Abstract
The facultative intracellular bacterial pathogen Brucella infects a wide range of warm-blooded land and marine vertebrates and causes brucellosis. Currently, there are nine recognized Brucella species based on host preferences and phenotypic differences. The availability of 10 different genomes consisting of two chromosomes and representing six of the species allowed for a detailed comparison among themselves and relatives in the order Rhizobiales. Phylogenomic analysis of ortholog families shows limited divergence but distinct radiations, producing four clades as follows: Brucella abortus-Brucella melitensis, Brucella suis-Brucella canis, Brucella ovis, and Brucella ceti. In addition, Brucella phylogeny does not appear to reflect the phylogeny of Brucella species' preferred hosts. About 4.6% of protein-coding genes seem to be pseudogenes, which is a relatively large fraction. Only B. suis 1330 appears to have an intact β-ketoadipate pathway, responsible for utilization of plant-derived compounds. In contrast, this pathway in the other species is highly pseudogenized and consistent with the “domino theory” of gene death. There are distinct shared anomalous regions (SARs) found in both chromosomes as the result of horizontal gene transfer unique to Brucella and not shared with its closest relative Ochrobactrum, a soil bacterium, suggesting their acquisition occurred in spite of a predominantly intracellular lifestyle. In particular, SAR 2-5 appears to have been acquired by Brucella after it became intracellular. The SARs contain many genes, including those involved in O-polysaccharide synthesis and type IV secretion, which if mutated or absent significantly affect the ability of Brucella to survive intracellularly in the infected host.
Brucellosis is a disease caused by bacteria of the genus Brucella. This disease is zoonotic and endemic in many areas throughout the world, causing chronic infections with common outcomes being abortion and sterility in infected animals. In humans, it is a severe acute febrile disease, producing focal lesions in bones, joints, the genitourinary tract, and other organs. Complications may include arthritis, sacroiliitis, spondylitis, and central nervous system effects. Brucella can cause abortions in women (as can other bacteria), mostly in the first and second trimesters of pregnancy (21, 27), and men can exhibit epididymo-orchitis (37).
Currently, there are nine recognized species of Brucella, based on host preferences and phenotypic differences. Six classically recognized species are Brucella abortus (cattle), Brucella canis (dogs), Brucella melitensis (sheep and goats), Brucella neotomae (desert wood rats), Brucella ovis (sheep), and Brucella suis (pigs, reindeer, and hares). These six species have been subdivided into 18 biovars based on a panel of culture and biochemical characteristics (41). Recently, three additional species have been identified, namely Brucella microti from voles (49), “Brucella pinnipediae” from pinnipeds, and Brucella ceti from cetaceans (20).
The genome from B. melitensis was the first to be sequenced (16), followed by those from strains of B. suis and B. abortus (9, 11, 24, 44). New genome sequences for B. canis, B. ceti, B. melitensis, and B. suis, as well as the recent release of the B. ovis genome, allow a more detailed look into this group. Furthermore, the increasing number of genomes for Brucella relatives from the order Rhizobiales allows examination of this genus in a broader context.
The main objectives of this study were to examine the phylogeny of Brucella, to examine differences among the different genomes and clades, and to do a detailed comparison between the Brucella genomes and those of their closest relatives in Rhizobiales. Techniques used to examine these differences included structural analysis of the Brucella chromosomes, an in-depth study of areas of possible horizontal transfer into the Brucella genomes, and a comparison of known genes and pseudogenes present in other Brucella genomes that correspond to them.
MATERIALS AND METHODS
Genome sequences and annotation.
Ten different strains from six of the Brucella species were used in this comparison. Three strains with complete genomes (B. canis ATCC 23365, B. melitensis ATCC 23457 [bv. 2], and B. suis ATCC 23445 [bv. 2]) were sequenced by Los Alamos National Labs and the Joint Genome Institute. They also sequenced B. ceti, which has an incomplete genome with seven contigs. All were given their primary annotation by PATRIC, which is the NIAID/PathoSystems Resource Integration Center, a major repository for Brucella genomic data (51). Six additional strains that had been annotated previously (B. abortus S19, B. abortus bv. 1 strain 9-941, B. melitensis 16 M, B. abortus 2308, B. ovis ATCC 25840, and B. suis 1330) were reannotated by PATRIC prior to the comparison to ensure uniformity.
Genome alignment.
Chromosomal DNA sequences from nine Brucella species (all except B. ceti) were aligned using Mauve 2.2.0 (14).
OG identification.
We used OrthoMCL (32) to create groups of orthologous proteins. To create a representative set of ortholog groups (OGs) for the order Rhizobiales, 37 complete or nearly complete genomes were used (Table 1), incorporating 8 of the 11 families in the order.
TABLE 1.
Rhizobiales genomes used in OG analysis
| Genome | Strain | GenBank accession number(s) | Family |
|---|---|---|---|
| Agrobacterium tumefaciens | C58 Cereon | NC_003062, NC_003063, NC_003064, NC_003065 | Rhizobiaceae |
| Azorhizobium caulinodans | ORS 571 | NC_009937 | Xanthobacteraceae |
| Bartonella bacilliformis | KC583 | NC_008783 | Bartonellaceae |
| Bartonella henselae | Houston-1 | NC_005956 | Bartonellaceae |
| Bartonella quintana | Toulouse | NC_005955 | Bartonellaceae |
| Bartonella tribocorum | CIP 105476 | NC_010161NC_010160 | Bartonellaceae |
| Bradyrhizobium | BTAi1 | NC_009475, NC_009485 | Bradyrhizobiaceae |
| Bradyrhizobium | ORS278 | NC_009445 | Bradyrhizobiaceae |
| Bradyrhizobium japonicum | USDA 110 | NC_004463 | Bradyrhizobiaceae |
| Brucella abortus | 9-941 | NC_006932, NC_006933 | Brucellaceae |
| Brucella abortus | S19 | NC_010742, NC_010740 | Brucellaceae |
| Brucella canis | ATCC 23365 | NC_010103, NC_010104 | Brucellaceae |
| Brucella ceti | Cudo | NZ_ACJD00000000 | Brucellaceae |
| Brucella melitensis | 16 M | NC_003317, NC_003318 | Brucellaceae |
| Brucella melitensis | ATCC 23457 | NC_012441, NC_012442 | Brucellaceae |
| Brucella abortus | 2308 | NC_007618, NC_007624 | Brucellaceae |
| Brucella ovis | ATCC 25840 | NC_009504, NC_009505 | Brucellaceae |
| Brucella suis | 1330 | NC_004310, NC_004311 | Brucellaceae |
| Brucella suis | ATCC 23445 | NC_010169, NC_010167 | Brucellaceae |
| Fulvimarina pelagi | HTCC2506 | NZ_AATP00000000 | “Aurantimonadaceae” |
| Hoeflea phototrophica | DFL-43 | NZ_ABIA00000000 | Phyllobacteriaceae |
| Mesorhizobium | BNC1 | NC_008254, NC_008242, NC_008243, NC_008244 | Phyllobacteriaceae |
| Mesorhizobium loti | MAFF303099 | NC_002678, NC_002679, NC_002682 | Phyllobacteriaceae |
| Methylobacterium extorquens | PA1 | NC_010172 | Methylobacteriaceae |
| Nitrobacter hamburgensis | X14 | NC_007959, NC_007960, NC_007961, NC_007964 | Bradyrhizobiaceae |
| Nitrobacter winogradskyi | Nb-255 | NC_007406 | Bradyrhizobiaceae |
| Ochrobactrum anthropi | ATCC 49188 | NC_009667, NC_009668, NC_009669, NC_009670, NC_009671, NC_009672 | Brucellaceae |
| Rhizobium etli | CFN 42 | NC_007761, NC_007762, NC_007763, NC_007764, NC_007765, NC_007766, NC_004041 | Rhizobiaceae |
| Rhizobium leguminosarum bv. viciae | 3841 | NC_008380, NC_008381, NC_008382, NC_008383, NC_008384, NC_008378, NC_008379 | Rhizobiaceae |
| Rhodopseudomonas palustris | BisA53 | NC_008435 | Bradyrhizobiaceae |
| Rhodopseudomonas palustris | BisB18 | NC_007925 | Bradyrhizobiaceae |
| Rhodopseudomonas palustris | BisB5 | NC_007958 | Bradyrhizobiaceae |
| Rhodopseudomonas palustris | CGA009 | NC_005297, NC_005296 | Bradyrhizobiaceae |
| Rhodopseudomonas palustris | HaA2 | NC_007778 | Bradyrhizobiaceae |
| Sinorhizobium medicae | WSM419 | NC_009620, NC_009621, NC_009622, NC_009636 | Rhizobiaceae |
| Sinorhizobium meliloti | 1021 | NC_003047, NC_003037, NC_003078 | Rhizobiaceae |
| Xanthobacter autotrophicus | Py2 | NC_009717, NC_009720 | Xanthobacteraceae |
Pseudogenes.
In this study, a pseudogene is defined as a gene containing one or more in-frame stop codons and/or frameshifts (FS) compared to those of its orthologs. Three methods were used to identify potential pseudogenes within Brucella. The first method was based on the program GenVar, an analytical pipeline used to examine closely related species or strains and identify missed gene calls as well as split genes or indels (62). The second method aligns neighboring pairs of protein predictions using BLASTP (3) against the National Center for Biotechnology Information (NCBI) nonredundant protein database. Neighbors with alignments to the same target sequence with an E value of <10−5 were further evaluated by manual curation. If the pseudogene prediction from either method proved to be correct upon manual examination, the original gene and coding sequence (CDS) features were deleted, and a new gene feature spanning both gene predictions was created and marked with the pseudogene qualifier.
Once a first set of pseudogenes was identified by the above-described two methods, a third method was used to identify additional pseudogenes based on the first set. The DNA sequences of pseudogenes in the first set were first aligned to the bacterial subdivision of NCBI's nonredundant protein database using BLASTX and subjected to cutoffs of 165 bits and an E value of 10−9 or, to ensure alignments to very short pseudogenes are not missed, greater than 85% identity (at the protein level) for 50% of the query length. For each pseudogene, the protein sequence with the highest-scoring alignment (by bit score) was retrieved for use in the next step. These retrieved protein sequences were used as queries in a TBLASTN search of the nine Brucella genomes to identify new genes or pseudogenes by orthology. The resulting alignments were processed to merge overlapping or nearby (within 30 bp) high-scoring segment pairs to form meta-alignments to determine the approximate coordinates of the new (pseudo)gene. To identify its endpoints more precisely and determine the number of FS and in-frame stop (nonsense) codons relative to the functional homolog used as a query, the program estwise from the Wise2.0 package (6) was used to generate an alignment spanning the FS and nonsense features. The command line option “-alg 333” was used to select the simplest FS-tolerant alignment algorithm instead of using the hidden Markov model, with states for intron identification (which is enabled by default and intended for processing eukaryotic sequences). Note that this third method in effect also computes groups of genes related by similarity. While our primary method for computing OGs was OrthoMCL, as noted above, we used the method described here to identify pseudogenes that are “genome specific.” A pseudogene is genome specific if it is the only pseudogene in a gene similarity group containing at least one other member.
Identification of anomalous regions and lateral transfer candidates.
We employed Alien Hunter (AH) (59), a program that identifies regions that may have been laterally transferred. These are regions that have unusual sequence composition in terms of k-mers for various values of k (called interpolated variable order motifs in the terminology used in reference 59). An anomalous region is one whose AH score is above a genome-dependent and automatically calculated threshold that takes into account the sequence composition of the whole genome (termed background composition). AH was run on all 10 Brucella genomes. We called the regions identified by AH anomalous regions.
Because AH has been noted to have low specificity (29), we applied additional filters to the regions detected by AH. Anomalous regions that contained syntenic protein-coding genes in different Brucella genomes as given by OrthoMCL ortholog data and double checked by BLAST2seq (56) were labeled shared anomalous regions (SARs). We then compared the SARs obtained to those of the Ochrobactrum anthropi genome using MUMmer, option PROmer, which compares translated nucleotide sequences in all six frames (28). Using a SAR as the query and the whole genome of O. anthropi as the subject, we computed the coverage of that SAR in the O. anthropi genome by adding up the total length of all matches found by PROmer, regardless of their location in the O. anthropi genome, and dividing the result by the SAR length. Note that this approach is conservative, because matches found by PROmer may be disjointed and therefore may not correspond to a contiguous region in the O. anthropi genome (as would be expected if O. anthropi did in fact share that region). SARs that were absent or less than 50% complete in O. anthropi were selected for further analysis. Finally, SARs were cross-referenced with previously published studies. Several of these interrupt a tRNA gene and were originally named (36) to designate the size of the region in kilobases and the tRNA identity (e.g., 8T is an 8-kb region that interrupts a tRNA that codes for a threonine). SARs are labeled by chromosome and region order within the chromosome (e.g., SAR 1-8 is the eighth shared anomalous region on chromosome 1).
Phylogenetic analysis.
Protein sequences for the 10 Brucella genomes and four outgroup species (Ochrobactrum intermedium [57], O. anthropi ATCC 49188, Bartonella quintana Toulouse, and Mesorhizobium loti MAFF 303099) were clustered by applying OrthoMCL (32) to all-versus-all BLAST data, yielding 2,246 protein families with one and only one representative from each Brucella genome. Each protein family was made representative for the outgroup strains by excluding strains with more than one member in the family, leaving O. anthropi represented in 1,970 families, O. intermedium in 1,924, B. quintana in 851, and M. loti in 1,699. The protein sequences from each family were aligned using MUSCLE (18), and ambiguous portions of the alignment were removed using Gblocks (8). The concatenation of these alignments contained 671,030 amino acid characters, though only 8,004 were Brucella informative (for which at least two Brucella genomes differed from the others or one Brucella genome differed from the others and an outgroup was present). RAxML (53) was used with the PROTGAMMAWAGF model to prepare a maximum likelihood tree and in its quick mode to prepare 100 bootstrap trees.
RESULTS
General features of the genomes.
All nine Brucella genomes studied have two circular chromosomes. Chromosome 1 is the larger chromosome, with a median length of 2.1 Mb, and chromosome 2 has a median length of 1.2 Mb. Both have similar G+C content, averaging 57.1% for chromosome 1 and 57.3% for chromosome 2. The total number of genes per genome (about 3,460) is very similar among the nine complete genomes studied, as is the number of protein-coding genes (about 3,180). These results are summarized in Table 2 on a per-genome and per-chromosome basis.
TABLE 2.
Summary of annotation information for 10 Brucella genomes
| Characteristica | Annotation information for:
|
Total characteristics
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
B. abortus strain
|
B. canis | B. cetib |
B. melitensis strain
|
B. ovis |
B. suis strain
|
Minimum | Maximum | Average | Median | |||||
| 9-941 | 2308 | S19 | 16 M | 23457 | 1330 | 23445 | ||||||||
| Chromosome 1 | ||||||||||||||
| Length | 2,124 | 2,121 | 2,122 | 2,105 | 1,905 | 2,117 | 2,125 | 2,111 | 2,107 | 1,923 | 1,923 | 2,125 | 2,095 | 2,117 |
| G+C% | 57.2 | 57.2 | 57.2 | 57.2 | NAc | 57.2 | 57.2 | 57.2 | 57.2 | 57.1 | 57.1 | 57.2 | 57.2 | 57.2 |
| Genes | 2,310 | 2,343 | 2,263 | 2,199 | NA | 2,268 | 2,236 | 2,289 | 2,276 | 2,006 | 2,006 | 2,343 | 2,243 | 2,268 |
| CDSs | 2,127 | 2,098 | 2,090 | 2,060 | NA | 2,049 | 2,078 | 2,093 | 2,126 | 1,876 | 1,876 | 2,127 | 2,066 | 2,090 |
| Pseudogenes | 81 | 81 | 79 | 83 | NA | 66 | 68 | 134 | 80 | 83 | 66 | 134 | 84 | 81 |
| P'gene fraction | 3.70% | 3.70% | 3.60% | 3.90% | NA | 3.10% | 3.20% | 6.00% | 3.60% | 4.20% | 3.10% | 6.00% | 3.90% | 3.70% |
| GS pseudogenes | 1 | 2 | 2 | 13 | NA | 7 | 4 | 59 | 8 | 18 | 1 | 59 | 13 | 7 |
| Chromosome 2 | ||||||||||||||
| Length | 1,162 | 1,156 | 1,161 | 1,206 | 1,483 | 1,177 | 1,185 | 1,164 | 1,207 | 1,400 | 1,156 | 1,400 | 1,202 | 1,177 |
| G+C% | 57.3 | 57.4 | 57.3 | 57.3 | NA | 57.3 | 57.4 | 57.2 | 57.3 | 57.3 | 57.2 | 57.4 | 57.3 | 57.3 |
| Genes | 1,222 | 1,232 | 1,171 | 1,224 | NA | 1,191 | 1,189 | 1,195 | 1,224 | 1,417 | 1,171 | 1,417 | 1,229 | 1,222 |
| CDSs | 1,108 | 1,083 | 1,063 | 1,133 | NA | 1,056 | 1,077 | 1,053 | 1,139 | 1,300 | 1,053 | 1,300 | 1,112 | 1,083 |
| Pseudogenes | 62 | 59 | 60 | 69 | NA | 59 | 65 | 112 | 70 | 85 | 59 | 112 | 71 | 65 |
| P'gene fraction | 5.30% | 5.20% | 5.30% | 5.70% | NA | 5.30% | 5.70% | 9.60% | 5.80% | 6.10% | 5.20% | 9.60% | 6.00% | 5.70% |
| GS pseudogenes | 0 | 1 | 3 | 9 | NA | 8 | 13 | 48 | 10 | 16 | 0 | 48 | 12 | 9 |
| Genome | ||||||||||||||
| Length | 3,286 | 3,277 | 3,283 | 3,311 | 3,388 | 3,294 | 3,310 | 3,275 | 3,314 | 3,323 | 3,275 | 3,323 | 3,297 | 3,294 |
| G+C% | 57.2 | 57.2 | 57.2 | 57.2 | 57.2 | 57.2 | 57.2 | 57.2 | 57.3 | 57.2 | 57.2 | 57.3 | 57.2 | 57.2 |
| Genes | 3,459 | 3,425 | 3,484 | 3,500 | 3,501 | 3,459 | 3,425 | 3,484 | 3,500 | 3,423 | 3,423 | 3,500 | 3,462 | 3,459 |
| CDSs | 3,235 | 3,181 | 3,153 | 3,193 | 3,293 | 3,105 | 3,155 | 3,146 | 3,265 | 3,176 | 3,105 | 3,265 | 3,179 | 3,176 |
| Pseudogenes | 143 | 140 | 139 | 152 | NA | 125 | 133 | 246 | 150 | 168 | 125 | 246 | 155 | 143 |
| P'gene fraction | 4.20% | 4.20% | 4.20% | 4.50% | NA | 3.90% | 4.00% | 7.30% | 4.40% | 5.00% | 3.90% | 7.30% | 4.60% | 4.20% |
| GS pseudogenes | 1 | 3 | 5 | 22 | NA | 15 | 17 | 107 | 18 | 34 | 1 | 107 | 25 | 17 |
| OGs | 3,118 | 3,068 | 3,024 | 3,086 | 3,125 | 3,089 | 3,039 | 3,098 | 3,117 | 3,060 | 3,024 | 3,118 | 3,078 | 3,086 |
P'gene fraction, pseudogene fraction. P'gene fractions are calculated as follows: pseudogene count/(pseudogene count + CDS count). GS pseudogenes, genome-specific pseudogenes. The number of GS pseudogenes indicates the number of cases in which the genome of interest contains a nonfunctional gene, but the gene is functional in all other genomes.
Brucella ceti assembly is incomplete; thus, all numbers are approximate.
NA, not applicable.
Genome alignment.
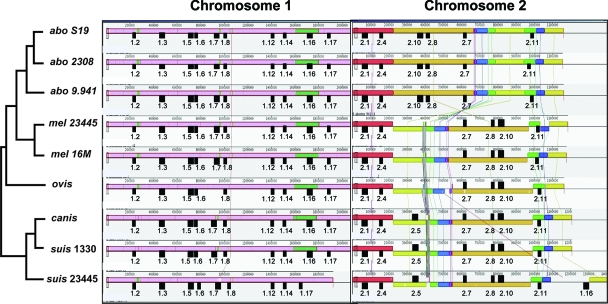
Multiple replicon alignments were done for 9 of the 10 genomes. (B. ceti was excluded because it is an unfinished genome.) Chromosome 1 is similarly arranged among all nine genomes, with the only major difference being the B. suis ATCC 23445 genome (Fig. 1). Examination of both chromosomes of this species indicated that a 210-kb segment of chromosome 1 has been translocated to chromosome 2. Chromosome 2 appears to be more plastic than chromosome 1, with more internal rearrangements. A segment of approximately 700 kb in chromosome 2 is a shared inversion among the three B. abortus genomes (Fig. 1), with respect to the others.
FIG. 1.
Mauve alignment of both chromosomes from the nine complete Brucella genomes. A phylogenetic map of the strains derived from the tree shown in Fig. 2 (topology only, not branch lengths) is on the left side (abo, B. abortus; mel, B. melitensis; ovis, B. ovis; canis, B. canis; suis, B. suis). SARs of interest are noted by filled boxes, with the names of those regions directly below them.
Phylogenetic analysis.
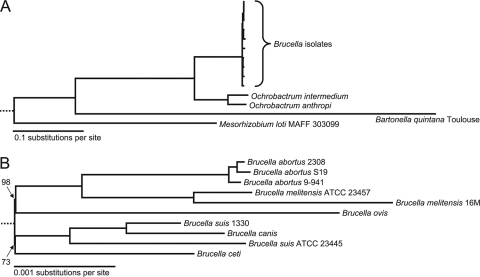
The results of a maximum likelihood phylogenetic analysis of the 10 Brucella strains plus four outgroup species are shown in Fig. 2. This tree sorts the Brucella genomes studied here into four clades, as follows: (i) the B. melitensis-B. abortus clade; (ii) the B. ovis clade; (iii) the B. suis-B. canis clade; and (iv) the B. ceti clade. Each node received 100% bootstrap support except for two extremely short internal branches. Although the tree is nominally bifurcating, the shortness and suboptimal support of those two branches suggest caution in assigning a strict evolutionary branching order to the four Brucella clades; they appear to have radiated explosively. The generated tree (Fig. 2A) also shows, as expected, that Ochrobactrum is the closest relative to Brucella (48).
FIG. 2.
Phylogenetic trees of 10 Brucella genomes with outgroups (A) and without outgroups (B). The maximum likelihood tree is based on a concatenated alignment of 2,246 protein families. (A) Full tree with outgroup species. (B) Brucella portion, only at a smaller scale. All nodes received 100% bootstrap support except the two very short ones indicated in panel B.
Anomalous regions, OGs, and lateral transfer analysis.
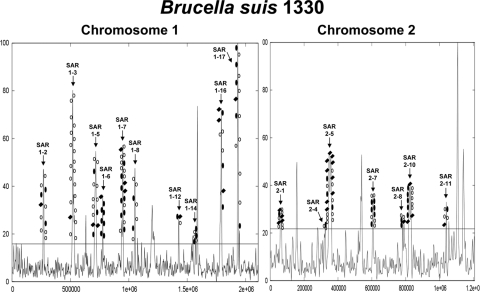
We identified an average of 40 anomalous regions in the Brucella strains (range, 32 to 51 regions). Chromosome 1 had an average of 17.4 regions (range, 13 to 21 regions), and chromosome 2 had an average of 23 (range, 14 to 38 regions). This variation is explained in part by variation in the genome-specific threshold score determined by AH, which was the main reason that led us to adopt the concept of the SAR. Seventeen SARs were absent or nearly absent in O. anthropi and were examined further (Table 3). These SARs ranged in size from 2 to 19 kb, with SAR 1-12 being the smallest and SAR 1-17 being the largest (see Table S1 in the supplemental material). Four of the 17 SARs showed the hallmark pattern of genomic islands, flanked on one side by an intact tRNA gene and on the other side by a fragment of that tRNA gene. Three of these, 8T (SAR 1-2), 15G (SAR 1-7), and 2I (SAR 1-12), have been described previously in chromosome 1 (36), and we identified a novel genomic island; SAR 2-10 is found in chromosome 2 and is 14 kb in length. This island is integrated into a tRNA-Thr and contains a type I restriction-modification system. For several additional SARs with tRNA gene neighbors, no tRNA fragment was identified at the other end (SARs 1-3, 1-5, 1-6, 1-8, 1-14, 1-16, 2-7, and 2-11). These may be older genomic islands that have lost the tRNA fragment, or the association with a tRNA gene may be accidental. It was more difficult to assign the endpoints of these SARs; the end of the sequence of the last shared ortholog identified by AH was used. Complete information on the 17 SARs examined, including the genes carried by them, is provided (see Table S1 in the supplemental material).
TABLE 3.
Seventeen specifically examined SARs and their presence/absence among the Brucella genomes and Ochrobactrum anthropia
| SAR | Other name | tRNA | Size (kb) | Presence/absence of SARs in:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
B. suis strain
|
B. canis | B. ceti | B. ovis |
B. melitensis strain
|
B. abortus strain
|
O. anthropi (%) | ||||||||
| 23445 | 1330 | 16 M | 23457 | 9941 | 2308 | S19 | ||||||||
| Chromosome 1 | ||||||||||||||
| SAR 1-2 | 8T | tRNA-Thr | 8,193 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 28 |
| SAR 1-3 | GI-1 | tRNA-Thr | 18,298 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 15 |
| SAR 1-5 | tRNA-Arg | 13,859 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 14 | |
| SAR 1-6 | tRNA-Val and tRNA-Asp | 4,038 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 49 | |
| SAR 1-7 | 15G | tRNA-Gly | 15,116 | √ | √ | √ | √ | abs | √ | √ | √ | √ | √ | 35 |
| SAR 1-8 | tRNA-Leu and tRNA-Leu | 7,213 | √ | √ | √ | √ | part | √ | √ | √ | √ | √ | 24 | |
| SAR 1-12 | 2I | tRNA-Ile | 2,007 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 0 |
| SAR 1-14 | tRNA-Gly | 4,199 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 37 | |
| SAR 1-16 | tRNA-Ser | 10,483 | √ | √ | √ | √ | part | √ | √ | √ | √ | √ | 36 | |
| SAR 1-17 | 19,446 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 44 | ||
| Chromosome 2 | ||||||||||||||
| SAR 2-1 | 11,103 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 15 | ||
| SAR 2-4 | 1,852 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 14 | ||
| SAR 2-5 | IncP | 19,305 | √ | √ | √ | √ | abs | abs | abs | abs | abs | abs | 4 | |
| SAR 2-7 | tRNA-Cys | 4,437 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 17 | |
| SAR 2-8 | 4,856 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 6 | ||
| SAR 2-10 | 14T | tRNA-Thr | 14,312 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 10 |
| SAR 2-11 | tRNA-Leu and tRNA-Ser | 3,864 | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | 39 | |
In the Brucella genomes, if the SAR is completely present, this is noted by a check mark; if it is only partially present, this is noted by part; and if it is completely absent, this is noted by abs. All of these SARs are at best only partially present in O. anthropi, and the last column provides the estimated coverage.
The translocation of a 210-kb segment from chromosome 1 to chromosome 2 in B. suis ATCC 23445 also moved SAR 1-16, and the inversion on chromosome 2 in the B. abortus genomes inverted SARs 2-8 and 2-10 (Fig. 1). Other SARs of interest that were not near tRNA genes include 1-17, 2-1, 2-4, 2-8, and the previously described IncP island (30), corresponding to SAR 2-5. Interestingly, SAR 1-2 (8T) contains a three-gene segment that is also found in SAR 1-8, including a resolvase family site-specific recombinase. Either these genes entered the Brucella twice independently or there was an insertion into one of the sites from either SAR 1-2 or SAR 1-8. SAR 2-5 is also interesting, as it had been noted previously that this region, the IncP island, was found only in B. suis, B. canis, B. neotomae, and in some of the marine strains (30). In this study, we found SAR 2-5 in B. ceti, but as noted previously, it is missing from B. ovis and from all the B. abortus and B. melitensis genomes.
SAR 1-7, first identified as 15G by Mantri and Williams (36) and later examined experimentally (45, 46), contains 15 genes, 2 of which (wboA and wboB) are of particular interest, since they help determine the smooth phenotype (see below).
We obtained 15,986 OGs from 37 Rhizobiales genomes (this number does not include singleton proteins that failed to group with others). Within the genus Brucella, there were 747 OGs that contained any combination of the 10 Brucella genomes but none of the other Rhizobiales genomes. Of these, 140 OGs had at least one representative from each of the 10 Brucella genomes (see Table S2 in the supplemental material). Using this set of 747 OGs, we identified a region that is found in all the Brucella genomes except for the three B. abortus genomes. This 23-kb segment contains a number of important genes, including those encoding glycosyl transferase and glycerol kinase (Table 4); this region was not identified as anomalous. It should be noted that the glycosyl transferase and glycerol kinases are the second copies of these genes. The B. abortus genomes have only a single copy of each gene.
TABLE 4.
Genes found in a 23-kb segment in Brucella suis 1330 and in all others, except for the three B. abortus genomes in which all genes are missinga
| RefSeq ID | PATRIC ID | Start (kb no.) | End (kb no.) | Size (bp) | Strand | Gene symbol | Description |
|---|---|---|---|---|---|---|---|
| BRA0418 | VBI0007BS2_0414 | 402846 | 403826 | 981 | − | GDP-L-fucose synthase 1 | |
| BRA0419 | VBI0007BS2_0415 | 403810 | 404880 | 1,071 | − | gmd | GDP-mannose 4,6-dehydratase |
| BRA0421 | VBI0007BS2_0417 | 406415 | 407650 | 1,236 | + | Hypothetical protein | |
| BRA0422 | VBI0007BS2_0418 | 407647 | 408843 | 1,197 | + | Hypothetical protein | |
| BRA0423 | VBI0007BS2_0419 | 408914 | 409636 | 723 | − | Hypothetical protein | |
| BRA0424 | VBI0007BS2_0420 | 410033 | 410647 | 615 | − | nodL | Nodulation protein L |
| BRA0426 | VBI0007BS2_0422 | 411918 | 412535 | 618 | − | Hypothetical protein | |
| BRA0427 | VBI0007BS2_0423 | 412532 | 413413 | 882 | − | waaE | LPS core biosynthesis glycosyl transferase waaE |
| BRA0428 | VBI0007BS2_0424 | 413410 | 414537 | 1,128 | − | rfe | Putative undecaprenyl-phosphate alpha-N-acetylglucosaminyl 1-phosphate transferase |
| BRA0429 | VBI0007BS2_0425 | 414830 | 416323 | 1,494 | + | Hypothetical protein | |
| BRA0430 | VBI0007BS2_0426 | 416339 | 417352 | 1,014 | + | hyaD | Hyaluronan synthase |
| BRA0431 | VBI0007BS2_0427 | 417308 | 418549 | 1,242 | − | Hypothetical protein | |
| BRA0432 | VBI0007BS2_0428 | 418816 | 420045 | 1,230 | + | Hypothetical protein | |
| BRA0433 | VBI0007BS2_0429 | 420083 | 421444 | 1,362 | + | hemL | Glutamate-1-semialdehyde 2,1-aminomutase |
| BRA0434 | VBI0007BS2_0430 | 421423 | 422757 | 1,335 | + | Hypothetical protein | |
| BRA0435 | VBI0007BS2_0431 | 422878 | 423939 | 1,062 | + | exoB | UDP-glucose 4-epimerase |
| BRA0436 | VBI0007BS2_0432 | 423978 | 425291 | 1,314 | + | Hypothetical protein | |
| BRA0437 | VBI0007BS2_0433 | 425254 | 425778 | 525 | − | rfbC | dTDP-4-dehydrorhamnose 3,5-epimerase |
| BRA0438 | VBI0007BS2_0434 | 426099 | 427400 | 1,302 | + | Hypothetical protein | |
| BRA0439 | VBI0007BS2_0435 | 427403 | 428212 | 810 | + | mpg1 | Mannose-1-phosphate guanyltransferase |
RefSeq, reference sequence; ID, identification.
A single protein representative from each of the 747 OGs was used to query the NCBI nonredundant protein database. Of these, 688 OGs had no BLASTP hits to any genome other than Brucella, accounting for 21.5% of all Brucella proteins (see Table S3 in the supplemental material). The majority of these Brucella-specific OGs are annotated as hypothetical proteins; over 50% have either 9 or all 10 of the genomes represented. There were 59 OGs with BLASTP hits (E value cutoff, >10−10) for Brucella and for genomes outside of Rhizobiales (see Table S4 in the supplemental material), indicating that the proteins with the nearest homology are not present in the closest relatives of Brucella; this is a small percentage (1.7%) of the Brucella proteins.
Notable among these 59 OGs are the components of the type IV secretion system (maps to SAR 2-1), tra genes (map to SAR 2-5), and the wbk gene cluster (maps to SAR 1-3), which was previously identified (22).
In a comparison of the AH and ortholog/BLASTP data, the observation that one of the regions contained a housekeeping gene led to the identification of SAR 1-17 as a composite. It contains a five-gene region shared with O. anthropi but flanked on both sides by genes unique to Brucella. Figure 3 shows an annotation of the 17 SARs in the B. suis 1330 genome.
FIG. 3.
AH analysis of B. suis 1330 chromosomes. The 17 shared SARs are annotated in this plot. Genes that were shared among the majority of the OGs are depicted on these peaks, with open circles indicating genes that are present in Brucella and among other members of Rhizobiales. Filled circles indicate genes that are found only among Brucella and share no homology outside this genus. Filled diamonds represent genes that are present in Brucella, absent among other Rhizobiales, but have significant BLASTP hits to genomes that are not members of the Rhizobiales order. The x axis represents the length of the chromosome, and the y axis represents the score range provided by the AH program.
Pseudogenes.
A total of 1,396 pseudogenes were identified (this analysis excludes the unfinished B. ceti genome). Of these, 222 were found to be genome specific. Many such genome-specific pseudogenes may simply be the result of a sequencing error. The other identified pseudogenes are members of 522 OGs (see Table S5 in the supplemental material). The ratio of pseudogenes to total genes carried by a genome (the pseudogene fraction) was used as a benchmark for comparison between organisms. Values ranged from a low of 3.9% for B. melitensis 16 M to a high of 7.3% for B. ovis. The average value for the nine complete Brucella genomes was 4.6%. The highest number of genome-specific pseudogenes is found in B. ovis, with 107 (Table 2). The next highest number is found in B. suis ATCC 23445, which has 34, followed by B. canis, with 22. Pseudogene fractions were also calculated on a per-chromosome basis. Chromosome 2 had a higher percentage of pseudogenes than chromosome 1 for all nine genomes studied; on average, the pseudogene fraction was 3.9% for chromosome 1 and 6.0% for chromosome 2.
β-Ketoadipate pathway.
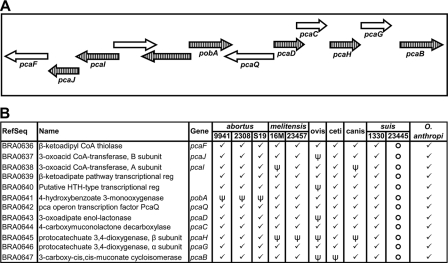
In the initial analysis of the B. suis 1330 genome sequence, Paulsen et al. (44) noted an unexpected capacity of this organism to use plant-derived compounds as an energy source. The β-ketoadipate pathway takes two aromatic compounds, protocatechuate and catechol, which are produced by the degradation of plant-derived molecules, and metabolizes them to intermediates that can enter the tricarboxylic acid cycle (34). There are 12 protein-coding genes that have been identified as being part of this pathway in B. suis 1330 (44); all of them are found on chromosome 2. In the case of Agrobacterium tumefaciens C58, the enzymes involved in this pathway are organized into two distinct operons (43); Brucella seems to have a similar arrangement, as do both Ochrobactrum genomes. Examination of all 10 Brucella genomes showed that at least 1 of the 12 genes carried by every genome except B. suis 1330 has become a pseudogene and that both of these operons are completely missing in B. suis ATCC 23445 (Fig. 4).
FIG. 4.
Pseudogenization of the β-ketoadipate pathway among the 10 different Brucella genomes and in Ochrobactrum anthropi in illustrated form (A) and tabular form (B). (A) Genes involved in this pathway are in two operons on opposite strands, and the individual genes are represented by arrows. Gene symbols, where available, are provided below the arrows. Hatched arrows indicate genes that are pseudogenized in one or more genomes. (B) Table showing the identification, name, gene symbol, and presence, absence, or pseudogenization of individual genes among the genomes. Checkmarks indicate a normal gene. Open circles denote the absence of this gene, and ψ indicates an apparent pseudogene.
DISCUSSION
The 10 different Brucella genomes examined here are quite similar in genome size and the numbers of genes and proteins. They are also similar in the structural organization of the chromosomes, with the exceptions being a 210-kb translocation seen in B. suis ATCC 23445 and a 700-kb inversion in chromosome 2 shared by the B. abortus genomes (Fig. 1).
The combined phylogenomic analysis of 2,377 ortholog families shows that the depth of divergence for these 10 Brucella strains is quite shallow (Fig. 2B). Despite this low level of divergence, with few characters differing among the genomes, the branching order seems to be clear and well supported, as reflected by support values. The major structure is a radiation producing a B. abortus-B. melitensis clade, a B. suis-B. canis clade, and B. ovis and B. ceti clades. A recent phylogenetic analysis (7) shows the same four subgroups of Brucella observed here, but our use of an outgroup further shows that these are four clades that radiated explosively. The B. abortus-B. melitensis clade segregated into two branches, one containing the B. abortus genomes and the other containing the B. melitensis genomes. B. canis nests within the B. suis clade, suggesting that there may have been a host switch. Genome sequences for B. neotomae and B. pinnipediae are not currently available, but previously presented evidence (7) indicates that a similar host switch may have occurred in these two species.
Assuming current knowledge of host preference is accurate, we can ask whether Brucella phylogeny reflects the phylogeny of their hosts. The mammalian taxa that have been identified as the preferred Brucella hosts belong to three distinct groups, all at the level of order in mammals. Neotoma is a genus of cricetid rodent found in the order Rodentia. The genus Canis (dogs, wolves, and coyotes) and the family Phocidae (seals) are in the order Carnivora. Bos (cattle and oxen), Ovis (sheep), Capra (goats), Sus (pigs), and the cetacean group (whales and dolphins) are all united in Cetartiodactyla. Humans (Primates) have also been infected but are not preferred hosts. These three mammalian orders representing the hosts are all well separated phylogenetically (42). The host and pathogen phylogenies are distinct and not similar. Although our phylogenetic data closely reflect the data found previously (7), our conclusions differ. The phylogeny of the Brucella isolates does not match that of their nominal mammalian hosts. This is especially clear from the inclusion of B. canis in our study. Considering the fact that most of the Brucella isolates have been identified in cetartiodactylid hosts, one could speculate that the ancestor of Brucella species infected a member of early cetartiodactylids and radiated within this group, with host switches to Carnivora and Rodentia occurring later.
The species concept in bacteria is a subject of debate (10, 52), as is the definition of different species within Brucella (40). The high degree of similarity of all these genomes, in comparison to other bacterial groups, suggests a close phylogenetic relationship. However, clear differences in host preference might still justify the separate species designations as they presently exist. For example, cattle have been described as the natural or primary hosts for B. abortus, and yet it has also been found in horses, pigs, sheep, goats, Bactrian camels, dromedary camels, water buffalo, yaks, elk, dogs (12), and humans (5, 55). It has also been isolated from rodents on occasion, although it was noted that these infections seem to be from areas where there was a large number of infected cattle (15). This list alone represents five different orders of mammalian hosts. A survey of the literature shows that the host range exhibited by B. abortus strains also extends to different degrees in the other Brucella clades. However, the isolation frequencies of different Brucella species from infected hosts are consistent with some type of host preference (58).
Genome reduction, or reductive evolution, involves gene loss through mutational inactivation and deletion (4, 19). It has been noted in a number of intracellular pathogenic bacteria, including Rickettsia prowazekii (4), Mycobacterium leprae (19), Shigella flexneri, and Salmonella enterica serovar Typhi (13). All of these bacteria are obligate intracellular pathogens, whereas Brucella is a facultative intracellular pathogen that can survive outside the host under certain conditions (12). Are the Brucella genomes undergoing reductive evolution? Based on genome size alone, the answer seems to be yes. Brucella genomes are all similar in size, with an average size of 3.29 Mb. Their nearest sequenced relatives are O. anthropi (5.22 Mb) and O. intermedium (4.6 Mb), which are both markedly larger. Pseudogene fractions can also be an indication of a genome reduction process. Excluding genome-specific pseudogenes, the average fraction determined here was 4.6%. This is low compared to the 50% estimate for Mycobacterium leprae (19), 24% for Rickettsia prowazekii (4), 15% for Shigella flexneri (13), 14% for Bartonella quintana (2), and 9% for Bartonella henselae (2). Of these, only the species of the Bartonella genus are in the order Rhizobiales. On the other hand, in three free-living Agrobacterium species, also in the order Rhizobiales, the fraction is less than 2% (50). Because the pseudogene fractions in these other studies were obtained using different methodologies, it is difficult to compare these numbers. However, using the general estimate that bacterial genomes have between 1 and 5% pseudogenes (33), the 4.6% fraction observed in Brucella can be considered relatively high and suggestive of genome degradation. Moreover, we did note more pseudogenes on chromosome 2 than on chromosome 1. Together with the higher degree of rearrangements observed on chromosome 2, this supports the conclusion that chromosome 2 is more dynamic, perhaps owing to its hypothesized origin as a plasmid (50).
The presence of many pseudogenes in the β-ketoadipate pathway is striking and reminiscent of the proposed “domino theory” of gene death (13), where after a crucial gene within a complex pathway becomes nonfunctional, a mass gene extinction is triggered. The B. suis genome is anomalous in retaining this gene cluster intact, as the cluster is entirely absent in the B. suis ATCC 23445 genome, and one or more of its genes have become pseudogenes in other genomes. It is likely that in its adaptation to an intracellular milieu, Brucella no longer requires this pathway that allows soil bacteria to break down plant compounds. We suspect that the preservation of this gene cluster in B. suis 1330 is anomalous and that over time it will succumb to pseudogenization; however, it is also possible that this particular strain (unlike other members of the B. canis-B. suis clade) makes use of these genes during periods of existence outside an animal host.
From examining the regions of potential lateral transfer, we note that many regions are unique to Brucella and not shared with Ochrobactrum. It is likely that these regions (Table 3; see also Table S1 in the supplemental material) entered Brucella after diverging from the ancestor it shared with Ochrobactrum, indicating that lateral transfer does happen despite intracellular preferences. Dobrindt et al. (17) suggest that horizontal transmission is more likely to occur in niches that contain diverse bacterial species and not as likely to occur in sparsely populated environments, which include intracellular niches like the host macrophage, the ultimate destination of Brucella (35). Of course, it is possible that these regions entered the genome at some point before Brucella committed to an intracellular preference. But when one considers that the journey to the macrophage takes Brucella through a complex series of environments that are inhabited by a wide variety of organisms with which they might interact, it seems plausible that Brucella has the opportunity to experience lateral transfer. The mammalian gut has been recognized as one of the most densely populated ecosystems on earth (38), and it has been documented that one of the most common means of transmission of Brucella involves ingestion of forage or water contaminated with genital discharge (54) or ingestion of raw milk or milk products (47). Passage through the gut would provide ample opportunity for different species of bacteria to interact, and it is plausible that Brucella experienced lateral transfer in this environment. In addition, Brucella might also interact with bacteria in the soil on which the blood, tissues, and aborted fetus of the host lies. Crawford et al. (12) report that Brucella can survive for up to 66 days in moist soil and up to 185 days in cold soil.
Our study contains strong indications that Brucella has acquired genes by lateral transfer. In particular, SAR 2-5, the IncP island, appears to have entered Brucella after it diverged from Ochrobactrum and after the individual species began to separate. This SAR contains the Tra proteins, known to be a type IV secretion system (31), and it is found in B. suis, B. canis, and B. neotomae (30) and is here identified in B. ceti. The fact that the phylogenetic tree places the B. suis-B. canis clade and the B. ceti clade cluster together makes it seem likely that SAR 2-5 was acquired by their common ancestor. A complete genome from B. neotomae is not yet available, but a previous study shows that this species is phylogenetically close to B. suis and the marine Brucella spp. Thus, we hypothesize that there was a common ancestor to these three clades and that SAR 2-5 was laterally transferred into it. The fact that it is in the same location in the genomes studied here gives further weight to a single, ancestral acquisition (Fig. 1). Because it is shared only among some of the Brucella genomes, it could be argued that it was acquired after the ancestor had begun living intracellularly, as it is unlikely that this type of lifestyle developed twice independently. However, it is also possible that the ancestor that gave rise to the B. ovis and the B. melitensis-B. abortus clades lost this region or that each of the clades lost it independently.
Some of the genes indicated as having been acquired by lateral transfer play an important role in the survival of this pathogen in its host. These include the enzymes involved in producing the smooth phenotype in Brucella (26, 45). Lipopolysaccharide (LPS) is the major structural component of the outer membrane of gram-negative bacteria. It is composed of a lipid core, a core oligosaccharide, and a distal O-polysaccharide (O-PS) side chain (22). A phenotypic characteristic used to distinguish between Brucella species is the presence of the O-PS. Isolates of B. abortus, B. suis, and B. melitensis have a smooth morphology with the O-PS intact, while B. canis and B. ovis are rough, as they have the lipid core and the core oligosaccharide but lack O-PS. The O-PS is a major contributor to the antigenic variation of the bacterial envelope as well as the ability of Brucella to survive in macrophages (26). Several studies have indicated specific genes as being important for the development of the smooth phenotype in Brucella (1, 22, 23, 25, 39, 61). Recently, Gonzalez et al. (23) looked at 19 genes that had been indicated as being important in producing smoothness and found that disruption of 13 genes (wboA, wboB, wa**, wbhE, manB, wbkA, gmd, per, wzm, wbkF, wbkD, prm, and manBcore) resulted in a rough phenotype in B. melitensis, with an additional 6 genes indentified as playing roles that were not fully determined. Rajashekara et al. (45) demonstrated that mutations of two genes, BMEI0997 (wboB) and BMEI0998 (wboA), resulted in a rough phenotype. Furthermore, they showed that BMEI0999, a hypothetical protein whose function is unknown, was necessary to restore a smooth LPS in rough strains. However, we have found that other smooth strains (B. abortus 2308 and B. melitensis ATCC 23457) are completely missing this hypothetical gene.
There are two well-established species of Brucella, B. canis and B. ovis, that are naturally rough and yet fully infective, and these 19 LPS-associated genes were specifically examined in these two genomes. B. ovis is missing two genes, wboA and wboB, that encode enzymes that polymerize N-formylperosamine (23), and without them, B. ovis is unable to complete the distal O-PS. Both of these genes reside in SAR 1-7 (15G) of B. suis 1330; their loss in B. ovis has been previously reported (46, 60). The B. ovis genome also has a truncated wzt; this gene encodes a protein that functions as a part of an ABC transporter, with its partner encoded by wzm. This specific enzyme (Wzt) is found in SAR 1-3 of B. suis 1330 and most likely entered Brucella by lateral transfer. This enzyme could be functional even if truncated. However, it could also indicate that the genes involved in LPS synthesis in SAR 1-3 are in a process of decay because the pathway is no longer complete in B. ovis. Only direct experimental evidence will determine if these genes are functional in B. ovis.
B. canis has truncations in 2 of the 19 LPS synthesis genes, wbkF, an undecaprenyl-glycosyltransferase, and wbkD, an epimerase/dehydratase. A truncated gene could still be functional, but the fact that B. canis is rough and that all other genes appear normal indicates that at least one of these genes is responsible for producing the rough phenotype. It is interesting that the rough phenotype results from different mutations in these two genomes, as follows: B. canis with mutations in wbkF and wbkD and B. ovis missing wboA and wboB and having a truncated wzt. It appears that roughness independently developed twice.
All known isolates of B. ceti are smooth (A. Whatmore, personal communication), and yet when the enzymes involved in LPS synthesis were examined in this species, manB, a phosphomannomutase whose function has not yet been determined (23), was found to be truncated due to a naturally occurring transposon insertion. Apparently, this does not affect the smooth phenotype of this organism. However, a rough phenotype was produced in B. melitensis when manB was experimentally mutated by a transposon (23).
Many of the 19 genes considered necessary for complete LPS synthesis are in SARs. Eighteen genes are located on chromosome 1, and one is found on chromosome 2. Of the genes found on chromosome 1, 12 are located on SAR 1-3 (Fig. 3), with one additional gene, wbkD (VBI0007BS1_0525), directly adjacent to this region. Two additional genes are found in SAR 1-7 (Fig. 3). These SARs are not adjacent on chromosome 1, with 402 kb between them, making it likely that these SARs represent genomic islands laterally transferred into Brucella genomes in separate events.
Genes of particular interest that we hypothesize to have entered the genome horizontally include the type IV secretion system, the tra genes, and the enzymes responsible for LPS synthesis that give Brucella a smooth phenotype. Mutations or the absence of these LPS genes is responsible for the rough phenotype of both B. ovis and B. canis. All these observations lead us to believe that Brucella, despite its preference for an intracellular milieu (e.g., phagocytic cells), has the ability and opportunity to interact with other bacteria in their environment and has acquired useful genes that facilitate its intracellular lifestyle.
Supplementary Material
Acknowledgments
We thank Sohan Nagrani (VBI) for his careful analysis of the literature and genes previously described in this genus.
This work is funded through NIAID contract HHSN266200400035C to Bruno Sobral. Funding to pay the Open Access publication charges for this article was provided by NIAID contract HHSN266200400035C to Bruno Sobral. We also thank Dennis Dean (Fralin Life Science Institute, Virginia Tech) for providing financial support to aid in resolving the number of contigs in the B. ceti genome.
Footnotes
Published ahead of print on 3 April 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, C. A., L. G. Adams, and T. A. Ficht. 1998. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 661008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canbäck, A. S. Eriksson, A. K. Näslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 1019716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson, S. G., A. S. Eriksson, A. K. Naslund, M. S. Andersen, and C. G. Kurland. 1996. The Rickettsia prowazekii genome: a random sequence analysis. Microb. Comp. Genomics 1293-315. [PubMed] [Google Scholar]
- 5.Arimi, S. M., E. Koroti, E. K. Kang'ethe, A. O. Omore, and J. J. McDermott. 2005. Risk of infection with Brucella abortus and Escherichia coli O157:H7 associated with marketing of unpasteurized milk in Kenya. Acta Trop. 961-8. [DOI] [PubMed] [Google Scholar]
- 6.Birney, E., M. Clamp, and R. Durbin. 2004. GeneWise and Genomewise. Genome Res. 14988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourg, G., D. O'Callaghan, and M. L. Boschiroli. 2007. The genomic structure of Brucella strains isolated from marine mammals gives clues to evolutionary history within the genus. Vet. Microbiol. 125375-380. [DOI] [PubMed] [Google Scholar]
- 8.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17540-552. [DOI] [PubMed] [Google Scholar]
- 9.Chain, P. S., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 738353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohan, F. M. 2002. Sexual isolation and speciation in bacteria. Genetica 116359-370. [PubMed] [Google Scholar]
- 11.Crasta, O. R., O. Folkerts, Z. Fei, S. P. Mane, C. Evans, S. Martino-Catt, B. Bricker, G. Yu, L. Du, and B. W. Sobral. 2008. Genome sequence of Brucella abortus vaccine strain S19 compared to virulent strains yields candidate virulence genes. PLoS ONE 3e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford, R. P., J. D. Huber, and B. S. Adams. 1990. Epidemiology and surveillance, p. 131-151. In N. Klaus and D. Robert (ed.), Animal brucellosis. CRC Press, Boca Raton, FL.
- 13.Dagan, T., R. Blekhman, and D. Graur. 2006. The “domino theory” of gene death: gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol. Biol. Evol. 23310-316. [DOI] [PubMed] [Google Scholar]
- 14.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 141394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, D. S. 1990. Brucellosis in wildlife, p. 321-334. In N. Klaus and D. Robert (ed.), Animal brucellosis. CRC Press, Boca Raton, FL.
- 16.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2414-424. [DOI] [PubMed] [Google Scholar]
- 18.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eiglmeier, K., J. Parkhill, N. Honore, T. Garnier, F. Tekaia, A. Telenti, P. Klatser, K. D. James, N. R. Thomson, P. R. Wheeler, C. Churcher, D. Harris, K. Mungall, B. G. Barrell, and S. T. Cole. 2001. The decaying genome of Mycobacterium leprae. Lepr. Rev. 72387-398. [PubMed] [Google Scholar]
- 20.Foster, G., B. S. Osterman, J. Godfroid, I. Jacques, and A. Cloeckaert. 2007. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 572688-2693. [DOI] [PubMed] [Google Scholar]
- 21.Franco, M. P., M. Mulder, R. H. Gilman, and H. L. Smits. 2007. Human brucellosis. Lancet Infect. Dis. 7775-786. [DOI] [PubMed] [Google Scholar]
- 22.Godfroid, F., A. Cloeckaert, B. Taminiau, I. Danese, A. Tibor, X. de Bolle, P. Mertens, and J. J. Letesson. 2000. Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151655-668. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, D., M. J. Grillo, M. J. De Miguel, T. Ali, V. Arce-Gorvel, R. M. Delrue, R. Conde-Alvarez, P. Munoz, I. Lopez-Goni, M. Iriarte, C. M. Marin, A. Weintraub, G. Widmalm, M. Zygmunt, J. J. Letesson, J. P. Gorvel, J. M. Blasco, and I. Moriyon. 2008. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS ONE 3e2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 1872715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 684102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez de Bagues, M. P., A. Terraza, A. Gross, and J. Dornand. 2004. Different responses of macrophages to smooth and rough Brucella spp.: relationship to virulence. Infect. Immun. 722429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan, M. Y., M. W. Mah, and Z. A. Memish. 2001. Brucellosis in pregnant women. Clin. Infect. Dis. 321172-1177. [DOI] [PubMed] [Google Scholar]
- 28.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langille, M. G., W. W. Hsiao, and F. S. Brinkman. 2008. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavigne, J. P., A. C. Vergunst, G. Bourg, and D. O'Callaghan. 2005. The IncP island in the genome of Brucella suis 1330 was acquired by site-specific integration. Infect. Immun. 737779-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2241-15. [DOI] [PubMed] [Google Scholar]
- 32.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 132178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y., P. M. Harrison, V. Kunin, and M. Gerstein. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacLean, A. M., G. MacPherson, P. Aneja, and T. M. Finan. 2006. Characterization of the beta-ketoadipate pathway in Sinorhizobium meliloti. Appl. Environ. Microbiol. 725403-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macrae, R. M., and H. Smith. 1964. The chemical basis of the virulence of Brucella abortus. VI. Studies on immunity and intracellular growth. Br. J. Exp. Pathol. 45595-603. [PMC free article] [PubMed] [Google Scholar]
- 36.Mantri, Y., and K. P. Williams. 2004. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 32D55-D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantur, B. G., M. S. Mulimani, S. S. Mangalagi, and A. V. Patil. 2001. Brucellar epididymoorchitis—report of five cases. Indian J. Med. Microbiol. 19208-211. [PubMed] [Google Scholar]
- 38.Marchesi, J., and F. Shanahan. 2007. The normal intestinal microbiota. Curr. Opin. Infect. Dis. 20508-513. [DOI] [PubMed] [Google Scholar]
- 39.McQuiston, J. R., R. Vemulapalli, T. J. Inzana, G. G. Schurig, N. Sriranganathan, D. Fritzinger, T. L. Hadfield, R. A. Warren, L. E. Lindler, N. Snellings, D. Hoover, S. M. Halling, and S. M. Boyle. 1999. Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect. Immun. 673830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno, E., A. Cloeckaert, and I. Moriyon. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90209-227. [DOI] [PubMed] [Google Scholar]
- 41.Morgan, W. J., and M. J. Corbel. 1976. Recommendations for the description of species and biotypes of the genus Brucella. Dev. Biol. Stand. 3127-37. [PubMed] [Google Scholar]
- 42.Murphy, W. J., E. Eizirik, W. E. Johnson, Y. P. Zhang, O. A. Ryder, and S. J. O'Brien. 2001. Molecular phylogenetics and the origins of placental mammals. Nature 409614-618. [DOI] [PubMed] [Google Scholar]
- 43.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J. Bacteriol. 1773808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 9913148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajashekara, G., J. Covert, E. Petersen, L. Eskra, and G. Splitter. 2008. Genomic island 2 of Brucella melitensis is a major virulence determinant: functional analyses of genomic islands. J. Bacteriol. 1906243-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajashekara, G., J. D. Glasner, D. A. Glover, and G. A. Splitter. 2004. Comparative whole-genome hybridization reveals genomic islands in Brucella species. J. Bacteriol. 1865040-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth, F., J. Zinsstag, D. Orkhon, G. Chimed-Ochir, G. Hutton, O. Cosivi, G. Carrin, and J. Otte. 2003. Human health benefits from livestock vaccination for brucellosis: case study. Bull. W. H. O. 81867-876. [PMC free article] [PubMed] [Google Scholar]
- 48.Scholz, H. C., S. Al Dahouk, H. Tomaso, H. Neubauer, A. Witte, M. Schloter, P. P. Kämpfer, E. Falsen, M. Pfeffer, and M. Engel. 2008. Genetic diversity and phylogenetic relationships of bacteria belonging to the Ochrobactrum-Brucella group by recA and 16S rRNA gene-based comparative sequence analysis. Syst. Appl. Microbiol. 311-16. [DOI] [PubMed] [Google Scholar]
- 49.Scholz, H. C., Z. Hubalek, I. Sedlacek, G. Vergnaud, H. Tomaso, S. Al Dahouk, F. Melzer, P. Kampfer, H. Neubauer, A. Cloeckaert, M. Maquart, M. S. Zygmunt, A. M. Whatmore, E. Falsen, P. Bahn, C. Gollner, M. Pfeffer, B. Huber, H. J. Busse, and K. Nockler. 2008. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 58375-382. [DOI] [PubMed] [Google Scholar]
- 50.Slater, S. C., B. S. Goldman, B. Goodner, J. C. Setubal, S. K. Farrand, E. W. Nester, T. J. Burr, L. Banta, A. W. Dickerman, I. Paulsen, L. Otten, G. Suen, R. Welch, N. F. Almeida, F. Arnold, O. T. Burton, Z. Du, A. Ewing, E. Godsy, S. Heisel, K. L. Houmiel, J. Jhaveri, J. Lu, N. M. Miller, S. Norton, Q. Chen, W. Phoolcharoen, V. Ohlin, D. Ondrusek, N. Pride, S. L. Stricklin, J. Sun, C. Wheeler, L. Wilson, H. Zhu, and D. W. Wood. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 1912501-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snyder, E. E., N. Kampanya, J. Lu, E. K. Nordberg, H. R. Karur, M. Shukla, J. Soneja, Y. Tian, T. Xue, H. Yoo, F. Zhang, C. Dharmanolla, N. V. Dongre, J. J. Gillespie, J. Hamelius, M. Hance, K. I. Huntington, D. Jukneliene, J. Koziski, L. Mackasmiel, S. P. Mane, V. Nguyen, A. Purkayastha, J. Shallom, G. Yu, Y. Guo, J. Gabbard, D. Hix, A. F. Azad, S. C. Baker, S. M. Boyle, Y. Khudyakov, X. J. Meng, C. Rupprecht, J. Vinje, O. R. Crasta, M. J. Czar, A. Dickerman, J. D. Eckart, R. Kenyon, R. Will, J. C. Setubal, and B. W. Sobral. 2007. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 35D401-D406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 521043-1047. [DOI] [PubMed] [Google Scholar]
- 53.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 222688-2690. [DOI] [PubMed] [Google Scholar]
- 54.Sutherland, S. S., and J. Searson. 1990. The immune response to Brucella abortus: the humoral response, p. 65-81. In N. Klaus and D. Robert (ed.), Animal brucellosis. CRC Press, Boca Raton, FL.
- 55.Swai, E. S., and L. Schoonman. 2008. Human brucellosis: seroprevalence and risk factors related to high risk occupational groups in Tanga municipality, Tanzania. Zoonoses Public Health. [Epub ahead of print.] doi: 10.1111/j.1863-2378.2008.01175.x. [DOI] [PubMed]
- 56.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174247-250. [DOI] [PubMed] [Google Scholar]
- 57.Velasco, J., C. Romero, I. Lopez-Goni, J. Leiva, R. Diaz, and I. Moriyon. 1998. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 48759-768. [DOI] [PubMed] [Google Scholar]
- 58.Verger, J. M., J. P. Duée, and M. Grayon. 1982. Brucella isolates in France: evaluation of 10 years of typing. Ann. Microbiol. (Paris) 133433-447. (In French.) [PubMed] [Google Scholar]
- 59.Vernikos, G. S., and J. Parkhill. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 222196-2203. [DOI] [PubMed] [Google Scholar]
- 60.Vizcaino, N., P. Caro-Hernandez, A. Cloeckaert, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6821-834. [DOI] [PubMed] [Google Scholar]
- 61.Winter, A. J., G. G. Schurig, S. M. Boyle, N. Sriranganathan, J. S. Bevins, F. M. Enright, P. H. Elzer, and J. D. Kopec. 1996. Protection of BALB/c mice against homologous and heterologous species of Brucella by rough strain vaccines derived from Brucella melitensis and Brucella suis biovar 4. Am. J. Vet. Res. 57677-683. [PubMed] [Google Scholar]
- 62.Yu, G. X., E. E. Snyder, S. M. Boyle, O. R. Crasta, M. Czar, S. P. Mane, A. Purkayastha, B. Sobral, and J. C. Setubal. 2007. A versatile computational pipeline for bacterial genome annotation improvement and comparative analysis, with Brucella as a use case. Nucleic Acids Res. 353953-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.