Abstract
Uropathogenic Escherichia coli (UPEC) strain CFT073 contains 13 large genomic islands ranging in size from 32 kb to 123 kb. Eleven of these genomic islands were individually deleted from the genome, and nine isogenic mutants were tested for their ability to colonize the CBA/J mouse model of ascending urinary tract infection. Three genomic island mutants (ΔPAI-aspV, ΔPAI-metV, and ΔPAI-asnT) were significantly outcompeted by wild-type CFT073 in the bladders and/or kidneys following transurethral cochallenge (P ≤ 0.0139). The PAI-metV mutant also showed significant attenuation in the ability to independently colonize the kidneys (P = 0.0011). Specific genes within these islands contributed to the observed phenotype, including a previously uncharacterized iron acquisition cluster, fbpABCD (c0294 to c0297 [c0294-97]), autotransporter, picU (c0350), and RTX family exoprotein, tosA (c0363) in the PAI-aspV island. The double deletion mutant with deletions in both copies of the fbp iron acquisition operon (Δc0294-97 Δc2518-15) was significantly outcompeted by wild-type CFT073 in cochallenge. Strains with mutations in a type VI secretion system within the PAI-metV island did not show attenuation. The attenuation of the PAI-metV island was localized to genes c3405-10, encoding a putative phosphotransferase transport system, which is common to UPEC and avian pathogenic E. coli strains but absent from E. coli K-12. We have shown that, in addition to encoding virulence genes, genomic islands contribute to the overall fitness of UPEC strain CFT073 in vivo.
Escherichia coli, a versatile microbe, can colonize the intestinal tract with no harmful effects to the host or can cause devastating and life-threatening disease (34). E. coli can be classified into one of three groups: commensal (nonpathogenic) E. coli strains that coexist with the host without causing overt disease, intestinal pathogenic (diarrheagenic) E. coli, and extraintestinal pathogenic E. coli (ExPEC). The latter category, ExPEC, was proposed in 2000 to classify E. coli isolates capable of causing disease outside of the intestinal tract, including uropathogenic E. coli (UPEC), sepsis-associated E. coli, and neonatal meningitis-associated E. coli (63). Within the human intestinal tract, ExPEC may colonize without causing disease. However, this subset of E. coli has the ability to disseminate to other sites of the body, including the urinary tract, bloodstream, and central nervous system, and elicit pathogenesis (77).
Urinary tract infections (UTIs), the most common type of bacterial infection (16), affect 11% of adult women every year, with an estimated one-third of women requiring antibiotic therapy for a clinician-diagnosed UTI by 24 years of age (17). Approximately 60% of all women will experience a UTI during their lifetime (17). Nearly 7 million physician office visits, 1 million emergency room visits, and 100,000 hospitalizations per year are attributed to UTIs, with women twice as likely as men to seek medical treatment for infections of the urinary tract (65). The estimated total cost (direct and indirect costs) of UTIs in the United States was $3.5 billion in 2000 (43).
UPEC strains cause at least 80% of all community-acquired UTIs (61, 73). A representative and prototypic UPEC strain, CFT073, isolated from blood from a hospitalized patient suffering from acute pyelonephritis, has been fully sequenced and annotated (46, 76). In this and other UPEC strains, an increasing repertoire of virulence factors have been identified, including pili (type I, P, S, and F1C), toxins (hemolysin [hly], cytotoxic necrotizing factor [cnf], vacuolating autotransporter toxin [vat], secreted autotransporter toxin [sat], and the autotransporter encoded by picU), and iron acquisition systems (enterobactin [ent], aerobactin [iuc], yersiniabactin [ybt], salmochelin [iro], heme uptake [chu and hma] [24], and ferrous iron transport [sit]) (77).
The prevalence of key virulence factors varies among UPEC strains (44, 77), and these differences are evident when comparing the three most well-studied UPEC isolates, CFT073, 536, and UTI89 (7, 11, 76, 77). For example, while all three strains express enterobactin, salmochelin, and heme uptake systems, only CFT073 produces aerobactin, and CFT073 and UTI89 carry the sit iron transport system, while strain 536 does not. All three strains carry the hly gene, with strain 536 containing two copies of this operon. Similarly, the P-fimbrial (pap) operon is present in all strains, with only strain CFT073 possessing two copies. Genes unique to one of these strains include sat (CFT073) and cnf1 (UTI89) (77).
Specific virulence factors are linked in UPEC isolates (6, 30, 45). Associated virulence genes in unrelated UPEC strains frequently map to a localized region of the chromosome, suggesting that these genes were acquired by horizontal transfer as a single event. For example, two studies (44, 45) revealed a strong correlation (75 to 87%) between the presence of cnf1, class III P-fimbrial adhesin (papG allele III), S-fimbrial adhesin (sfa), and hly genes; interestingly, this trend was observed only for isolates from patients with cystitis, not for isolates from patients with pyelonephritis (44). Additionally, the genes encoding three virulence factors, hly, prs (P fimbriae), and cnf1, are linked on the chromosome of UPEC strain J96 (6) and localize to a region corresponding to pathogenicity island II of strain J96. Localization of virulence factors to specific regions of the chromosome suggests that these genes were acquired simultaneously.
Bacterial chromosomes are dynamic, with the genomic content in a constant state of flux (70). The major driving forces for microbial evolution are point mutations, insertions, deletions, and genome rearrangements (3, 70) and their natural selection. Although point mutations result in genetic variability, these events generate microbial diversity at a much lower rate. In contrast, the acquisition (insertion) or loss (deletion) of DNA through horizontal gene transfer may result in genetic quantum leaps (15, 21) in bacterial evolution. Horizontal gene transfer, the process by which genetic material is transferred between bacterial species independent of cell division (39), likely remains the most powerful and rapid means of prokaryotic evolution. Acquisition of genetic material from closely related or distant species generates genetic variability, permits adaptation to life in a specialized niche, and serves as a source of potential virulence or fitness factors. These large regions of mobile genomic material are referred to as genomic islands (GIs).
GIs containing one or more virulence genes have been further classified as pathogenicity islands (PAIs). PAIs, first reported in the late 1980s in UPEC strain 536 (23, 36), meet most, if not all, of the following criteria: contain virulence genes; are present in pathogenic strains but are absent or rare in nonpathogenic strains of the same or related species; consist of large genomic regions ranging from 10 kb up to 200 kb; are relatively unstable; often have different G+C contents relative to that of the core genome; are often associated with tRNA genes; frequently contain mobile genetic elements, such as insertion sequences, transposons, integrases, and bacteriophage DNA; are often flanked by direct repeat sequences and regularly have a mosaic-like structure, composed of smaller segments of DNA, possibly acquired at different stages via horizontal gene transfer (20, 22, 67).
PAIs encode virulence factors, including, but not limited to, adhesins, bacterial secretion systems, invasins, toxins, proteases, lipases, and iron uptake systems (22). Obviously, bacterial species cannot sustain uncontrolled genome growth; integration of new PAIs may necessitate gene loss from elsewhere in the genome (67). If newly acquired genes confer a fitness advantage or contribute to virulence, chromosomal stabilization may occur through inactivation or deletion of mobility genes associated with the PAI (22). Retention of beneficial DNA promotes bacterial evolution and increases the probability that resident genes of lower selective value will be lost (41, 51, 67).
We recently identified 10 novel GIs in UPEC strain CFT073 (44) and hypothesized that these GIs along with three previously described islands (19, 52, 57) contribute to virulence. To test this hypothesis, 11 of the 13 GIs were individually deleted from strain CFT073 using the lambda red recombinase system (12), and nine of these mutants were assessed for virulence in the CBA/J mouse model of ascending UTI. Three GI mutants, PAI-CFT073-aspV, PAI-CFT073-metV, and PAI-CFT073-asnT (high-pathogenicity island [HPI]) were significantly outcompeted by wild-type CFT073 in the bladders and/or kidneys following transurethral cochallenge, and the contribution of specific blocks of genes within these islands to colonization of the urinary tract was studied in greater detail.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli CFT073 was isolated from blood from a patient admitted to the University of Maryland Medical System for the treatment of acute pyelonephritis (46). This strain is highly virulent in the CBA/J mouse model of ascending UTI (47), is cytotoxic for cultured human renal proximal tubular epithelial cells (46), and has been sequenced and annotated (76).
For growth on solid medium, bacterial strains were streaked onto LB agar plates (10 g tryptone, 5 g yeast extract, 10 g NaCl, 15 g agar [all per liter]) and incubated at 37°C for 18 h. For growth in liquid culture, strains were inoculated into LB broth (10 g tryptone, 5 g yeast extract, 10 g NaCl [all per liter]) and incubated at 37°C for 18 h with aeration (200 rpm). Kanamycin (25 μg/ml), chloramphenicol (20 μg/ml), or ampicillin (100 μg/ml) was added as appropriate. Strains to be tested in the mouse model of ascending UTI were cultured overnight in the absence of antibiotic selection (with the exception of strains containing a vector for in vivo complementation), resuspended in phosphate-buffered saline (PBS), and adjusted to 4.0 × 109 CFU/ml.
For growth on CAS agar (69) containing 2 mM FeCl3, bacterial strains were cultured overnight in LB broth and standardized in PBS to 1 × 109 CFU/ml. A sample (20 μl) of the suspension was spotted onto CAS agar plates, and the plates were incubated at room temperature overnight. Siderophore production was indicated by orange halos around the bacterial growth. The enterobactin/aerobactin (entF::kan iucB::cam) siderophore-deficient strain of CFT073 (75) was kindly provided by Alfredo Torres, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston. For growth under iron-limiting conditions, strains were cultured overnight in LB broth containing 200 μM 2,2′-dipyridyl and washed twice in PBS. LB broth or M9 minimal medium containing 200, 300, 400, 500, or 600 μM 2,2′-dipyridyl was inoculated (in triplicate) with 1 × 108 CFU/ml of bacterial suspension. Each well contained 300 μl of medium, corresponding to 3 × 107 CFU/well. Growth curves were performed using the Bioscreen growth curve analyzer (Growth Curves USA) with growth at 37°C with continuous shaking.
Identification of genomic islands of E. coli CFT073.
GIs of E. coli CFT073 were identified and defined using comparative genomic hybridization microarray analysis of seven UPEC strains and three fecal/commensal E. coli strains as described previously (44). The presence of several of these GIs was demonstrated by sequence comparison with E. coli 536 (7).
Construction of genomic island mutants.
Isogenic mutants in E. coli CFT073 were constructed using the lambda red recombinase system (12). Briefly, primers homologous to sequences within the 5′ and 3′ ends of the target regions were designed (H1 and H2 primers, respectively; Table 1) and were used to replace these genes with a nonpolar kanamycin or chloramphenicol resistance cassette derived from plasmid pKD4 or pKD3, respectively (12). Less than 10% of the targeted gene sequence (with the exception of Δc0363, at 13.4%) remained after homologous recombination. Kanamycin or chloramphenicol was used for selection of all deletion constructs. GI deletions ranged from 32 kb to 123 kb, and individual gene and potential operon deletions were also constructed using this method. The boundaries of each of the 11 GI mutants are defined in Table 2.
TABLE 1.
Primers used in this study
| Mutant | Primers used to construct isogenic mutants
|
Primers used to confirm isogenic mutants
|
||
|---|---|---|---|---|
| Primer typea | Primer sequence (5′-3′) | Primer directionb | Primer sequence (5′-3′) | |
| ΔPAI-CFT073-pheV | H1 | ACACAGCGATAAAGTACTCAAAAGCCTCGAGACTCACG | Fwd | CAGTCGGTAGAGCAGGGGATTGAA |
| H2 | TATTGCCATTTCCTTAACCCCACCTGATAACCCTTAGC | Rev | AGCGACTGGAGTTTGGGCGGGGGTAGG | |
| ΔPAI-CFT073-pheU | H1 | CAGGCTGATGGTACATGCTCTGAAACTGGCTGCAGGATACG | Fwd | GCACAGAAGGAAAGTACCTGGCTATTA |
| H2 | TCGCTTTTACTGAAATTAGGTTGACGAGATGTGCAGATTACG | Rev | GGAGATGGTTGCTGAACGTGTGGATTA | |
| ΔPAI-CFT073-aspV | H1 | GTGCAGTTCCCTTCTGAAAATACTTAATCACAAACATCTCA | Fwd | TCAGACAACTCACTCACCTCTCATCTC |
| H2 | ATAACCCATCAGCCCGCTTCTGTAATACCTCCATTCGTTCTA | Rev | TGAAATTATACTGAACGGATACAAGAC | |
| Δφ-CFT073-b0847 | H1 | GGGCCTCTATCTTCAATCTGTTCGACTAACCCCTCCTCT | Fwd | CTTTGTCGCCACTTGTTTTACCTTAGA |
| H2 | AGAATCATTCCATTTCGAAATCATTAATCTTCACTTCAAG | Rev | CGATGGCGACAAATTGGCGGCAGAGTC | |
| ΔPAI-CFT073-serX | H1 | GTATTGCTGAAGCTGCACGTACTGCCCGGATAATGCGAGAG | Fwd | GTAAAGGGGCGGGGGAAATGGGTTTTT |
| H2 | CTTTTCCCTGAAGAGACCGGATGTGATCGTCCAGATGAATAG | Rev | ACAGGAACAACAATTTGGTGAGGTGTC | |
| Δφ-CFT073-potB | H1 | CTTCTTTTAACGTTATCCCAATGGCATCACAGCGTAGTGTAA | Fwd | TTATAGCGATCGGTGGTTGCCTGGACT |
| H2 | GGCTTCTTCCAGTGGTACGTAATTTTCTCCGTTTCCCGATAC | Rev | ACGTGGAACAAATAGACACAAGAAATA | |
| ΔPAI-CFT073-asnT | H1 | CCTTACCGACGCAAAAATCCGCACCCTCAAGCCTTCTGATAA | Fwd | GCCCCGTTCTCACGATTCCTCTGTAGT |
| H2 | CAGCGTGATTCTTGCGGTACCGAAGCGGCTTAACCAGTCTGT | Rev | GCATTCGTGACGTTCGGCACATAGTTC | |
| ΔGI-CFT073-asnW | H1 | ACCCCCATATGTCCCTTAACGACGCAAAAATCCGTAGTCTCA | Fwd | GCATCGCTAATATTCGCCTCGTTCTCA |
| H2 | GGAAGCGCTGATCCTCTCCCCTAGTGGAACTGTGTCTAAAGZ | Rev | CGGGAATGCCTGTGCAAATTAGTTCTG | |
| ΔGI-CFT073-cobU | H1 | GGTTGACCTAAGGTAGCAGTTTATCCTGATGCGCTGAGATTT | Fwd | GCTGACATCATCAAGAATAAAAAGGTT |
| H2 | CACGGAAACAGAAGGTGTGGTGGAATTATGCGAAGAGGTT | Rev | TTCCGGATGTTGCAGGGCGTAAT | |
| ΔPAI-CFT073-metV | H1 | AGTAAACCGTTAATATCCCTCCATCAAAGCCATCCATCTTAT | Fwd | TTTTTCGTTTTTACGCTTCCTTAC |
| H2 | TTGTAACTGTTAAATCAGGCAAGGCAATGTTTGAAGTAGT | Rev | TATTAGACGCTGGTTTTGTGACTGATG | |
| ΔGI-CFT073-selC | H1 | AAAAACTGATCTGGGGGATGTAGAAACTCAAGGAAGTAG | Fwd | TCCTTGATGCTATAGGGGTGCTGAGAC |
| H2 | ATGACGGTGAGGGAGTAGAGTAATCAATCAGTTTTAGTGAAT | Rev | ACCCATTTTTCCCTCTGCATACTGTTT | |
| ΔPAI-CFT073-aspV1 | H1 | GGAGCGGTAGTTCAGTCGGTTAGAATACCTGCCTGTCA | Fwd | AACAGCAACAAGGTGAAACAACAAT |
| H2 | CGAAAAATCACTAACGAAACATTGGATCCCCATTGTTGC | Rev | CTGAGAGCGAGGAGCGGAAGTAAG | |
| ΔPAI-CFT073-aspV2 | H1 | CAGCTTTCTGACAACCCGGCCGCTCCTGCTTTCAACAA | Fwd | TACATGCTTACTTCCGCTCCTCGCTCTC |
| H2 | GGTTTTTAATGCACACTGGATAACAACCGGACGAATCT | Rev | TAGTCTGATATGGCTTTTGTCGCTGTCC | |
| ΔPAI-CFT073-aspV3 | H1 | TGATAAGCCAAATTGATAAGCTGGAATATGTGATGAAAGTGC | Fwd | CAGCGCCAGTGATATTTGAAGATTCGTC |
| H2 | CTTGTCATATCCGGATAAAATCACCCTCTGGTAATACTCTTA | Rev | TGCACATCGCACAAGTGATTATGAACAG | |
| ΔPAI-CFT073-aspV4 | H1 | GAATTAAGCGCCAGACGTATGGTCAAAAGTAGTGGAGTAGAA | Fwd | TTATTGTTACCTTCTTTTGTTGTGATGA |
| H2 | CATGGCGGGATGCGGATGAGTTTAGGTTGCTGTGAGTG | Rev | TTGATGCGGAGTTGTCGATGGCTGTATT | |
| ΔcdiA (c0345) | H1 | AGCCTCCCGTTCGCTTCACTTACCGCCTGCTGAGTTAC | Fwd | TTCACTGCCGGACTGCCTCTGGTT |
| H2 | CCGTAGAAAGCCCCACTGCACCAACGCCAAAACCACCATTTA | Rev | AGCGCAAGCATCAATAAAAATAGT | |
| ΔpicU (c0350) | H1 | GGGACTTATTGTTGTCTCTGAACTTGCCAGCAGGGTA | Fwd | ACATCATGGAGAGTCCGCAGTGAA |
| H2 | CGCATCACGCAGTACCGTCTCACCATTATTCAGTAGG | Rev | GCTGACTTCTCAAACTCCAGACCA | |
| Δc0294-97 (fbpABCD) | H1 | GCTCCTCGCTCTCAGCGGACCTTCAGCTCAGTGATATCG | Fwd | GGGGATCCAATGTTTCGTTAGTGA |
| H2 | CCTGAGGCTGAAGCAACCACGTTAATCCGACTATTTTTCG | Rev | TGGTAGGCGGATAGATAATAGAAA | |
| Δc0363 #1c | H1 | CTGTCGGAGGTCGTCATACAGTTAAAGCACAG | Fwd | TACCGATGGTGATGGTGCAACAGA |
| H2 | GTTAATATAGCCCTGATTACCGGAGGAGGTGGAGTC | Rev | ATGGATACTTTACCGGCAGCCACT | |
| Δc0363 #2 | H1 | AGGAAGAAGATACTGAGGTCCGGATAGAGGGATTCTGG | Fwd | TGTCGGAGGTCGTCATACAGTT |
| H2 | CCGTTAATATAGCCCTGATTACCGGAGGAGGTGGAGTC | Rev | TAACAGTTGTTGCCGTTGCCGT | |
| ΔPAI-CFT073-metV1 | H1 | AAGGATATGGCCGACAGTTTCCAGAATGAAGTTCCCGC | Fwd | ATTCTCTCACCAGATAATGCCGCC |
| H2 | TATTCCTTCTGGGCGCGAACGATAGCCTGTATAAAGCG | Rev | ATCAACAGACGAAGCCAGACAGCA | |
| ΔPAI-CFT073-metV2 | H1 | CTTTTGGGTGTGGGGTTACTGCTCCTTGTTGTGTTGTTGC | Fwd | CATTGCCAGGCATCGTCTTTGGTT |
| H2 | CAATGAATTTATATTTCGTTGAATAGATAACATTTACC | Rev | ACAGGCTGGGTTTGCCGTACTAAA | |
| Δc3391-92 (Hcp, ClpB) | H1 | AGAAGGCGGTGCTTCAATCACACTAACAAGGAGAGTAA | Fwd | CAGTCAACCGGCGTGTCGAAATCAGTCT |
| H2 | GTGGTGTGCGTGGTCGAAGAACTGTACGTTCATAAGAG | Rev | CTCGCGGCCTTCAAAGGTCAGCACATCC | |
| Δc3398-c3404 | H1 | GCAGAACGAAGCCTCGACGATATCACTATACGCTCAACC | Fwd | TGGGGTTACTGCTCCTTGTTGTG |
| H2 | CTGATTTAACCGGGTATCAATTTGCGTCAACAGCGTTGGC | Rev | GCGGTTTGTCAGCATTCTAA | |
| Δc3405-c3410 | H1 | TTTTGTGACTGATGTCGGATATTTGAATGTCGGCTTG | Fwd | GACGCTGGCGAGAAGGGGATAA |
| H2 | ATCTCCCTTCCTGCGAAGTAATCAATTATCGACTGGG | Rev | ATTTCGGTAGATAGCTTGGGTTCG | |
Primers homologous to sequences within the 5′ and 3′ ends of the target regions were designed (H1 and H2 primers, respectively) and were used to replace these genes with a nonpolar kanamycin or chloramphenicol resistance cassette derived from plasmid pKD4 or pKD3, respectively (12).
Forward (Fwd) and reverse (Rev) genomic primers.
Two independently constructed Δc0363 mutants were tested in this study. Δc0363 #1 is referred to as Δc0363 throughout the article.
TABLE 2.
Characteristics of genomic and pathogenicity island deletion constructs in E. coli CFT073
| Genomic island | Size (kb) | CDSa in island | Region replaced in λ red mutant | tRNA gene present in mutant?b |
|---|---|---|---|---|
| PAI-CFT073-pheV (PAI I) | 123 | c3556-kpsM | Nucleotide 383 of c3556 to 1134 nucleotides downstream of kpsM (intergenic region) | Yes (pheV) |
| PAI-CFT073-pheU (PAI II) | 52 | c5143-c5216 | Nucleotide 1319 of c5144 to 290 nucleotides upstream of c5216 | No (pheU) |
| PAI-CFT073-aspV (PAI III) | 100 | c0253-c0368 | Nucleotide 221 of c0253 to 9 nucleotides upstream of c0368 | Yes (aspV) |
| φ-CFT073-b0847 | 33 | intT-ogrK | 206 nucleotides downstream of ybjK to 94 nucleotides upstream of ogrK | NA |
| PAI-CFT073-serX | 113 | c1165-c1293 | Nucleotide 232 of c1165 to nucleotide 664 of c1292 | Yes (serX) |
| φ-CFT073-potBc | 44 | c1400-c1474 | Nucleotide 772 of c1400 to nucleotide 242 of c1506 | NA |
| PAI-CFT073-asnT (HPI) | 32 | c2418-c2437 | 172 nucleotides upstream of c2418 to nucleotide 657 of c2437 | Yes (asnT) |
| GI-CFT073-asnW | 54 | c2449-c2475 | Nucleotide 35 of c2449 to 23 nucleotides upstream of c2475 | Yes (asnW) |
| GI-CFT073-cobU | 44 | c2482-c2528 | 969 nucleotides upstream of c2482 to 50 nucleotides downstream of c2528 | NA |
| PAI-CFT073-metV | 32 | c3385-c3410 | 475 nucleotides upstream of c3385 to 53 nucleotides upstream of c3410 | Yes (metV) |
| GI-CFT073-selC | 68 | intC-c4581 | 432 nucleotides upstream of intT to 65 nucleotides downstream of c4581 | No (selC) |
CDS, coding sequence.
PAIs are commonly inserted adjacent to tRNA genes; deletion of individual GIs generally did not disrupt the adjacent tRNA gene. NA, not applicable (GI is not located near a tRNA gene).
PAI-CFT073-potB was originally designated as c1400-c1507, so this region was replaced in the GI isogenic mutant. However, the boundaries of the island have since been redefined as c1400-c1474, since c1475-c1507 was a separate island (<30 kb), so it was excluded from our analysis.
Genotypic analysis of mutants.
To verify whether the kanamycin or chloramphenicol resistance cassette recombined within the target gene site, primers that flank target GI sequence were designed (Table 1). Both wild-type (where possible) and mutant gene sequences were amplified with genomic confirmation primers by PCR using Taq DNA polymerase (New England Biolabs). Additional confirmation that the desired genomic replacements had occurred was obtained by digesting each confirmation PCR product with the restriction enzyme EagI (New England Biolabs) for mutants containing a kanamycin resistance cassette or with EaeI (New England Biolabs) for mutants containing a chloramphenicol resistance cassette. The kanamycin resistance cassette contains a single EagI restriction site, and the chloramphenicol resistance cassette contains a single EaeI restriction site, confirming replacement of the GI with the antibiotic resistance cassette if bands of the predicted size were observed. PCR products and restriction enzyme digests were electrophoresed on a 1% agarose gel for visualization of the amplified and digested DNA. The 1-kb+ DNA ladder (Invitrogen) was used to estimate DNA fragment size.
Growth rates of genomic island mutants.
All mutants were tested for their ability to grow in of LB broth (10 g/liter NaCl) or sterile, pooled human urine using the Bioscreen growth curve analyzer (Growth Curves USA) to determine whether deletion mutants showed a growth defect in vitro. Growth rates of all deletion mutants were measured prior to in vivo testing of the isogenic mutants in the murine model of ascending UTI.
Murine model of ascending UTI.
An adaptation (32) of the CBA/J mouse model of ascending UTI, originally developed by Hagberg et al. (25), was used to assess virulence of E. coli CFT073 and its deletion constructs. Female 6- to 8-week-old CBA/J mice (Harlan Sprague-Dawley [Indianapolis, IN] or Jackson Laboratory [Bar Harbor, ME]) were anesthetized with 100 mg ketamine and 10 mg xylazine per kg body weight and inoculated transurethrally with a 50-μl bacterial suspension delivering 2 × 108 CFU per mouse. A sterile polyethylene catheter (inner diameter, 0.28 mm; outer diameter, 0.61 mm) connected to an infusion pump (Harvard Apparatus) was used to deliver the inoculum over a 30-second period. For independent challenges, mice were transurethrally inoculated with 2 × 108 CFU of a single strain per mouse as described above. For cochallenge studies, two cultures were prepared as described above, mixed in a 1:1 ratio, and used to deliver a total of 2 × 108 CFU per mouse. The two strains consisted of either wild-type CFT073 and an isogenic mutant or two different isogenic mutants. To determine the input CFU/ml for each strain, dilutions of each inoculum were plated on LB agar plates containing an antibiotic where required using an Autoplate 4000 (Spiral Biotech). Bacterial counts were determined using a Q-Count machine and accompanying software (Spiral Biotech). Mice were sacrificed at 48 h postinoculation (hpi); the bladder and kidneys were aseptically removed, weighed, and homogenized in 3 ml sterile PBS. Homogenized tissue samples were plated onto LB plates with or without antibiotic (as required) to determine the output CFU/g of tissue for each strain. Wild-type bacterial counts were obtained by subtracting the CFU/ml of the antibiotic-containing plates from the CFU/ml of the plain LB plates. The lower limit of detection of this assay is 102 CFU/g of tissue. Thus, for statistical analysis, this value (102 CFU/g of tissue) was assigned to samples with an undetectable level of colonization. All animal protocols were approved by the University Committee on Use and Care of Animals at the University of Michigan Medical School.
Cloning of the fbp locus into pGEN-MCS.
One of the ferric binding protein (fbp) loci (c0294 to c0297 [c0294-97]) was PCR amplified using primers 5′-NNNNNNTACGTAATGAAAACTCAAATAACTTTCGCTGCG-3′ (to introduce a 5′ SnaBI site) and 5′-NNNNNNGAGCTCCTATTTTTCGATTGCACCATCC-3′ (to introduce a 3′ SacI site) and cloned into the pGEN-MCS vector (38) under the control of the em7 promoter. The pGEN-luxCDABE (Pem7) (38) construct, containing the luciferase (lux) genes under the control of the em7 promoter, was digested with SnaBI and SacI to replace the lux cassette with the c0294-97 genes. Total membrane preparations were isolated using a modified version of the protocol of Hagan and Mobley (24) to visualize expression of the fbp locus. Briefly, 50-ml overnight E. coli TOP10/pGEN and TOP10/pGENfbp cultures were pelleted by centrifugation (8,000 × g, 10 min, 4°C), resuspended in 7 ml of 10 mM HEPES, and treated with 10 μl benzonase nuclease (Sigma; diluted to 10 U/μl in 50% glycerol, 20 mM Tris [pH 8], 2 mM MgCl2, and 20 mM NaCl). Cells were lysed by two passages through a French pressure cell (20,000 lb/in2), and total membrane was isolated from the cleared lysate by ultracentrifugation (100,000 × g, 30 min, 4°C). Total membrane preparations were resuspended in 100 μl of 10 mM HEPES, solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, and electrophoresed on a 10% polyacrylamide gel.
Statistical analysis.
Median CFU/g tissue was reported for all independent and cochallenge experiments to reflect the nonparametric distribution of the in vivo data. Statistically significant differences in colonization, defined as a P value of <0.05, were determined using InStat software (Graphpad, San Diego, CA). Independent challenges were analyzed using the unpaired, nonparametric Mann-Whitney test, and cochallenge infections were analyzed using the nonparametric Wilcoxon matched-pair test.
RESULTS
Isogenic mutants lacking PAI-CFT073-aspV, PAI-CFT073-metV, and PAI-CFT073-asnT are outcompeted by wild-type strain CFT073 in vivo.
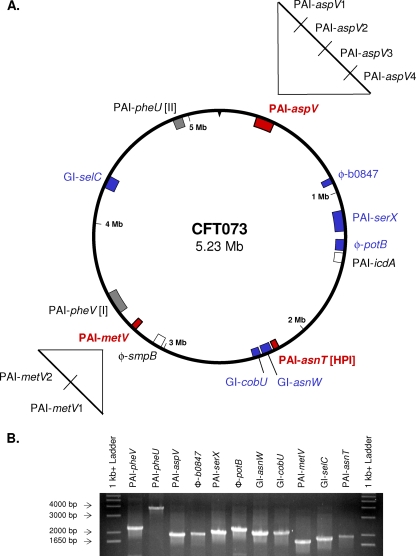
Eleven genomic islands (Fig. 1A), ranging from 32 kb to 123 kb, were individually deleted from E. coli CFT073 using the lambda red recombinase system. Deletions were verified by PCR analysis (Fig. 1B). In each case, PCR analysis confirmed that PAI sequences were absent (i.e., had been successfully deleted) and that flanking PAI boundaries were now close enough to each other to allow PCR amplification of the intervening sequence. The growth rates of the 11 GI mutants and wild-type CFT073 in LB broth or sterile, pooled human urine were comparable (see Fig. S1 in the supplemental material). However, the PAI-metV mutant entered stationary phase earlier than did strains with the other constructs when the strains were grown in LB broth (see Fig. S1A in the supplemental material), and the PAI-aspV mutant demonstrated a slight lag after 2 h of growth in human urine but had reached a similar A600 after 8 h of growth (see Fig. S1B in the supplemental material).
FIG. 1.
(A) Thirteen genomic islands of >30 kb in E. coli CFT073. Isogenic mutants were constructed in 11 GIs (shaded) using the lambda red recombinase system. Deletion mutants of nine of these mutants (shaded blue and red) were tested in cochallenge with wild-type strain CFT073 in the CBA/J mouse model of ascending UTI. Deletion mutants of six islands shown in blue did not show levels of colonization that were statistically different from that of CFT073. Mutants with deletion of genes in the three islands shown in red (PAI-aspV, PAI-metV, and PAI-asnT) were significantly outcompeted by wild-type strain CFT073 in the bladders and/or kidneys (P < 0.05). Smaller deletion mutants spanning PAI-aspV (PAI-aspV1, PAI-aspV2, PAI-aspV3, and PAI-aspV4) and PAI-metV (PAI-metV1 and PAI-metV2) were also constructed and tested in cochallenge with strain CFT073. Isogenic mutants in GIs PAI-icdA and PAI-smpB were not created (white). (B) Confirmation of homologous recombination and subsequent replacement of each GI with a kanamycin resistance cassette. PCR amplification of isogenic mutants with primers flanking the targeted GIs (32 to 123 kb) resulted in PCR products with the predicted sizes: ΔPAI-pheV (2,204 bp), ΔPAI-pheU (3,820 bp), ΔPAI-aspV (1,883 bp), φΔ-b0847 (1,938 bp), ΔPAI-serX (2,061 bp), φΔ-potB (2,290 bp), ΔGI-asnW (2,113 bp), ΔGI-cobU (2,122 bp), ΔPAI-metV (1,746 bp), ΔGI-selC (1,850 bp), and ΔPAI-asnT (1,943 bp). The estimated sizes of PCR products were consistent with the predicted sizes.
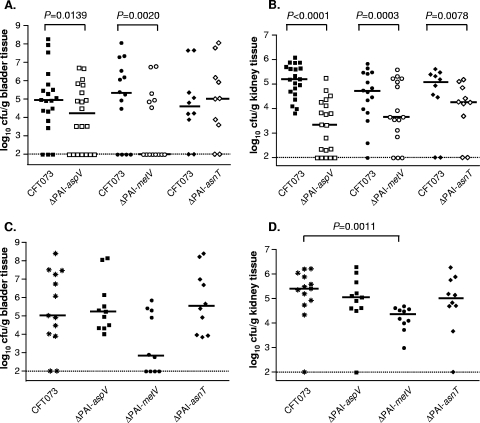
Nine isogenic GI mutants were tested for the ability to colonize the CBA/J mouse model of ascending UTI at 48 h after transurethral inoculation (Table 3). Two islands (PAI-CFT073-pheV and PAI-CFT073-pheU) were not considered further, as these islands had been characterized previously (19, 57). Three mutants, ΔPAI-CFT073-aspV, ΔPAI-CFT073-metV, and ΔPAI-CFT073-asnT (subsequently referred to as ΔPAI-aspV, ΔPAI-metV, and ΔPAI-asnT, respectively) were significantly outcompeted in vivo by wild-type strain CFT073. The ΔPAI-aspV and ΔPAI-metV mutants were significantly outcompeted by strain CFT073 in the bladders (Fig. 2A; P = 0.0139 and P = 0.0020, respectively) and in the kidneys (Fig. 2B; P < 0.0001 and P = 0.0003, respectively), while the ΔPAI-asnT mutant was outcompeted in the kidneys (Fig. 2B; P = 0.0078), indicating that these islands contribute to the fitness of CFT073 in vivo.
TABLE 3.
Colonization of CBA/J mice transurethrally inoculated with wild-type E. coli CFT073 and its genomic island deletion mutants in cochallenge and independent challenge experiments
| Genomic islanda | Island size (kb) | Cochallengeb
|
Independent challengec
|
||||
|---|---|---|---|---|---|---|---|
| No. of miced |
P valuese
|
No. of mice |
P values
|
||||
| Bladder | Kidneys | Bladder | Kidneys | ||||
| PAI-CFT073-aspV(PAI III) | 100 | 20 | 0.0139 | <0.0001 | 11 | 0.8201 | 0.3692 |
| φ-CFT073-b0847 | 33 | 8 | 0.2969 | 0.0781 | ND | ND | ND |
| PAI-CFT073-serX | 113 | 8 | 0.6406 | 0.1563 | ND | ND | ND |
| φ-CFT073-potB | 44 | 9 | 0.9102 | 0.0781 | ND | ND | ND |
| PAI-CFT073-asnT(HPI) | 32 | 10 | 0.1641 | 0.0078 | 10 | 0.8793 | 0.3211 |
| GI-CFT073-asnW | 54 | 13 | 0.7869 | 0.5879 | 13 | >0.9999 | 0.3622 |
| GI-CFT073-cobU | 44 | 10 | 0.4922 | 0.4922 | ND | ND | ND |
| PAI-CFT073-metV | 32 | 16f | 0.0020 | 0.0003 | 10 | 0.0668 | 0.0011 |
| GI-CFT073-selC | 68 | 13 | 0.2163 | 0.4648 | 13 | 0.6810 | 0.6139 |
PAI-CFT073-pheV (PAI I) (123 kb) and PAI-CFT073-pheU (PAI II) (52 kb) mutants were not tested in vivo, as these PAIs have already been published.
In cochallenge infection experiments, mice were inoculated with a 1:1 ratio of standardized cultures of wild-type CFT073 and a deletion mutant.
In independent challenge experiments, mice were inoculated separately with standardized cultures of either wild-type CFT073 or the deletion mutant. ND, not determined.
Number of mice with detectable colonization in bladder and/or kidneys.
P values comparing the ability of the mutants with the indicated genomic island to colonize the bladder and kidneys of mice shown in bold type were statistically significant (P < 0.05). P values for cochallenge infections were determined using the Wilcoxon matched-pair test. P values for independent challenges were determined using the Mann-Whitney test.
Sixteen mice were used in the PAI-metV cochallenge experiments, although only 14 data points were usable for the bladder data.
FIG. 2.
Colonization levels in the bladder and kidneys of CBA/J mice at 48 hpi. Cochallenge of wild-type CFT073 with mutant strains or ΔPAI-aspV (n = 20), ΔPAI-metV (n = 14), and ΔPAI-asnT (n = 10) in the bladder (A) and kidneys (B). Independent challenges of wild-type CFT073 (n = 13) with mutant strains ΔPAI-aspV (n = 11), ΔPAI-metV (n = 10), and ΔPAI-asnT (n = 10) in the bladder (C) and kidneys (D). Bars indicate the median level of colonization (CFU/g tissue). A P value of <0.05 was considered statistically significant. P values for cochallenge infections were determined using the Wilcoxon matched-pair test. P values for independent challenges were determined using the Mann-Whitney test.
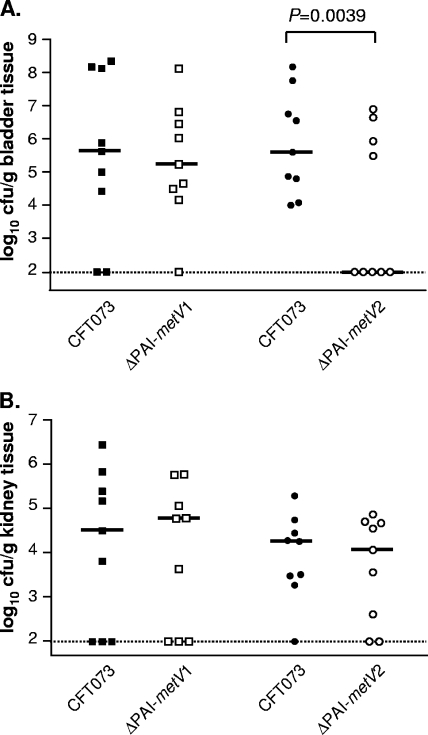
GI mutants ΔPAI-aspV, ΔPAI-metV, and ΔPAI-asnT were also assessed in independent challenges in the CBA/J mouse model of ascending UTI to determine whether these PAIs were required for colonization. Levels of colonization (CFU/g tissue) for ΔPAI-aspV and ΔPAI-asnT mutants were not statistically significantly different from that of wild-type strain CFT073 in the bladder (Fig. 2C) or kidneys (Fig. 2D). The ΔPAI-metV mutant colonized the kidneys (Fig. 2D) at a significantly lower level than that of strain CFT073 (P = 0.0011) and tended to show attenuated colonization in the bladder (P = 0.0668) (Fig. 2C). These data indicate that genes present on PAI-aspV and PAI-asnT contribute to the fitness of UPEC in vivo, while a gene(s) on PAI-metV is required for colonization of the kidneys and possibly the bladder.
PAI-aspV region.
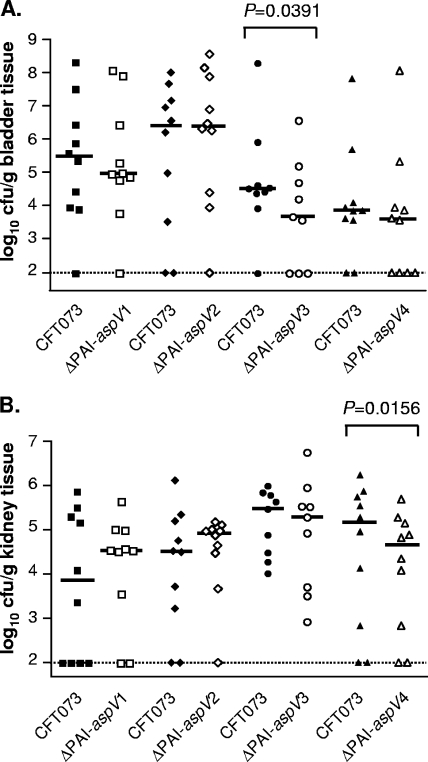
The aspV GI is 99.7 kb in length, and its deletion attenuated the ability of UPEC strain CFT073 to colonize. To localize the observed phenotype of the ΔPAI-aspV mutant to a specific region, four smaller deletion mutants, ΔPAI-aspV1 (25.0 kb), ΔPAI-aspV2 (26.4 kb), ΔPAI-aspV3 (30.0 kb), and ΔPAI-aspV4 (17.4 kb), were constructed to cover the entire genomic island (Fig. 1A) (see Fig. S2A in the supplemental material). Each of the four isogenic deletion mutants was tested in vivo by cochallenge with wild-type CFT073. The ΔPAI-aspV3 mutant was significantly outcompeted by wild-type strain CFT073 in the bladders of mice (P = 0.0391) (Fig. 3A), while the ΔPAI-aspV4 mutant was significantly outcompeted in the kidneys (P = 0.0156) (Fig. 3B).
FIG. 3.
Colonization levels in the bladder (A) and kidneys (B) of CBA/J mice at 48 hpi during cochallenge of wild-type CFT073 with mutant strains ΔPAI-aspV1 (n = 10), ΔPAI-aspV2 (n = 10), ΔPAI-aspV3 (n = 9), and ΔPAI-aspV4 (n = 10).
Several genes with known or predicted roles in the pathogenicity of UPEC reside within the aspV GI, including the contact-dependent inhibition gene cdiA (2) and the autotransporter gene picU (29). One gene of interest, c0363, appears to be part of an operon consisting of open reading frames (ORFs) c0360 to c0363 (c0360-c0363). c0363 is annotated as putative RTX family exoprotein A gene, and on the basis of results of in silico analysis (52), the c0360-63 locus has been reported to encode a type one secretion (tos) system, with c0363 designated as tosA. Additionally, the UPEC-specific operon (c0294-97), identified in our comparative genomic hybridization study of strain CFT073 (44), is present in the aspV pathogenicity island. Two identical copies of the c0294-97 operon exist in strain CFT073, one of which is located in PAI-aspV and the second in GI-CFT073-cobU (c2518-15). The PAI-aspV2 island contains c0294-97, PAI-aspV3 island contains cdiA (c0345) and picU (c0350), and PAI-aspV4 contains c0363 (see Fig. S2A in the supplemental material). Isogenic mutants were constructed for cdiA (Δc0345), picU (Δc0350), one copy of the iron acquisition operon (Δc0294-97), and the type one secretion system gene tosA (Δc0363).
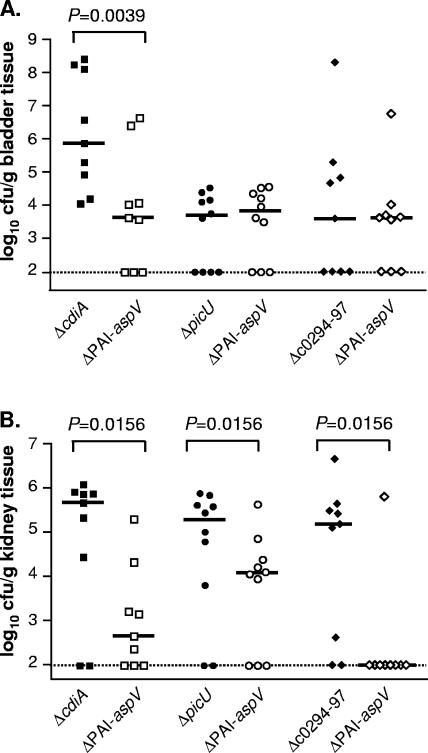
The ΔcdiA, ΔpicU and Δc0294-97 mutants were tested in cochallenge with the ΔPAI-aspV mutant (Fig. 4) to identify the contribution of these known or putative virulence genes to the ΔPAI-aspV phenotype. The ΔPAI-aspV mutant was significantly outcompeted by ΔcdiA in the bladder (P = 0.0039) (Fig. 4A) and kidneys (P = 0.0156) (Fig. 4B). There was no significant difference in the levels of colonization between ΔpicU and Δc0294-97 and ΔPAI-aspV mutants in the bladders of mice (Fig. 4A), indicating that picU and c0294-97 contribute to the ΔPAI-aspV phenotype in the bladder. However, the ΔPAI-aspV mutant was outcompeted by ΔpicU and Δc0294-97 mutants in the kidneys (P = 0.0156 and P = 0.0156, respectively) (Fig. 4B).
FIG. 4.
Colonization levels in the bladder (A) and kidneys (B) of CBA/J mice at 48 hpi during cochallenge of strains with mutations in individual genes of known function against the entire PAI mutant in which the gene is located. Cochallenge of the ΔcdiA mutant with the ΔPAI-aspV mutant (n = 9), ΔpicU mutant with ΔPAI-aspV mutant (n = 10), and Δc0294-97 mutant with ΔPAI-aspV mutant (n = 9).
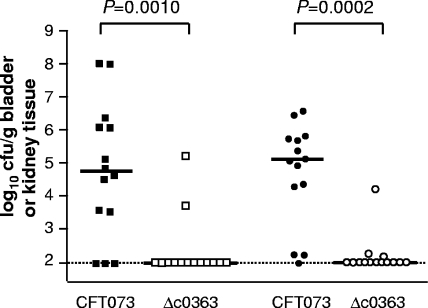
A Δc0363 mutant was tested in cochallenge with wild-type CFT073 (Fig. 5). Deletion of c0363 resulted in statistically significant outcompetition of the Δc0363 mutant by strain CFT073 in both the bladder (P = 0.0010) and kidneys (P = 0.0002). To confirm this observation and minimize the possibility that a secondary mutation contributed to the attenuation of colonization, a second Δc0363 mutant (Δc0363 2, constructed independently using different primers) was tested in cochallenge with CFT073, also resulting in statistically significant outcompetition of the mutant by CFT073 in both the bladder (P = 0.0156) and kidneys (P = 0.0078) (data not shown). These data indicate that c0363 contributes to the fitness of CFT073 in the urinary tract.
FIG. 5.
Colonization levels in the bladder and kidneys of CBA/J mice at 48 hpi during cochallenge of wild-type CFT073 with the Δc0363 (tosA) RTX toxin mutant (n = 14). The colonization levels in bladders (squares) and kidneys (circles) are indicated.
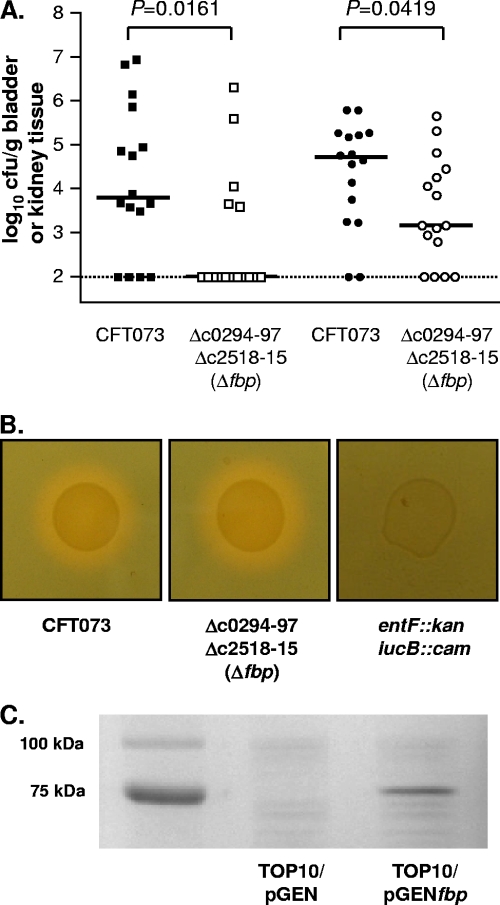
A mutant in which both copies of the putative iron acquisition system (Δc0294-97 Δc2518-15) had been deleted was tested in cochallenge with wild-type strain CFT073. The Δc0294-97 Δc2518-15 mutant was significantly attenuated in both the bladder (P = 0.0161) and kidneys (P = 0.0419) (Fig. 6A) of mice during cochallenge with strain CFT073. Wild-type CFT073, the Δc0294-97 Δc2518-15 mutant, and a siderophore-deficient strain of CFT073 (entF::kan iucB::cam) were grown on CAS siderophore agar. No difference in halo formation was observed between wild-type CFT073 and the Δc0294-97 Δc2518-15 mutant (Fig. 6B), indicating that both strains are producing siderophores. The fbp locus (c0294-97) was cloned into pGEN-MCS under the control of the constitutive em7 promoter, and protein expression was confirmed examining whole-membrane preparations of E. coli TOP10/pGEN and TOP/pGENfbp (Fig. 6C). In order to restore the wild-type phenotype of the fbp (Δc0294-97 Δc2518-15) double mutant, in vivo complementation was attempted using cochallenge of CFT073/pGEN with Δfbp/pGENfbp. However, CFT073/pGEN significantly outcompeted Δfbp/pGENfbp in the kidneys (P = 0.0156), while median levels of bladder colonization were at the limit of detection for both strains (P > 0.9999) (data not shown). No difference in growth was observed between wild-type CFT073 and the Δc0294-97 Δc2518-15 double mutant under iron-limiting conditions in LB broth or M9 minimal medium containing 200, 300, 400, 500, or 600 μM 2,2′-dipyridyl.
FIG. 6.
(A) Colonization levels in the bladder and kidneys of CBA/J mice at 48 hpi during cochallenge of wild-type CFT073 with the double iron system Δc0294-97 Δc2518-15 (Δfbp) mutant (n = 16). The colonization levels in bladders (squares) and kidneys (circles) are indicated. (B) Growth of wild-type CFT073, Δc0294-97 Δc2518-15 (Δfbp) mutant, and an enterobactin/aerobactin-negative (entF::kan iucB::cam) strain of CFT073 on CAS siderophore agar. Siderophore production is indicated by orange halos around the bacterial growth. (C) Expression of the cloned fbp locus under the control of a constitutive em7 promoter (E. coli TOP10/pGENfbp) and the vector-only negative control (pGEN-MCS). The predicted molecular mass of the c0294 protein is 78.4 kDa.
PAI-metV region.
Since deletion of PAI-metV attenuated colonization in strain CFT073, two smaller mutants in PAI-metV were constructed to examine this 32.1-kb GI in greater detail (Fig. 1A) (see Fig. S2B in the supplemental material). Wild-type CFT073 was used in separate cochallenge experiments with strains carrying ΔPAI-metV1 (16.1 kb) and ΔPAI-metV2 (15.8 kb) (Fig. 7). The ΔPAI-metV2 mutant was significantly outcompeted by CFT073 in the bladder (P = 0.0039) (Fig. 7A), with the median level of colonization of the deletion mutant at the limit of detection. In the bladder, the ΔPAI-metV2 mutant showed the same phenotype as the ΔPAI-metV whole-GI knockout, with both mutants showing a 3-log-unit decrease in colonization levels from the colonization level of CFT073. There was no significant difference in the levels of colonization between CFT073 and ΔPAI-metV1 mutant in the bladder (Fig. 7A), and the median CFU/g kidney tissue for the ΔPAI-metV1 or ΔPAI-metV2 mutant was not statistically significantly different from that of wild-type CFT073 (Fig. 7B).
FIG. 7.
Colonization levels in the bladder (A) and kidneys (B) of CBA/J mice at 48 hpi during cochallenge of wild-type CFT073 with the ΔPAI-metV1 mutant (n = 9) and ΔPAI-metV2 mutant (n = 9).
The only two genes in PAI-metV with a predicted role in virulence encode the secreted protein Hcp (c3391) and the ClpB protein (c3392). Cochallenge of the ΔPAI-metV mutant with the Δc3391-92 mutant revealed that the ΔPAI-metV mutant colonized the bladder to significantly lower levels (P = 0.0156) than the Δc3391-92 mutant did. A similar trend was seen in the kidneys of mice, although this difference was not statistically significant (P = 0.0781) (data not shown).
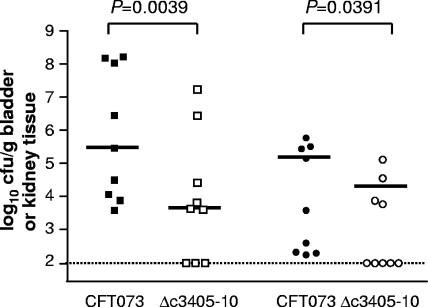
The ΔPAI-metV2 mutant, which showed a 3-log-unit drop in colonization of the bladder from the colonization level of strain CFT073, was further characterized by constructing a strain with mutations in genes c3398-c3404, representing the first half of the genes in PAI-metV2. In cochallenge with CFT073, the Δc3398-c3404 mutant showed similar levels of colonization to the wild-type strain (data not shown), and these differences were not statistically significant. The remaining genes in the PAI-metV2 region, c3405-10, were deleted and tested in cochallenge with CFT073. The ability of the Δc3405-10 deletion mutant to colonize was significantly attenuated in both the bladder (P = 0.0039) and kidneys (P = 0.0391) than in wild-type CFT073, as shown in (Fig. 8).
FIG. 8.
Colonization levels in the bladder and kidneys of CBA/J mice at 48 hpi during cochallenge of wild-type CFT073 with the Δc3405-10 mutant (n = 9). The colonization levels in bladders (squares) and kidneys (circles) are indicated.
DISCUSSION
Uropathogenic E. coli strains, as exemplified by strain CFT073, have acquired pathogenicity islands that contribute to fitness in the urinary tract. In this study, three of the nine genomic island deletion mutants tested were significantly outcompeted by wild-type CFT073 in the bladders or kidneys of CBA/J mice during experimental cochallenge. The PAI-aspV and PAI-metV mutants showed attenuation in the ability to colonize bladders and kidneys, while the PAI-asnT mutant showed attenuation in the kidneys only. In contrast, all mutants were able to colonize the urinary tracts of mice to levels similar to that of the wild-type strain during independent challenges of mice with the exception of the PAI-metV mutant, which was attenuated in the kidneys (and tended toward attenuation in the bladder [P = 0.0668]). The ability of these mutants to colonize the murine urinary tract to a level comparable to that of the wild type when not in a competitive setting demonstrates that while the genes contained in PAIs contribute to the overall fitness of the strain, they are generally not essential for survival in the host.
The pangenome of E. coli was recently described by Rasko and colleagues (58). Analysis of 17 sequenced E. coli genomes, including commensal and pathogenic strains, revealed the genome size of E. coli to be 5,020 ± 446 genes (mean ± standard deviation) with a “conserved core” genome size (genes that are highly conserved in all 17 isolates) of 2,344 ± 43 genes (58). This points to the extraordinary plasticity of strains equipped to colonize a variety of body sites and the potential to cause different disease syndromes. Eleven sequenced, assembled, and annotated E. coli genomes range in size from 4.64 Mb (E. coli K-12) to 5.53 Mb (E. coli O157:H7 EDL933) with a median genome size of 5.08 Mb (58).
E. coli CFT073, the subject of this study, has the largest genome (5.23 Mb) of the three sequenced and annotated UPEC strains and is 592 kb larger than E. coli K-12 (76). Two independent studies have now estimated that 17 to 18% of all ORFs in E. coli strain K-12 MG1655 were horizontally acquired (40, 50). Nakamura and colleagues (50) compared 116 sequenced prokaryotic genomes and suggested that 21.4% of the E. coli CFT073 genome was horizontally acquired, placing it sixth among all isolates studied. The average proportion of horizontally acquired genes from this group, consisting of 16 archaebacteria and 100 eubacteria, was estimated at 12.4% (50).
Pathogenic strains of E. coli generally have larger genomes than commensal E. coli isolates, with the majority of this difference attributed to the insertion of relatively few large chromosomal regions (59). These GIs are acquired by horizontal gene transfer (51) and comprise 12.8% of the CFT073 genome (44). GIs contribute to bacterial fitness (21) by conferring new properties that increase the adaptability of the organism and may also encode genes involved in pathogenicity. Strain CFT073 has 13 genomic or pathogenicity islands (Fig. 1).
PAI-aspV.
We observed that individual deletion of three PAIs attenuated the ability of wild-type strain CFT073 to colonize in cochallenge with the wild-type strain. PAI-aspV showed a median level of colonization that was about 0.5 log unit less than CFT073 in the bladder and 2 log units less in the kidneys. Construction of mutants in smaller regions of two PAIs allowed us to localize the observed attenuation phenotype to specific genes or operons, often with organ-specific effects. For example, the aspV PAI was further characterized by construction of four mutants, each deleting smaller sections of the PAI (PAI-aspV1, PAI-aspV2, PAI-aspV3, and PAI-aspV4) (Fig. 1) (see Fig. S2A in the supplemental material). Testing the smaller mutants in cochallenge revealed that PAI-aspV3, containing the contact-dependent inhibition gene cdiA and the autotransporter protease gene picU, was important for colonization of the bladder, whereas PAI-aspV4, containing the RTX family exoprotein A gene, c0363, previously described as tosA for type 1 secretion (52), was significantly outcompeted in the kidneys of mice.
Testing mutants in three genes of known function located within PAI-aspV in cochallenge against the whole-island mutant demonstrated an organ-specific phenotype for two of the mutants, indicating that some genes play a more crucial role in either the bladder or the kidneys, but not necessarily in both organs. Mutants with deletions in picU and c0294-97 showed levels of colonization in the bladder similar to that of the PAI-aspV whole-island mutant, indicating that these two genes are at least partially responsible for the attenuation observed in the bladder with the ΔPAI-aspV mutant. However, neither mutant was attenuated in their ability to colonize the kidneys. Interestingly, deletion of c0363, the putative RTX toxin (tosA) gene, resulted in statistically less colonization than wild-type CFT073 in both the bladder and kidneys of mice at 48 hpi, despite this trend not reaching significance in the bladder for the PAI-aspV4 mutant lacking the c0363 gene. Taken together, the high degree of attenuation resulting from the deletion of PAI-aspV is likely the result of several virulence genes that, although not required for colonization, provide a clear fitness advantage to the strain.
PAI-metV.
Relative to wild-type CFT073, deletion of PAI-metV resulted in more than a 3-log-unit reduction in colonization in the bladder and an approximately 1.5-log-unit reduction in the kidneys during cochallenge. The presence of specific genes may explain this attenuation. For example, the recently described (54) type VI secretion system (T6SS) provides a mechanism for gram-negative bacteria to export proteins across the cell envelope (4). Homologs of T6SS have been identified in Vibrio cholerae (53, 54), Pseudomonas aeruginosa (49), Burkholderia pseudomallei (70), Burkholderia mallei (66), and Edwardsiella tarda (56). T6SSs are usually encoded within PAIs (10).
Eighteen proteins are involved in T6SS in V. cholerae (54), many of which have been designated vas for virulence-associated secretion. T6SSs encode between 12 and 20 proteins, although the composition and organization of this gene cluster vary considerably between species (10). Shrivastava and Mande identified 15 orthologs of T6SS in strain CFT073 (71). Fourteen of the 15 orthologous genes in CFT073 are located within PAI-metV: c3385-88, c3391-93, and c3398-c3403 (71). We have identified two additional VgrG homologs (c1883 and c1888) (see Fig. S3 in the supplemental material). The two newly identified VgrG homologs in CFT073 are located in a small (11.5-kb) genomic island (c1881-c1893), identified during our comparative genomic hybridization study of uropathogenic and fecal E. coli strains (44).
Two T6SSs, sci-1 and sci-2, have been identified within a pheU PAI in enteroaggregative E. coli (EAEC) strain 042 (13). The smaller of these two T6SSs (sci-1), consisting of ORFs Ec042-4524 to Ec042-4544, shows homology to the metV ORFs c3385-c3401, with only the region corresponding to c3394-97 differing between the two E. coli strains. Interestingly, Hancock and colleagues examined 16 sequenced E. coli and Shigella strains, including four UPEC strains, and determined that the genes c3394-96 are present only in UPEC strain CFT073 and asymptomatic bacteriuria E. coli strain 83972 (28). However, the functions of these genes remain unknown but suggest a role in colonization or persistence within the urinary tract. SciN, an outer membrane lipoprotein present in the sci-1 T6SS of EAEC strain 042 that is required for type VI secretion (3), is 43.8% identical at the amino acid level to c3401 of strain CFT073 (data not shown). The c3401 protein contains the N-terminal feature characteristic of lipoproteins, the lipoprotein signal sequence box (L-A/S-G/A) followed by a cysteine residue (3, 74) (data not shown).
The hcp (hemolysin-coregulated protein) and vgrG (valine-glycine repeat) genes are often located distally to the T6SS gene cluster (15), as was observed with the two newly identified vgrG genes (c1883 and c1888) in strain CFT073. As shown in Fig. S3 in the supplemental material, the two newly identified VgrG homologs in strain CFT073, c1883 and c1888, are closely related to the VgrG proteins of E. coli O157:H7 (713 amino acids) and Shigella sonnei (713 amino acids), in both length and amino acid sequence.
Many of the species containing T6SS homologs closely associate with eukaryotic cells (10). Indeed, disruption of the T6SS or its effectors displays the most prominent phenotype when the bacterial organism is studied intracellularly or while in close contact with eukaryotic cells. Examples include induction of the T6SS of Burkholderia pseudomallei upon invasion of macrophages (70), requirement of the type VI secretion cluster for survival of V. cholerae within Dictyostelium amoebae (54), and a reduction in the ability of the fish pathogen Edwardsiella tarda to multiply inside phagocytic cells following mutation of T6SS genes (56, 79).
Deletion of hcp and clpB (c3391-92) or the T6SS gene cluster c3398-c3404 does not attenuate strain CFT073 in the urinary tract. Similarly, no virulence defect was observed in EAEC strain 042 when mutations were introduced into the sci-2 T6SS gene (13). However, the PAI-metV2 genomic island mutant (c3398-c3410 deleted) was severely attenuated in its ability to colonize the bladders of mice (P = 0.0039). Further characterization of this region using deletion mutant Δc3398-c3404 did not, however, show a trend toward attenuation in vivo. This suggests the major defect of PAI-metV in the bladder may be due to one or more genes within the gene cluster c3405-10. Deletion of c3405-10 caused significant attenuation of the mutant relative to wild-type CFT073, demonstrating that this operon was responsible for the observed phenotype with the entire ΔPAI-metV deletion.
The genes c3405-09 appear to constitute an operon and are annotated as 2-hydroxyacid dehydrogenase (c3405); phosphosugar isomerase (c3406); beta-cystathionase (c3407); phosphotransferase system, maltose- and glucose-specific IIABC component (c3408); and antiterminator (c3409). The c3405-09 gene cluster is also present in uropathogenic E. coli strains UTI89 (11) and 536 (7) and avian pathogenic E. coli (APEC) strain O1 (33) but is absent from E. coli K-12 (5). Indeed, we recently identified c3405 and c3408 as UPEC specific (44). APEC strains, the primary cause of colibacillosis in the poultry industry, are also members of the ExPEC family of E. coli. Similarities between APEC and UPEC strains include overlapping O serotypes and phylogenetic groups and shared virulence factors (60), which has raised concerns over the ability of APEC to cause disease in humans.
A signature-tagged mutagenesis screen of APEC strain O1 identified 28 genes that were required for septicemia in chickens (42). Strains with mutations in three of these genes, located at the right junction of a metV PAI, showed attenuation in the ability to colonize during in vivo competition experiments. The only other bacterial species shown to contain these genes was UPEC strain CFT073 (c3406, c3407, and c3408) (42), where they are also located within a metV PAI. The identification of genes that are important in vivo in both UPEC and APEC strains further supports the concept of shared virulence genes in ExPEC, particularly since these pathotypes cause disease in different niches and hosts.
Iron acquisition system double mutant.
The urinary tract is an iron-limiting environment (62, 64, 72), and uropathogens must be well-equipped to sequester the limited iron available in the host environment. E. coli CFT073 contains six iron acquisition and transport systems, five of which are upregulated during UTI (72). The operon c0294-97, annotated in the CFT073 genome as four open reading frames with hypothetical functions, is 100% identical at the nucleotide level to c2518-15, annotated in strain CFT073 as a TonB receptor; periplasmic binding protein; ABC transporter, FecCD transport family; and putative ABC transporter, respectively. In strain CFT073, these operons are present in two different genomic islands (c0294-97 in PAI-aspV and c2518-15 in GI-cobU). This operon was recently identified as UPEC specific (44), and a single, identical, copy of this operon is present in UPEC strains 536 (ECP_2036 to ECP_2033) (7) and UTI89 (UTI89_C2266 to UTI89_C2263) (11). On the basis of in silico analysis, Parham and colleagues (52) designated the c0294-97 operon the ferric binding protein (Fbp) locus, containing genes fbpA, fbpB, fbpC, and fbpD, respectively. Consequently, we have designated the duplicated operon in CFT073 fbpA, fbpB, fbpC, and fbpD for c0294-97 and fbpA_2, fbpB_2, fbpC_2, and fbpD_2 for c2518-15, respectively.
Deletion of these operons individually does not substantially attenuate the ability of the strain to colonize during cochallenge with wild-type strain CFT073, with the partially attenuated phenotype of the PAI-aspV mutant in the bladder attributed to multiple genes. However, the Δc0294-97 Δc2518-15 double mutant was significantly outcompeted by wild-type CFT073 in both the bladder and kidneys at 48 hpi. Thus, strain CFT073 can function adequately with a single copy of this operon but is attenuated if it loses both copies, suggesting that the fbp iron acquisition system is required for virulence in the urinary tract. Interestingly, both UPEC strain 536 and UTI89 contain only one copy of this iron acquisition system, suggesting that CFT073 has acquired a fitness advantage over these strains by the acquisition of the additional copy on a separate genomic island. Despite cloning the fbp locus under the control of an em7 promoter and confirming protein expression, complementation of the Δc0294-97 Δc2518-15 double mutant in vivo was not achieved. One explanation could be that overexpression of an outer membrane protein slowed the growth rate enough for the strain to be outcompeted by the wild type. Siderophore production of the Δc0294-97 Δc2518-15 double mutant was indistinguishable from that of wild-type CFT073. This finding was not surprising, since we do not propose that fbp genes encode a siderophore system. Rather, we believe this locus encodes an iron transport system that does not involve carrier molecules such as siderophores. Furthermore, growth of the Δc0294-97 Δc2518-15 double mutant was indistinguishable from that of wild-type CFT073 under all levels of iron limitation tested, indicating that we have not yet identified the mechanism of iron acquisition conferred by this system.
In addition to the fbp_2 iron transport operon, the genomic island GI-cobU also contains the recently described heme acquisition protein Hma (24). Although the GI-cobU mutant was not outcompeted by wild-type CFT073 at 48 hpi, Hagan and Mobley (24) did not observe attenuation of an hma mutant in vivo until 72 hpi. Taken together, it can be seen that the cobU GI also functions as a fitness island, encoding two additional iron acquisition systems and conferring an advantage to strains containing these genes, while not essential for colonization of the urinary tract.
PAI-asnT.
The high-pathogenicity island, encoding the yersiniabactin biosynthesis system, was originally discovered in Yersinia enterocolitica (9) but has since been identified in many other bacterial species, including Klebsiella pneumoniae (37), Enterobacter spp. (48), Citrobacter spp. (48), and multiple pathotypes of E. coli (18, 31, 35, 68). The widespread prevalence of the HPI in a range of bacterial species capable of causing disease in various niches and through different pathogenic mechanisms support the concept that HPI contributes to the overall fitness of the organism rather than to its pathogenic potential (21, 35, 67). FyuA, the outer membrane receptor for yersiniabactin and the bacteriocin pesticin (14, 55), is one of the most highly upregulated genes in biofilm formation in human urine (27). Mutation of fyuA does not affect growth but results in significantly reduced capacity for biofilm formation in human urine (26).
The Yersinia pseudotuberculosis HPI was shown to insert into any of the three chromosomal asn tRNA genes (8). E. coli CFT073 contains the HPI, also located at an asn tRNA gene (PAI-asnT) (44), but it does not produce detectable yersiniabactin due to in-frame stop codons in the yersiniabactin biosynthesis genes (7, 31). However, the fyuA yersiniabactin receptor is 99.9% identical at the protein level to the fyuA gene of UPEC strains 536 and UTI89 and the ability of the asnT PAI (HPI) mutant to colonize is significantly attenuated in the kidneys in vivo (P = 0.0078). During cochallenge with CFT073, the PAI-asnT mutant exhibited a 1-log-unit decrease in colonization of the kidneys relative to the wild-type strain. This interesting phenotype warrants further examination.
We have demonstrated that genomic and pathogenicity islands contribute to both virulence and fitness of uropathogenic E. coli in vivo. Multiple virulence factors and metabolic genes encoded on genomic islands provide an advantage to UPEC in vivo and allow adaptation to niches unable to be colonized by commensal E. coli strains. The major genomic difference between E. coli K-12 and pathogenic strains is the acquisition of large chromosomal islands (59), suggesting that these GIs confer the ability of UPEC to successfully transition from the intestinal tract to the urinary tract. Therefore, deletion of multiple GIs in a single CFT073 backbone would demonstrate the importance of GIs to UPEC in terms of virulence and fitness and would highlight the differences between commensal and uropathogenic E. coli. Indeed, UPEC may lose their ability to colonize the urinary tract following sequential removal of the major GIs. Other groups have shown that deletion of GIs or PAIs can reduce the virulence of an ExPEC strain (7, 78).
The increasing body of evidence that virulence factors are shared between ExPEC strains (60) capable of causing disease in extraintestinal sites (urinary tract, bloodstream, bone, lungs, and abdomen) (63) reinforces the idea that the same virulence factors may provide different benefits depending on the environment. Redundancy is characteristic of UPEC, particularly for iron acquisition systems (1). This was clearly demonstrated with the fbp operon, which is present in two copies in E. coli CFT073, was acquired independently on separate GIs, and permits survival in vivo following the loss of one copy of this operon. The more GIs a strain contains, the more flexibility it may possess, including genes that confer an organ-specific phenotype, such as bladder or kidney colonization. Horizontally acquired islands contribute significantly to the ability of UPEC to thrive in the urinary tract. Often, deletion of single genes or small chromosomal regions does not elicit a strong phenotype in vivo but contributes through the accumulation of multiple factors increasing the overall fitness of that strain.
Supplementary Material
Acknowledgments
We thank Sara Smith, Erin Hagan, and Chelsea Lane for invaluable assistance with the CBA/J mouse model of UTI, Stephanie Himpsl for providing the CAS agar plates, and Erin Hagan for assistance with cell fractionation and whole-membrane preparation.
This work was supported in part by Public Health Services grant AI43363 from the National Institutes of Health.
Footnotes
Published ahead of print on 27 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alteri, C. J., and H. L. Mobley. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 752679-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 3091245-1248. [DOI] [PubMed] [Google Scholar]
- 3.Aschtgen, M. S., C. S. Bernard, S. De Bentzmann, R. Lloubes, and E. Cascales. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 1907523-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingle, L. E., C. M. Bailey, and M. J. Pallen. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 113-8. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Blum, G., V. Falbo, A. Caprioli, and J. Hacker. 1995. Gene clusters encoding the cytotoxic necrotizing factor type 1, Prs-fimbriae and alpha-hemolysin form the pathogenicity island II of the uropathogenic Escherichia coli strain J96. FEMS Microbiol. Lett. 126189-195. [DOI] [PubMed] [Google Scholar]
- 7.Brzuszkiewicz, E., H. Bruggemann, H. Liesegang, M. Emmerth, T. Olschlager, G. Nagy, K. Albermann, C. Wagner, C. Buchrieser, L. Emody, G. Gottschalk, J. Hacker, and U. Dobrindt. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA 10312879-12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchrieser, C., R. Brosch, S. Bach, A. Guiyoule, and E. Carniel. 1998. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 30965-978. [DOI] [PubMed] [Google Scholar]
- 9.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J. Bacteriol. 1786743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascales, E. 2008. The type VI secretion toolkit. EMBO Rep. 9735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley, E. G., N. R. Thomson, J. Parkhill, N. P. Morin, and J. P. Nataro. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 611267-1282. [DOI] [PubMed] [Google Scholar]
- 14.Fetherston, J. D., J. W. Lillard, Jr., and R. D. Perry. 1995. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J. Bacteriol. 1771824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filloux, A., A. Hachani, and S. Bleves. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 1541570-1583. [DOI] [PubMed] [Google Scholar]
- 16.Foxman, B. 2003. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis. Mon. 4953-70. [DOI] [PubMed] [Google Scholar]
- 17.Foxman, B., R. Barlow, H. D'Arcy, B. Gillespie, and J. D. Sobel. 2000. Urinary tract infection: self-reported incidence and associated costs. Ann. Epidemiol. 10509-515. [DOI] [PubMed] [Google Scholar]
- 18.Gophna, U., T. A. Oelschlaeger, J. Hacker, and E. Z. Ron. 2001. Yersinia HPI in septicemic Escherichia coli strains isolated from diverse hosts. FEMS Microbiol. Lett. 19657-60. [DOI] [PubMed] [Google Scholar]
- 19.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 664411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 231089-1097. [DOI] [PubMed] [Google Scholar]
- 21.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54641-679. [DOI] [PubMed] [Google Scholar]
- 23.Hacker, J., S. Knapp, and W. Goebel. 1983. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 1541145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagan, E. C., and H. L. Mobley. 2009. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 7179-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagberg, L., I. Engberg, R. Freter, J. Lam, S. Olling, and C. Svanborg Edén. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock, V., L. Ferrieres, and P. Klemm. 2008. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology 154167-175. [DOI] [PubMed] [Google Scholar]
- 27.Hancock, V., and P. Klemm. 2007. Global gene expression profiling of asymptomatic bacteriuria Escherichia coli during biofilm growth in human urine. Infect. Immun. 75966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock, V., A. S. Seshasayee, D. W. Ussery, N. M. Luscombe, and P. Klemm. 2008. Transcriptomics and adaptive genomics of the asymptomatic bacteriuria Escherichia coli strain 83972. Mol. Genet. Genomics 279523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heimer, S. R., D. A. Rasko, C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2004. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect. Immun. 72593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houdouin, V., S. Bonacorsi, P. Bidet, M. Bingen-Bidois, D. Barraud, and E. Bingen. 2006. Phylogenetic background and carriage of pathogenicity island-like domains in relation to antibiotic resistance profiles among Escherichia coli urosepsis isolates. J. Antimicrob. Chemother. 58748-751. [DOI] [PubMed] [Google Scholar]
- 31.Hu, J., B. Kan, Z. H. Liu, and S. Y. Yu. 2005. Enteroaggregative Escherichia coli isolated from Chinese diarrhea patients with high-pathogenicity island of Yersinia is involved in synthesis of siderophore yersiniabactin. World J. Gastroenterol. 115816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, D. E., C. V. Lockatell, M. Hall-Craigs, H. L. Mobley, and J. W. Warren. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J. Urol. 138632-635. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, T. J., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. J. Johnson, C. Doetkott, J. A. Skyberg, A. M. Lynne, J. R. Johnson, and L. K. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 1893228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 35.Karch, H., S. Schubert, D. Zhang, W. Zhang, H. Schmidt, T. Olschlager, and J. Hacker. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect. Immun. 675994-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knapp, S., I. Then, W. Wels, G. Michel, H. Tschape, J. Hacker, and W. Goebel. 1985. Analysis of the flanking regions from different haemolysin determinants of Escherichia coli. Mol. Gen. Genet. 200385-392. [DOI] [PubMed] [Google Scholar]
- 37.Koczura, R., and A. Kaznowski. 2003. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb. Pathog. 35197-202. [DOI] [PubMed] [Google Scholar]
- 38.Lane, M. C., C. J. Alteri, S. N. Smith, and H. L. Mobley. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. USA 10416669-16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence, J. G. 2005. Horizontal and vertical gene transfer: the life history of pathogens. Contrib. Microbiol. 12255-271. [DOI] [PubMed] [Google Scholar]
- 40.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 959413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence, J. G., and J. R. Roth. 1999. Genomic flux: genome evolution by gene loss and acquisition, p. 263-290. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. ASM Press, Washington, DC.
- 42.Li, G., C. Laturnus, C. Ewers, and L. H. Wieler. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 732818-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Litwin, M. S., and C. S. Saigal. 2007. Introduction, p. 3-7. In M. S. Litwin and C. S. Saigal (ed.), Urologic diseases in America. National Institutes of Health publication 07-5512:3-7. U.S. Department of Health and Human Services, Public Health Service, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Washington, DC.
- 44.Lloyd, A. L., D. A. Rasko, and H. L. Mobley. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J. Bacteriol. 1893532-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsumori, K., A. Terai, S. Yamamoto, and O. Yoshida. 1998. Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol. Med. Microbiol. 21261-268. [DOI] [PubMed] [Google Scholar]
- 46.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 581281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10143-155. [DOI] [PubMed] [Google Scholar]
- 48.Mokracka, J., R. Koczura, and A. Kaznowski. 2004. Yersiniabactin and other siderophores produced by clinical isolates of Enterobacter spp. and Citrobacter spp. FEMS Immunol. Med. Microbiol. 4051-55. [DOI] [PubMed] [Google Scholar]
- 49.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 3121526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura, Y., T. Itoh, H. Matsuda, and T. Gojobori. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36760-766. [DOI] [PubMed] [Google Scholar]
- 51.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405299-304. [DOI] [PubMed] [Google Scholar]
- 52.Parham, N. J., S. J. Pollard, R. R. Chaudhuri, S. A. Beatson, M. Desvaux, M. A. Russell, J. Ruiz, A. Fivian, J. Vila, and I. R. Henderson. 2005. Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 432425-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA 10415508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rakin, A., E. Saken, D. Harmsen, and J. Heesemann. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13253-263. [DOI] [PubMed] [Google Scholar]
- 56.Rao, P. S., Y. Yamada, Y. P. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53573-586. [DOI] [PubMed] [Google Scholar]
- 57.Rasko, D. A., J. A. Phillips, X. Li, and H. L. Mobley. 2001. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. J. Infect. Dis. 1841041-1049. [DOI] [PubMed] [Google Scholar]
- 58.Rasko, D. A., M. J. Rosovitz, G. S. Myers, E. F. Mongodin, W. F. Fricke, P. Gajer, J. Crabtree, M. Sebaihia, N. R. Thomson, R. Chaudhuri, I. R. Henderson, V. Sperandio, and J. Ravel. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 1906881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rode, C. K., L. J. Melkerson-Watson, A. T. Johnson, and C. A. Bloch. 1999. Type-specific contributions to chromosome size differences in Escherichia coli. Infect. Immun. 67230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 1512097-2110. [DOI] [PubMed] [Google Scholar]
- 61.Ronald, A.2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am. J. Med. 113(Suppl. 1A)14S-19S. [DOI] [PubMed] [Google Scholar]
- 62.Roos, V., and P. Klemm. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect. Immun. 743565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 1811753-1754. [DOI] [PubMed] [Google Scholar]
- 64.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 707156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schappert, S. M. 1999. Ambulatory care visits to physician offices, hospital outpatient departments, and emergency departments: United States, 1997. Vital Health Stat 13i-iv, 1-39. [PubMed] [Google Scholar]
- 66.Schell, M. A., R. L. Ulrich, W. J. Ribot, E. E. Brueggemann, H. B. Hines, D. Chen, L. Lipscomb, H. S. Kim, J. Mrazek, W. C. Nierman, and D. Deshazer. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 641466-1485. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 1714-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schubert, S., B. Picard, S. Gouriou, J. Heesemann, and E. Denamur. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect. Immun. 705335-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 16047-56. [DOI] [PubMed] [Google Scholar]
- 70.Shalom, G., J. G. Shaw, and M. S. Thomas. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 1532689-2699. [DOI] [PubMed] [Google Scholar]
- 71.Shrivastava, S., and S. S. Mande. 2008. Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS ONE 3e2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 726373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stamm, W. E., and T. M. Hooton. 1993. Management of urinary tract infections in adults. N. Engl. J. Med. 3291328-1334. [DOI] [PubMed] [Google Scholar]
- 74.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 16935-13. [DOI] [PubMed] [Google Scholar]
- 75.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 696179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wiles, T. J., R. R. Kulesus, and M. A. Mulvey. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp. Mol. Pathol. 8511-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie, Y., V. Kolisnychenko, M. Paul-Satyaseela, S. Elliott, G. Parthasarathy, Y. Yao, G. Plunkett III, F. R. Blattner, and K. S. Kim. 2006. Identification and characterization of Escherichia coli RS218-derived islands in the pathogenesis of E. coli meningitis. J. Infect. Dis. 194358-364. [DOI] [PubMed] [Google Scholar]
- 79.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 661192-1206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.