provided to L.R.C.-V.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Child, S. J., M. K. Miller, and A. P. Geballe. 1999. Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem. 27424335-24341. [DOI] [PubMed] [Google Scholar]
- 2.Cruz-Vera, L. R., M. Gong, and C. Yanofsky. 2006. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. USA 1033598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz-Vera, L. R., A. New, C. Squires, and C. Yanofsky. 2007. Ribosomal features essential for tna operon induction: tryptophan binding at the peptidyl transferase center. J. Bacteriol. 1893140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz-Vera, L. R., S. Rajagopal, C. Squires, and C. Yanofsky. 2005. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell 19333-343. [DOI] [PubMed] [Google Scholar]
- 5.Cruz-Vera, L. R., and C. Yanofsky. 2008. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of tna operon expression. J. Bacteriol. 1904791-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeley, M. C., and C. Yanofsky. 1982. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J. Bacteriol. 151942-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, P., C. C. Spevak, C. Wu, and M. S. Sachs. 2004. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. USA 1014059-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong, F., K. Ito, Y. Nakamura, and C. Yanofsky. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNAPro. Proc. Natl. Acad. Sci. USA 988997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong, F., and C. Yanofsky. 2002. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 27717095-17100. [DOI] [PubMed] [Google Scholar]
- 10.Gong, F., and C. Yanofsky. 2002. Instruction of translating ribosome by nascent peptide. Science 2971864-1867. [DOI] [PubMed] [Google Scholar]
- 11.Gong, F., and C. Yanofsky. 2001. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 2761974-1983. [DOI] [PubMed] [Google Scholar]
- 12.Hofstee, B. H. 1959. Non-inverted versus inverted plots in enzyme kinetics. Nature 1841296-1298. [DOI] [PubMed] [Google Scholar]
- 13.Korostelev, A., H. Asahara, L. Lancaster, M. Laurberg, A. Hirschi, J. Zhu, S. Trakhanov, W. G. Scott, and H. F. Noller. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. USA 10519684-19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krol, A., and P. Carbon. 1989. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 180212-227. [DOI] [PubMed] [Google Scholar]
- 15.Lovett, P. S. 1996. Translation attenuation regulation of chloramphenicol resistance in bacteria—a review. Gene 179157-162. [DOI] [PubMed] [Google Scholar]
- 16.Moore, P. B., and T. A. Steitz. 2003. After the ribosome structures: how does peptidyl transferase work? RNA 9155-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatogawa, H., and K. Ito. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108629-636. [DOI] [PubMed] [Google Scholar]
- 18.Porse, B. T., S. V. Kirillov, M. J. Awayez, H. C. Ottenheijm, and R. A. Garrett. 1999. Direct crosslinking of the antitumor antibiotic sparsomycin, and its derivatives, to A2602 in the peptidyl transferase center of 23S-like rRNA within ribosome-tRNA complexes. Proc. Natl. Acad. Sci. USA 969003-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raney, A., G. L. Law, G. J. Mize, and D. R. Morris. 2002. Regulated translation termination at the upstream open reading frame in S-adenosylmethionine decarboxylase mRNA. J. Biol. Chem. 2775988-5994. [DOI] [PubMed] [Google Scholar]
- 20.Rodnina, M. V., M. Beringer, and W. Wintermeyer. 2007. How ribosomes make peptide bonds. Trends Biochem. Sci. 3220-26. [DOI] [PubMed] [Google Scholar]
- 21.Sato, N. S., N. Hirabayashi, I. Agmon, A. Yonath, and T. Suzuki. 2006. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc. Natl. Acad. Sci. USA 10315386-15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmeing, T. M., K. S. Huang, S. A. Strobel, and T. A. Steitz. 2005. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438520-524. [DOI] [PubMed] [Google Scholar]
- 23.Schuwirth, B. S., M. A. Borovinskaya, C. W. Hau, W. Zhang, A. Vila-Sanjurjo, J. M. Holton, and J. H. Cate. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310827-834. [DOI] [PubMed] [Google Scholar]
- 24.Steitz, T. A. 2008. Structural insights into the functions of the large ribosomal subunit, a major antibiotic target. Keio J. Med. 571-14. [DOI] [PubMed] [Google Scholar]
- 25.Steitz, T. A. 2008. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 9242-253. [DOI] [PubMed] [Google Scholar]
- 26.Stewart, V., and C. Yanofsky. 1985. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 164731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vazquez-Laslop, N., C. Thum, and A. S. Mankin. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell 30190-202. [DOI] [PubMed] [Google Scholar]
- 28.Weixlbaumer, A., H. Jin, C. Neubauer, R. M. Voorhees, S. Petry, A. C. Kelley, and V. Ramakrishnan. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winzer, K., K. R. Hardie, and P. Williams. 2002. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5216-222. [DOI] [PubMed] [Google Scholar]
- 30.Yanofsky, C., V. Horn, and P. Gollnick. 1991. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 1736009-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youngman, E. M., J. L. Brunelle, A. B. Kochaniak, and R. Green. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117589-599. [DOI] [PubMed] [Google Scholar]
- 32.Youngman, E. M., M. E. McDonald, and R. Green. 2008. Peptide release on the ribosome: mechanism and implications for translational control. Annu. Rev. Microbiol. 62353-373. [DOI] [PubMed] [Google Scholar]
- 33.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292883-896. [DOI] [PubMed] [Google Scholar]