Abstract
The group B streptococcus type I pullulanase (SAP) is a class 13 glycoside hydrolase that is anchored to the bacterial cell surface via a conserved C-terminal anchoring motif and involved in α-glucan degradation. Recent in vitro functional studies have shown that SAP is immunogenic in humans and that anti-SAP sera derived from immunized animals impair both group A and group B streptococcus pullulanase activities, suggesting that in vivo immunization with this antigen could prevent streptococcal colonization. To further investigate the putative role of SAP in bacterial pathogenesis, we carried out functional studies and found that recombinant SAP binds to human cervical epithelial cells. Furthermore, with a view of using SAP as a vaccine candidate, we present high-resolution crystal structure analyses of an N-terminally truncated form of SAP lacking the carbohydrate binding module but containing the catalytic domain and displaying glycosidase hydrolase activity, both in its apo form and in complex with maltotetraose, at resolutions of 2.1 and 2.4 Å, respectively.
Bacterial infections represent a constant threat to human health on a global scale, with present treatments largely dependent on antibiotic administration. Such a diffuse approach is of great concern, given that resistance has been observed for all existing antibiotics available and considering that they do not prevent reinfection. It is therefore evident that to avoid future epidemics, alternative therapies must be sought, and in order to achieve this goal, it is necessary to understand the mechanisms of bacterial colonization.
An accessible carbon source is essential for bacteria to colonize the host and potentially cause disease in humans; in particular, highly polymerized α-glucan polysaccharides, such as starch and glycogen, are the most common substrates in environmental niches. Indeed, it is known that diet-derived starches are abundant in the human colon, while glycogen is deposited in large amounts in the vaginal epithelium during periods of high estrogen availability (3, 10, 21). Due to the structural organization of highly polymerized α-glucans, bacteria require appropriate combinations of enzymes for depolymerization of the more complex polysaccharides to oligo- and monosaccharides. Among these enzymes are the pullulanases, which have glycosidase hydrolase activity toward α-glucan polysaccharides, considered to be key extracellular components in bacterial metabolism. Recently, group A streptococcus (GAS) and Streptococcus pneumoniae pullulanases, named PulA and SpuA, respectively, were demonstrated to be anchored to the cell surface via a conserved C-terminal LPXTG anchoring motif and to possess strepadhesin activities, in addition to their classical starch-hydrolyzing activities (15, 35). Glycogen granules present in lung type II alveolar cells have been confirmed to be the substrate for two S. pneumoniae (SpnDX) and Streptococcus pyogenes (SpyDX) pullulanases, whose family 41 carbohydrate binding module (CBM) structures in complex with glycogen have been solved (34).
We recently identified a novel surface-exposed α-glucan-degrading enzyme, named SAP, belonging to the streptococcal family of pullulanases (31). The sap gene is highly conserved among group B streptococcus (GBS) strains; homologous genes, such as those for PulA and SpuA, are present in other pathogenic streptococci. SAP is a member of the class 13 glycoside hydrolase (GH13; α-amylase) family and is a type I pullulanase; in vitro studies have shown that recombinant SAP can degrade α-glucans such as pullulan, glycogen, and starch (31). Furthermore, fluorescence-activated scanning analysis and confocal imaging studies performed on whole bacteria indicate that the presence of α-glucan polysaccharides in culture medium upregulates the expression of SAP on the bacterial surface (31). As reported for other streptococcal pullulanases, we found specific anti-SAP antibodies in human sera from healthy volunteers. Investigation of the functional role of anti-SAP antibodies revealed that incubation of GBS in the presence of sera from animals immunized with SAP reduced the capacity of the bacterium to degrade pullulan. Interestingly, anti-SAP sera, although to a lesser extent, also inhibited GAS pullulanase activity, suggesting the use of SAP as a potential vaccine component to induce functional cross-reacting antibodies that interfere with streptococcal infections.
In this paper, we provide further information on the functional role of SAP by reporting the ability of SAP protein recombinant forms to bind to human epithelial cells. Furthermore, in order to investigate the structural characteristics of SAP, thus improving our understanding of this potential GBS vaccine candidate, we undertook the crystallographic analysis of its three-dimensional (3D) structure. We report here the expression, purification, and crystallization of SAP-L, which lacks the N1 domain containing CBM41 while retaining catalytic activity toward α-glucan substrates, and we present high-resolution crystal structures of its apo form (2.1-Å resolution) and of a complex with the substrate analog maltotetraose (MTT) (2.4-Å resolution).
MATERIALS AND METHODS
Expression and confocal imaging of SAP. (i) Expression and purification.
SAP was cloned into pET21-b and expressed as a C-terminal His-tagged fusion protein in BL21(DE3) Escherichia coli cells, as previously described (32). Bacterial cells were harvested and lysed in buffer A (50 mM sodium phosphate [pH 8.0], 300 mM sodium chloride [NaCl], and 10% glycerol, containing 10 mM imidazole) containing lysozyme (0.25 mg/ml), DNases, and Complete EDTA-free protease inhibitors (Roche). Following sonication and centrifugation, the soluble fraction was loaded onto a 1-ml HisTrap FF column (GE Healthcare) preequilibrated with buffer A. The protein was eluted using a step gradient, achieved by mixing buffer A with buffer B (same as buffer A but with 0.5 M imidazole). Clean fractions, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were pooled, concentrated, and exchanged into crystallization buffer (0.1 M HEPES [pH 7.0], 200 mM NaCl, 5% glycerol, and 2 mM dithiothreitol) using a PD-10 desalting column (GE Healthcare). The crystallization buffer was selected according to thermal shift experiments using an MJ Mini personal thermocycler (Bio-Rad Laboratories), following the fluorescence increase upon thermal denaturation, using the probe Sypro Orange (Sigma-Aldrich) according to standard protocols.
(ii) Confocal immunofluorescence microscopy.
For the confocal imaging of recombinant SAP binding to epithelial cells, ME180 epithelial cells were grown to confluence on Falcon transwell filters (pore size, 3 mm) in 24-well dishes. Cells were then incubated with recombinant forms of SAP at a concentration of 10 μg/ml for 2 h on ice. After washing, cells were fixed in 1% paraformaldehyde for 1 h at 4°C. The monolayers were then blocked with 3% bovine serum albumin and incubated with rabbit anti-SAP polyclonal antibody diluted in 1% bovine serum albumin for 1 h at room temperature. Samples were then stained for 1 h at room temperature with goat anti-rabbit Alexa Fluor (Molecular Probes)-conjugated antibody (excitation wavelength, 488 nm). F-actin was stained with Alexa Fluor 622-conjugated phalloidin. A Slow Fade reagent kit (Molecular Probes) was used to mount coverslips.
Crystal structure of SAP-L. (i) Crystallization.
Crystallization trials were set up in 96-well sitting-drop plates (Greiner) using the Orxy 8.0 crystallization robot (Douglas Instruments) at a protein concentration of 40 mg/ml. Crystals grew after 2 weeks at 20°C under JBScreen 2 (Jena Biosciences) condition D2 (30% polyethylene glycol [PEG] 4000, 0.2 M calcium chloride [CaCl2], and 0.1 M HEPES [pH 7.0]) in a 0.4-μl drop containing 60% protein and 40% well solution. Crystals also grew under alternative conditions, i.e., PEG 3000 (24 to 10%), 0.2 M CaCl2, 50 mM CHES [2-(cyclohexylamino)ethanesulfonic acid] (pH 9.6 to 9.8), and CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] (pH 10). Crystals were flash frozen in liquid nitrogen in the crystallization solution with increased precipitant concentration (35% PEG 3000). For inhibitor-bound SAP-L, crystals grown at pH 9.8 were transferred to 10-μl manual sitting drops for stepwise soaking in MTT solution until a final concentration of 5 mM was reached (10 mM stock, prepared in crystallization solution) and allowed to incubate for 15 min at 20°C prior to cryocooling as before.
(ii) Data collection and processing.
Diffraction data were collected at resolutions of 2.1 Å (unbound) and 2.4 Å (MTT bound) at the European Synchrotron Radiation Facility (Grenoble, France; beam line ID23-1). Both data sets were processed using Mosflm and SCALA and assigned to the orthorhombic P212121 space group (9, 20). One SAP-L chain was present per asymmetric unit, and the solvent content was estimated to be 44.5% (Table 1).
TABLE 1.
Data collection and refinement parameters of SAP-L crystals
| Parametera | Valueb for:
|
|
|---|---|---|
| SAP-L | SAP-L + MTT | |
| Data collection statistics | ||
| Space group | P212121 | P212121 |
| Unit cell dimensions | ||
| a, b, c (Å) | 48.9, 102.8, 172.4 | 48.2, 102.9, 171.7 |
| α = β = γ (°) | 90 | 90 |
| No. of unique reflections | 49,187 | 34,193 |
| Average I/σ (I) | 8.5 (3.0) | 11.9 (2.7) |
| Completeness (%) | 95.6 (98.1) | 99.6 (99.9) |
| Redundancy | 2.9 (2.8) | 3.5 (3.5) |
| Rmerge (%) | 0.117 (0.258) | 0.077 (0.351) |
| Refinement statistics | ||
| Resolution range (Å) | 2.1-40.0 | 2.4-40.0 |
| R (%) | 19.4 | 22.4 |
| Rfree (%) | 23.5 | 28.6 |
| RMSD | ||
| Bond length (Å) | 0.006 | 0.007 |
| Bond angle (°) | 0.981 | 1.017 |
| No. of: | ||
| Molecules/a.u. | 1 | 1 |
| Water molecules | 438 | 64 |
| Calcium ions | 4 | 4 |
| Chloride ions | 5 | 1 |
| Ramachandran plot (%) | ||
| Favored regions | 96.5 | 95.5 |
| Allowed regions | 100 | 100 |
Rmerge = Σ|I − (I)|/Σ I × 100, where I is the intensity of a reflection and (I) is the average intensity; Rfree was calculated from 5% of randomly selected data for cross-validation; R = Σ|Fo − Fc|/Σ|Fo| × 100. The MolProbity program (http://molprobity.biochem.duke.edu/) was used to define the favored and allowed regions of the Ramachandran plot.
The values in parentheses represent the highest-resolution shell for apo-SAP (2.10 to 2.21 Å) and MTT-bound SAP (2.4 to 2.53 Å).
(iii) Molecular replacement, model building, and refinement.
The 3D structure of apo-SAP-L was solved by molecular replacement using the structure of Klebsiella pneumoniae pullulanase (PulKp) (Protein Data Bank [PDB] code 2FGZ), from which the N1 and N2 domains had been deleted, as a search model (25). Despite the relatively low primary sequence identity between SAP-L and PulKp (29%), covering N3 and A domain residues 481 to 1022, a molecular replacement solution was found with R factor and Rfree values of 47.3% and 53.8%, respectively, at 3-Å resolution. The N2 domain, corresponding to residues 382 to 475, was subsequently built into the remaining visible electron density using the web-based Arp/warp 7.0.1 program (27) and a composite omit map (8). The MTT-bound structure was solved using the apo structure model via molecular replacement. Both structures were refined using REFMAC 5.4 (26) and fitted to generated electron density maps using Coot (13). All data were refined to satisfactory final R and Rfree factors and geometric parameters (Table 1).
(iv) Quality of the structural data.
The electron density is well defined for residues 382 to 1159 (apo form) and 383 to 1160 (MTT-bound form), except for several short stretches corresponding to flexible loops. The electron density is absent for the initial two (apo) and three (MTT-bound) N-terminal residues, while the last 61 C-terminal residues, including the His tag, are lost to the solvent. The geometric parameters were checked using the MolProbity program (http://molprobity.biochem.duke.edu/), and all residues were in the Ramachandran plot allowed or favorably allowed regions (12, 22).
Protein structure accession numbers.
The atomic coordinates and structure factors for the apo and MTT-bound SAP-L structures (codes 3FAW and 3FAX, respectively) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics (RCSB), Rutgers University, New Brunswick, NJ (http://www.rcsb.org) (5).
RESULTS
Primary structure analysis.
In addition to the signal peptide (residues 1 to 41), full-length SAP (residues 1 to 1252) contains five highly conserved domains common to pullulanases: the N1 domain comprising two identical CBMs (CBM41-1 [residues 41 to 264] and CBM41-2 [residues 265 to 380]), a pullulanase-associated domain (N2 domain, residues 380 to 471) of unknown function, an isoamylase N-terminal-like domain (N3 domain, residues 472 to 590), the catalytic A domain (residues 591 to 1056), and the C domain (residues 1057 to 1252). A BLAST search against proteins in the RCSB Protein Data Bank reveals highest sequence identity with PulKp (accession no. 2FGZ_A) and Bacillus subtilis pullulanase (PulBs) (accession no. 2E9B_A) (5). The degree of sequence conservation within each domain varies, with the highest sequence homology being in the N1, N3, and A domains (see Fig. SA1 in the supplemental material).
Expression and purification.
The SAP gene (TIGR accession no. SAG_1216), corresponding to residues 41 to 1214, was amplified, cloned, and expressed in bacterial cells as described in Materials and Methods. As previously reported, two expression forms are evident following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, with two bands visible at 98 kDa and 139 kDa, corresponding to a N-terminally truncated form (SAP-L) and a full-length form (SAP-H), respectively (31). SAP-L accounts for 80 to 90% of the total expression and arises due to a second translation start site at Val346 (GTG), a phenomenon also observed for the type I pullulanase from Anaerobranca gottschalkii (6, 31). No attempt was made to separate these two forms for subsequent crystallization trials.
Recombinant SAP binds human cervical epithelial cells.
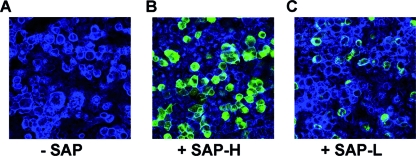
It has been previously reported that PulA is likely to be involved in bacterial adhesion to host cells (16). We also found that both recombinant forms of SAP appear to bind to ME180 cervical epithelial cells. Indeed, confocal imaging studies of differentiated ME180 cells incubated for 1 h in the presence of 10 μg/ml of SAP-H or SAP-L revealed binding to the cell surface by both SAP forms (Fig. 1B and C), although SAP-L was much less adhesive (Fig. 1C). These findings suggest that the carbohydrate binding motif present in SAP-H gives a pivotal contribution to the adhesiveness of SAP to target host cells. To evaluate whether the SAP adhesive phenotype contributed to in vivo GBS binding to host cells, we compared the capacities of COH1 wild-type and COH1 Δsap strains to associate with ME180 cells. Given that SAP expression is inhibited by the presence of glucose in the infection medium, we grew bacteria in a pullulan-containing complex medium to allow pullulanase expression. Association assays and confocal microscopy imaging revealed that although the COH1 wild-type strain, but not the COH1 Δsap strain, positively stained for SAP, both strains adhered to ME180 cells in a similar fashion (results not shown). These data suggest that although recombinant SAP can bind epithelial cells, in vitro expression on the bacterial surface is not sufficient to affect the GBS adhesive phenotype. This may be due a redundancy of GBS bacterial adhesins or to a low expression of specific ligands.
FIG. 1.
Confocal imaging analysis of ME180 epithelial cells sham incubated with SAP (A) or incubated with 10 μg/ml of recombinant SAP-H (B) or SAP-L (C). Cellular F-actin was stained with Alexa Fluor 647-phalloidin (blue) and SAP with rabbit polyclonal anti-SAP antibodies (green). Magnification, ×63.
Crystallization.
Crystals of SAP were obtained as described in Materials and Methods. Although crystals grew over a wide pH range, with hits observed from pH 4.6 to 11.0, the largest crystals were selected for diffraction studies (pH 9.6 to 9.8). A critical parameter for optimum growth was the presence of calcium chloride in the crystallization solution. In the absence of this divalent cation, fine needle bundles rather than good-quality crystals grew (results not shown). Unit cell parameters, data collection statistics, and crystallographic refinement statistics are displayed in Table 1.
N-terminal sequencing.
In order to confirm which expression form of SAP was present in the crystals grown, approximately 30 crystals were thoroughly washed, and run on a 12% Bis-Tris NuPage gel (Invitrogen), and transferred to a polyvinylidene difluoride Immobilon-P membrane (Millipore) for N-terminal sequencing (Gabriella Tedeschi, Department of Animal Pathology, Hygiene and Pubic Veterinary Health, University of Milan, Italy). The crystals were in fact confirmed to contain SAP-L; however, unexpectedly, the N terminus initiated with Tyr379, not at the second translation site (Val346) (results not shown). Following sequence analysis for common proteolysis sites using the PeptideCutter tool (www.expasy.org), it is proposed that residues 346 to 378 of SAP-L are cleaved during expression by diverse bacterial proteases, given that this region is flexible and that Tyr379 is immediately preceded by Tyr378, thus yielding a common cleavage site for several proteases, including chymotrypsin, proteinase K, and thermolysin.
Overall structure.
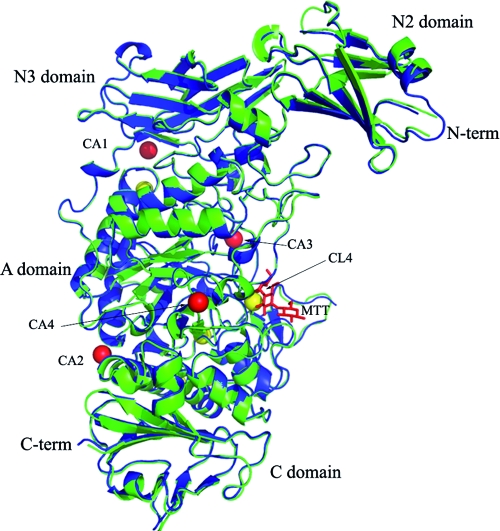
In agreement with other pullulanases, the overall tertiary structure results in an extended (approximately 98 Å in length), crescent-shaped protein moiety, where discrete domains can be easily distinguished (Fig. 2). The electron density corresponding to the main chain and side chains of both apo and MTT-bound SAP-L is well defined for residues 474 to 1159, encompassing the N3, A, and C-terminal domains, with the exception of several flexible-loop regions. The N2 domain is less well defined relative to the other domains, which is not unexpected given that it is largely solvent exposed and isolated with respect to the main body of the protein, as seen in the crystal structures of both B. subtilis and K. pneumoniae pullulanases (Fig. 2). In the inhibitor-bound structure, one molecule of MTT is bound, with electron density for three of the four sugar units being fully interpretable (Fig. 2; see Fig. 4A).
FIG. 2.
Ribbon secondary structure representation of the global structures of apo (green) and MTT-bound (blue) SAP-L, highlighting the four pullulanase domains, the N and C termini, calcium ions (red spheres), chloride ions (yellow spheres), and bound MTT (red sticks). For simplicity, only the chloride ion present in the binding site of the apo structure (CL4) is labeled. This figure was generated using Pymol version 1.1r1 (www.pymol.org).
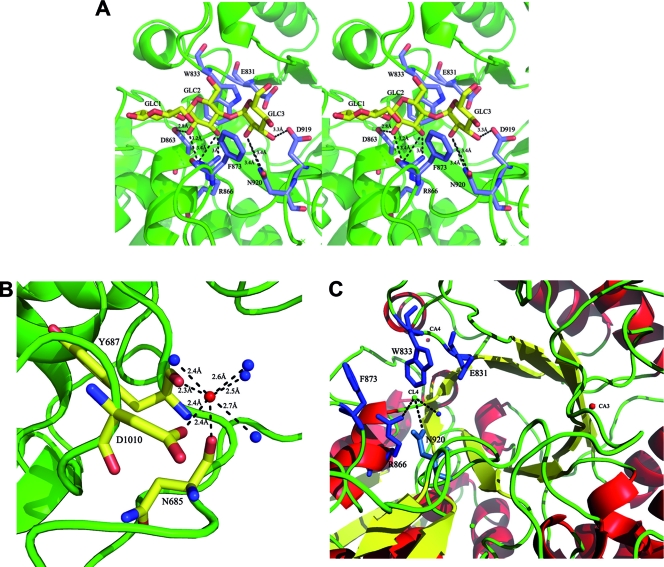
FIG. 4.
(A) The binding of the three sugar units (GLC1 to -3; yellow sticks) of MTT to protein residues is shown in wall-eyed stereo view. (B) The coordination interactions between the third, conserved Ca2+ ion (red sphere) in the substrate binding site, residues, and water molecules (blue spheres) in apo SAP-L are shown. (C) The locations of the conserved chloride (CL4) and calcium (CA3 and CA4) ions can be seen in relation to the canonical α-amylase TIM barrel and the substrate binding site. Bound calcium (red) and chloride (green) ions are shown as spheres, and residues involved in binding MTT are shown in blue stick representation. Hydrogen bonds (dashes) are shown with their respective distances (Å). The figure was generated using Pymol version 1.1r1 (www.pymol.org).
The N2 domain.
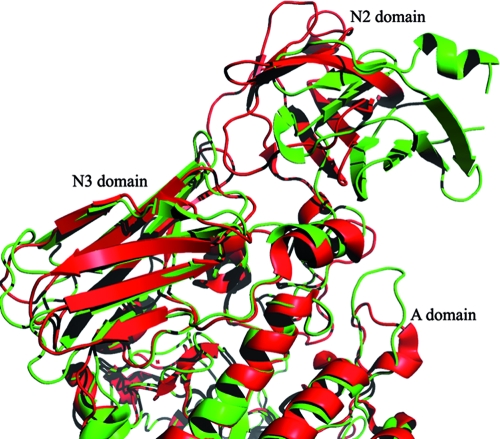
The N2 domain (residues 382 to 473), generally flanked by the N1 and N3 domains (as seen in PulKp), is an as-yet functionally uncharacterized domain, being typically referred to as a “pullulanase-associated domain.” The electron density for the main-chain atoms of this domain is satisfactory; however, the density for the side chains of many residues is scarce, particularly at the N terminus and in a loop region which is followed by a short α-helix (residues 400 to 410). Superimposition of the apo structures of SAP-L and the B. subtilis and K. pneumoniae pullulanases highlights the significantly diverse positioning of the N2 domain with regard to the overall structure (Fig. 3). The positions of the two N2 domains of SAP-L and PulBs are related by a rigid-body movement that includes a slight rotation and translation (approximately 10Å). Despite the diverse orientation of this domain, the overall fold of the N2 domain is similar, being made up of a β-barrel comprising seven β-strands and two α-helices (Fig. 3). According to the interactive secondary structure matching service available from the European Bioinformatics Institute (www.ebi.ac.uk) (18), this domain fold is similar to that of human β2-microglobulin protein (PDB code 2d4d) (17), a ubiquitous fold found in many receptor molecules. Despite low (18%) primary sequence identity, 88% of residues matched, with a root mean square deviation (RMSD) in atomic positions of 2.54 Å.
FIG. 3.
Ribbon secondary structure representation of the apo forms of SAP-L (green) and PulBs (red), superimposed using the C alpha match program (http://bioinfo3d.cs.tau.ac.il/c_alpha_match/). Although the N2 domain of PulKpA is also diversely positioned, for simplicity in the illustration it was not included in the superimposition. This figure was generated using Pymol version 1.1r1 (www.pymol.org).
There is no evident contact interface between the N2 and N3 domains; instead, they are connected via an α-helix (residues 467 to 474) which is stabilized by both aromatic stacking interactions and hydrogen bonds with the side chains of residues from the A domain. The aromatic stacking interactions occur between Trp467 and His776, and there is a hydrogen bond between Trp467(NE1) and Asp839(OD2) at a distance of 3.2 Å. There are also additional hydrogen bonds between Glu468(OE1) and Lys842(NZ) at a distance of 2.8 Å and between Lys470(NZ9) and (i) Glu750(OE2) and (ii) Pro751(O) at distances of 2.9 Å and 2.8 Å, respectively. There is a hydrogen bond between Asp471(OD1) and Tyr753(OH) at a distance of 2.6 Å. Further computational analyses were carried out to investigate the possible function of this domain using the Profunc server, also available at the European Bioinformatics Institute; this is a program developed to help assign a likely biochemical function of a protein based on its 3D structure. Interestingly, the highest score was observed for a collagen binding domain from a Staphylococcus aureus adhesin (PDB code 1amx) (33), with which the N2 domain exhibits 26.4% primary sequence identity; structure superimposition gave an RMSD of 1.7 Å (covering 55 out of the 84 residues). Such structural identity occurs between four β-strands of the N2 domain (β1, β2, β4, and β5) that together comprise one β-sheet of the β-barrel (see Fig. SA1 in the supplemental material). Although the N2 domain is not directly implicated in bacterial adhesion, our functional data confirm such a role for the full-length protein (SAP-H), as discussed in the following section.
The N3 and A domains.
Based on structural considerations and on primary sequence alignments with PulBs and PulKp, the N3 domain comprises residues 474 to 590. It is highly conserved within the α-amylase family and possesses a β-sandwich fold, formed by eight antiparallel β-strands. Unlike the interaction with the N2 domain, the SAP-L N3 domain is connected to the largest and most functionally relevant domain, the A domain, via a significant interface involving both hydrophobic interactions and hydrogen bonds.
The catalytic A domain (residues 591 to 1056) possesses the canonical TIM barrel fold (eight-stranded β-barrel, surrounded by eight α-helices), typical of the GH13 family (Fig. 2). The substrate binding site is housed in this domain and is formed by several loops, which together form a shallow crevice on the protein surface (Fig. 2 and 4A and C). The catalytic residues are highly conserved throughout the pullulanase and GH13 family in general. In comparison with PulKp and PulBs, the positions of the main and side chains of the catalytically essential residues His739, Asp802, and His807 are identical, as are other invariant residues involved in substrate binding (Tyr691, Asp734, Arg800, Asp802, His918, Asp919, and Glu831) (25) (Fig. 4A). In comparison with PulKp, the side chain of Glu831, the catalytic acid/base residue in the hydrolysis reaction, is oriented differently, thus forcing, due to steric hindrance, repositioning of the side chain of Trp833, which is involved in substrate binding.
The Cα carbons of the A domain of SAP-L were superimposed on the corresponding domains of PulKp and PulBs using the C-alpha match program (http://bioinfo3d.cs.tau.ac.il/c_alpha_match/). From all structures scored as highly similar, based on the resulting RMSD values, the highest homology was found between SAP-L and PulBs (apo-enzyme PDB code 2E8Y).
Comparing the SAP-L, PulKp, and PulBs structures, there are two evident structurally diverse regions. The first is a short 14-residue loop region present in SAP-L (residues 672 to 686) and also PulBs, which is replaced with an extra 66-residue domain, called the L2 region, in PulKp (residues 488 to 554). The second is an additional domain (residues 974 to 1004) present in SAP-L but completely absent in both PulKp and PulBs. This domain, in fact, is in close vicinity to the position normally occupied by the L2 region in PulKp. In SAP-L, the consensus sequence YNWGYDP (residues 687 to 693), typically found in pullulanase type I enzymes, is present and contains the tyrosine residue (Y691) involved in substrate binding. In the structure, this zone corresponds to the region immediately after the L2 loop in PulKp. Additional structural differences were observed in several disordered regions, with loops varying in both length and conformation.
The C-terminal domain.
Electron density is well defined for residues 1057 to 1159 (apo) and residues 1057 to 1160 (MTT) of the C-terminal domain; however, the last 61 residues (1161 to 1214), including the His tag, are lost to the solvent. The C-terminal domain is a β-sandwich comprising eight antiparallel β-strands and one α-helix (residues 1056 to 1062), which connects it to the A domain (Fig. 2). The principal interdomain interactions comprise three hydrogen bond interactions: two formed between Asp1061(OD1), in the C domain, and Gln899(OE1) and Lys897(NZ) present in a loop region of the A domain at distances of 2.9 Å and 2.8 Å, respectively, and Tyr1084(OH) with Thr955(OG1), at a distance of 2.8 Å.
The structure of the C domain is highly conserved throughout the family, and C-alpha match alignments carried out with the corresponding domains in PulKp and PulBs gave RMSD values of 1.5 Å for PulBs (covering 63 residues) and of 1.7 Å and 1.5 Å for 83 and 70 residues, respectively, of PulKp. The most divergent regions correspond to flexible loops (residues 1069 to 1080 and 1088 to 1092). The most diverse loop spans residues 1122 to 1145, and in PulKp the corresponding region forms an α-helix. In addition, there is an additional α-helix (residues 1114 to 1118) that connects two C domain β-strands in SAP, which are not found in the homolog proteins.
Calcium ions.
As mentioned above, CaCl2 was found to be essential for crystallization; the absence of this ion resulted in needle bundle growth rather than single crystals (results not shown). Correspondingly, in six data sets collected to date, in both the presence and absence of MTT, there is electron density representing four calcium (Ca2+) ions, which appear to have structural roles and stabilize potentially mobile regions of the protein structure (Fig. 2). The first Ca2+ ion is present between the hinge of the N2 and N3 domains, being coordinated by interactions with Tyr547(OH), Lys510(O), Lys708(O), and four water molecules. The second is located between two helices in the A domain and is particularly solvent exposed, interacting only with the carbonyl oxygen of residue Lys652 and four water molecules. The third is of particular interest, as it is conserved in PulKp and faces the substrate binding site, interacting with the carbonyl oxygens of Asn685 and Tyr687, the side chain oxygen (OD1) of Asp1010, and four water molecules (Fig. 4B). The fourth interacts with Thr855(O), Asp910(OD1 and OD2), Asp856(OD1), Asp906(OD1), Met852(O), and one water molecule. All Ca2+ coordination interactions have bond lengths that range from 2.2Å to 2.8Å.
Chloride ions.
In addition to the bound Ca2+ ions, chloride anions (Cl−) were bound to both apo and inhibitor-bound SAP-L structures (Fig. 2 and 4C). Cl− ions are not present in PulBs and PulKp; however, they have been reported in the crystal structures of several α-amylases (1, 14, 24). In fact, several reports describe the identification of Cl−-dependent α-amylases from three gram-negative bacteria, invertebrates, and mammals; such enzymes have been found to also possess additional structural features specific to Cl−-dependent activity (2, 11). Typically one or two Cl− ions are bound, one of which is highly conserved and located in proximity to the active site and the active-site Ca2+. However, in the SAP-L structures, five Cl− ions are bound in the apo-enzyme, and only one in the complex. With regard to apo-SAP-L, the fourth anion is of particular interest. Although it is not located in the precise location as the conserved anion in the Cl−-bound α-amylase structures, it is bound in the substrate binding site, and it is replaced with a glucose unit in the MTT-bound SAP-L structure. It is held by electrostatic interactions and by loose hydrogen bonds with Asn920(ND2), Arg866(NH1), and a water molecule (Fig. 4C). As expected, due to its slightly diverse position, this Cl− anion does not interact with the equivalent residues in the α-amylase structures; however, its overall location in the substrate binding site is consistent with that in other α-amylases, including human and porcine pancreatic α-amylases (7, 19, 28).
MTT-bound SAP-L.
An MTT-bound crystal structure is available for PulKp (PDB code 2FHF), in which all pullulanase domains, including the N1 domain, are defined (25). In PulKp, there are two MTT molecules bound to the active-site crevice of the A domain and a maltose unit (two glucose units) bound at the N1 domain, which is oriented to face the A domain. As for the other pullulanases, the substrate binding site of SAP-L is formed by several loop regions that form a shallow crevice on the surface of the protein. In two data sets of MTT-bound SAP-L collected to date, there is only one molecule of MTT bound to the substrate binding site present in the A domain (Fig. 4A). Electron density corresponding to only three (GLC1, -2, and -3) of the four MTT glucose units is visible. In fact, maltotriose is the final product of type I pullulanase activity; therefore, it is possible that the fourth unit of the supplied MTT was cleaved off.
MTT binding does not induce any global conformational changes. The 3D structures of apo and MTT-bound SAP-L are highly similar; superimposition of the main-chain C-alpha atoms yields an RMSD value of 0.41 Å, spanning 640 residues. The ligand interacts principally via hydrogen bonds between GLC2 and GLC3 and A domain residues Asp863, Asp919, and Asn920, as in PulKpA, although they are slightly longer in SAP-L (≥2.8 Å) than in the homolog (Table 2). GLC2 and GLC3 are also stabilized by sugar-aromatic stacking interactions with Trp833 and Phe873, respectively (Fig. 4A and Table 2). GLC1 does not interact with protein residues. One interesting point to note is that Arg866, Phe873, and Trp833 are not conserved in human α-amylase; therefore, they might be key residues to exploit for designing specific inhibitors.
TABLE 2.
Hydrogen bond and aromatic-sugar interactions between maltotetraose and SAP-L protein atomsa
| Source ligand atom and interaction type | Target atom | Bond distance (Å) |
|---|---|---|
| Hydrogen bond | ||
| GLC2(O2) | Arg866(NH2) | 3.2 |
| Asp863(OD2) | 2.8 | |
| GLC2(O3) | Arg866(NH1) | 3.0 |
| Arg866(NH2) | 3.4 | |
| GLC3(O3) | Asn920(OD1) | 3.4 |
| Asn920(ND2) | 3.4 | |
| Asp919(OD2) | 3.3 | |
| Aromatic-sugar (C-H-π) interaction | ||
| GLC2 | Trp833 | ≤4.0 |
| GLC3 | Phe873 | ≤4.0 |
Bond distances between the individual sugar units (GLC1 to -3) of MTT and the target protein residues are shown, together with the participating atoms.
DISCUSSION
The increasing requirement for new vaccine-based preventive strategies, supplementing the antibiotic prophylaxis used for eradicating GBS colonization of the genital tract in pregnant women, has recently led to identification of antigens conferring broad protection in mice (23). The discovery of novel immunogenic virulence factors has also opened new perspectives to tackle GBS-associated infections (4, 32). In this context, we considered the characterization of the structural properties of the GBS genes involved in adaptive metabolism of the bacterium to be a necessary step. Indeed, the mechanisms underlying the capacity of GBS to use complex carbon sources available at site of colonization are largely undefined.
The confirmed role of pullulanases in their ability to recognize and bind to host cell surface glycans, in particular glycogen, is an essential process for bacterial virulence (35). Our recent functional studies have shown that SAP is highly immunogenic in humans and that antibodies raised against recombinant SAP prevented glycogen degradation (31), a crucial in vivo carbon source for the establishment of GBS colonization. The precise epitopes that confer immunogenicity to SAP remain to be identified, and consequently, functional experiments are in progress to isolate the specific monoclonal antibodies that inhibit SAP catalytic activity. In light of its potential as an antigen target for vaccine development, the X-ray structural characterization of SAP-L is a further step toward fulfillment of a reverse vaccinology study (29, 30) applied to GBS.
SAP was expressed as a His-tagged fusion protein in bacterial cells in two forms, the truncated form (SAP-L) and the full-length form (SAP-H), with the former being expressed to a greater extent (>80%). As mentioned above, this truncation is due to the presence of a second translation site in the nucleotide sequence, which, when specifically mutated, resulted in exclusively SAP-H expression. Thus, SAP-L lacks the N1 domain containing the CBMs, which are involved in substrate recognition and binding, but contains the four remaining domains common to pullulanases: an N-terminal pullulanase-associated domain (N2 domain), an N-terminal isoamylase-like domain (N3 domain), an α-amylase-like domain (A domain) containing the active site, and a C-terminal β-sandwich domain (C domain).
Despite relatively low primary sequence identity, the global structure of SAP-L is highly similar to that of the K. pneumoniae and B. subtilis pullulanases, displaying an extended, multidomain fold. Superimposition of the crystal structures of SAP-L, PulKpA, and PulBs illustrated that the N2 domain is the most diverse domain in its overall orientation with respect to the remaining domains, which are highly similar in the three structures, with regard to both secondary and tertiary structure. The precise function of the N2 domain, which is particularly isolated with respect to the rest of the protein, remains to be elucidated. In fact, the highest sequence identity is observed in the N3 and A domains, the latter of which contains the active site and a TIM barrel fold, conserved in α-amylases from microbes to mammals. The last domain, located at the C terminus, is a β-sandwich, also highly conserved within the pullulanase and α-amylase family. Such structural similarity is of great advantage, as a potential vaccine raised against this antigen would not be limited to only GBS infection.
In addition to the apo form of the enzyme, we solved the structure of SAP-L in complex with one molecule of MTT, a four-saccharide cleavage product of its glycogen substrate. The 3D structures of apo and MTT-bound SAP-L are highly similar, and there is no evident conformational change upon binding of MTT, which is not unexpected given that the substrate binding site is a shallow crevice formed by several surface loops, rather than an internal cavity. In the inhibitor-bound structure, only three of the four MTT sugar units were visible. This has been observed in all MTT-bound SAP-L data sets collected to date, and therefore it is proposed that the fourth ring is cleaved off, given that maltotriose is the final product.
The presence of Ca2+ ions bound to the α-amylase domains of the crystal structures of pullulanases and several other α-amylase members is a recurring observation. Typically, one or two Ca2+ ions are bound in the structures containing an α-amylase fold known to date. Although the precise location of these cations differs in our structures, being located at opposite ends of the TIM barrel, during crystallization trials the presence of Ca2+ was found to be essential for crystal growth of SAP-L, and four ions were bound in identical positions in both apo and MTT-bound structures. The four Ca2+ ions reported in both apo and MTT-bound SAP-L clearly play a structural role, stabilizing the interaction between the N2 and N3 domains and the various loops in the A domain. The Ca2+ ion present just below the TIM barrel is also conserved in apo and inhibitor-bound PulKpA structures; it stabilizes two loops building the substrate binding site, thus being instrumental for the stabilization of the overall architecture.
In addition to Ca2+ ions, Cl− ions were also found in both structures; this is an observed feature in some α-amylases but not pullulanases. The binding of one Cl− ion, in particular, has been demonstrated to be important for overall catalytic activity and for the pH optima of some α-amylase members. In one case, Cl− ions were shown to act as allosteric activators providing the electrostatic environment to the positively charged residues in the substrate binding site (11). In Cl−-dependent α-amylases, this anion is bound close to the center of the active-site TIM barrel and next to the conserved Ca2+ binding site, stabilized by a conserved three-residue motif (Arg-Asn-Arg/Lys); enzymes which contain a lysine as the third residue usually have a lower affinity for chloride and lower catalytic efficiency. Following primary and tertiary sequence alignments with porcine α-amylase, SAP-L was confirmed not to host such a triad, therefore suggesting that it does not belong to the Cl−-dependent α-amylase family. With regard to SAP, the precise role of Cl− ions remains to be clarified; however, as suggested in other reports, Cl− anions could be involved in creating a suitable electrostatic environment for substrate binding (2, 11).
With regard to SAP function, it is important to underline that both recombinant GAS and S. pneumoniae pullulanases specifically target, in vitro, a subset of glycogen-rich alveolar cells, suggesting that they recognize glycogen and not a ligand associated with the host cell membrane (35). Interestingly, our study reported that the recombinant form of SAP was able to bind to human epithelial cells of cervical origin mainly through the CBM-containing portion. Although this phenotype was not evident by comparing the capacities of COH1 wild-type and COH1 Δsap mutant strains to adhere to host cells, it cannot be excluded that binding of SAP to carbohydrate substrates present on the surfaces of host tissues may contribute to the establishment of GBS colonization.
We have recently reported that incubation of GBS in the presence of sera from animals immunized with SAP impaired the capacity of the bacterium to degrade alpha-glucans in vitro (31). Based on these data and the evidence reported in this paper that recombinant SAP binds to host cells, we propose to use SAP as a potential vaccine component. Indeed, we hypothesize that anti-SAP functional antibodies may reduce bacterial fitness in vivo and interfere with the capacity of GBS to colonize host tissues. To this end, work is in progress to isolate monoclonal antibodies that specifically inhibit SAP activity. The confirmed high structural identity observed between SAP-L and other bacterial pullulanases reinforces its use as an antigen to target diverse bacterial infections. In addition to attempts to crystallize SAP-H, in the context of epitope determination, the 3D structure of apo-SAP-L is presently being used to help develop software to predict the locations of potential epitopes. Such an endeavor will be guided by comparison with experimental data that will follow inhibitory monoclonal antibody isolation. Such data taken together will greatly assist in the development of novel vaccine strategies.
Supplementary Material
Acknowledgments
This European Community FP6 project, “Assessment of Structural Requirements in Complement-Mediated Bactericidal Events: Toward a Global Approach to the Selection of New Vaccine Candidates” (BacAbs), is gratefully acknowledged (contract LSH-2005-037325). Part of this work was funded by Progetto FIRB “Biologia Strutturale” RBLA03B3KC_005.
We are grateful to the European Synchrotron Radiation Facility (Grenoble, France) and to Gabriella Tedeschi (University of Milan, Italy) for N-terminal sequencing of SAP crystals.
Footnotes
Published ahead of print on 27 March 2009.
Supplemental material for this article is available at http://jb.asm.org/.
REFERENCES
- 1.Aghajari, N., G. Feller, C. Gerday, and R. Haser. 1998. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 7564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aghajari, N., G. Feller, C. Gerday, and R. Haser. 2002. Structural basis of alpha-amylase activation by chloride. Protein Sci. 111435-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, I. H., A. S. Levine, and M. D. Levitt. 1981. Incomplete absorption of the carbohydrate in all-purpose wheat flour. N. Engl. J. Med. 304891-892. [DOI] [PubMed] [Google Scholar]
- 4.Baron, M. J., D. J. Filman, G. A. Prophete, J. M. Hogle, and L. C. Madoff. 2007. Identification of a glycosaminoglycan binding region of the alpha C protein that mediates entry of group B Streptococci into host cells. J. Biol. Chem. 28210526-10536. [DOI] [PubMed] [Google Scholar]
- 5.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoldo, C., M. Armbrecht, F. Becker, T. Schafer, G. Antranikian, and W. Liebl. 2004. Cloning, sequencing, and characterization of a heat- and alkali-stable type I pullulanase from Anaerobranca gottschalkii. Appl. Environ. Microbiol. 703407-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brayer, G. D., Y. Luo, and S. G. Withers. 1995. The structure of human pancreatic alpha-amylase at 1.8 A resolution and comparisons with related enzymes. Protein Sci. 41730-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, J. H., E. W. Pomare, W. J. Branch, C. P. Naylor, and G. T. Macfarlane. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 281221-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Amico, S., C. Gerday, and G. Feller. 2000. Structural similarities and evolutionary relationships in chloride-dependent alpha-amylases. Gene 25395-105. [DOI] [PubMed] [Google Scholar]
- 12.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Feller, G., O. Bussy, C. Houssier, and C. Gerday. 1996. Structural and functional aspects of chloride binding to Alteromonas haloplanctis alpha-amylase. J. Biol. Chem. 27123836-23841. [DOI] [PubMed] [Google Scholar]
- 15.Hytonen, J., S. Haataja, and J. Finne. 2003. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect. Immun. 71784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hytonen, J., S. Haataja, and J. Finne. 2006. Use of flow cytometry for the adhesion analysis of Streptococcus pyogenes mutant strains to epithelial cells: investigation of the possible role of surface pullulanase and cysteine protease, and the transcriptional regulator Rgg. BMC Microbiol. 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara, M., E. Chatani, K. Iwata, K. Yamamoto, T. Matsuura, A. Nakagawa, H. Naiki, and Y. Goto. 2006. Conformation of amyloid fibrils of beta2-microglobulin probed by tryptophan mutagenesis. J. Biol. Chem. 28131061-31069. [DOI] [PubMed] [Google Scholar]
- 18.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 602256-2268. [DOI] [PubMed] [Google Scholar]
- 19.Larson, S. B., A. Greenwood, D. Cascio, J. Day, and A. McPherson. 1994. Refined molecular structure of pig pancreatic alpha-amylase at 2.1 A resolution. J. Mol. Biol. 2351560-1584. [DOI] [PubMed] [Google Scholar]
- 20.Leslie, A. G. 2006. The integration of macromolecular diffraction data. Acta Crystallogr. D 6248-57. [DOI] [PubMed] [Google Scholar]
- 21.Levitt, M. D., P. Hirsh, C. A. Fetzer, M. Sheahan, and A. S. Levine. 1987. H2 excretion after ingestion of complex carbohydrates. Gastroenterology 92383-389. [DOI] [PubMed] [Google Scholar]
- 22.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins 50437-450. [DOI] [PubMed] [Google Scholar]
- 23.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. Nardi Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maurus, R., A. Begum, H. H. Kuo, A. Racaza, S. Numao, C. Andersen, J. W. Tams, J. Vind, C. M. Overall, S. G. Withers, and G. D. Brayer. 2005. Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase. Protein Sci. 14743-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikami, B., H. Iwamoto, D. Malle, H. J. Yoon, E. Demirkan-Sarikaya, Y. Mezaki, and Y. Katsuya. 2006. Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site. J. Mol. Biol. 359690-707. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 27.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6458-463. [DOI] [PubMed] [Google Scholar]
- 28.Qian, M., R. Haser, and F. Payan. 1993. Structure and molecular model refinement of pig pancreatic alpha-amylase at 2.1 A resolution. J. Mol. Biol. 231785-799. [DOI] [PubMed] [Google Scholar]
- 29.Rappuoli, R. 2000. Reverse vaccinology. Curr. Opin. Microbiol. 3445-450. [DOI] [PubMed] [Google Scholar]
- 30.Rappuoli, R., and A. Covacci. 2003. Reverse vaccinology and genomics. Science 302602. [DOI] [PubMed] [Google Scholar]
- 31.Santi, I., A. Pezzicoli, M. Bosello, F. Berti, M. Mariani, J. L. Telford, G. Grandi, and M. Soriani. 2008. Functional characterization of a newly identified group B streptococcus pullulanase eliciting antibodies able to prevent alpha-glucan degradation. PLoS One 3e3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santi, I., M. Scarselli, M. Mariani, A. Pezzicoli, V. Masignani, A. Taddei, G. Grandi, J. L. Telford, and M. Soriani. 2007. BibA: a novel immunogenic bacterial adhesin contributing to group B Streptococcus survival in human blood. Mol. Microbiol. 63754-767. [DOI] [PubMed] [Google Scholar]
- 33.Symersky, J., J. M. Patti, M. Carson, K. House-Pompeo, M. Teale, D. Moore, L. Jin, A. Schneider, L. J. DeLucas, M. Hook, and S. V. Narayana. 1997. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol. 4833-838. [DOI] [PubMed] [Google Scholar]
- 34.van Bueren, A. L., and A. B. Boraston. 2007. The structural basis of alpha-glucan recognition by a family 41 carbohydrate-binding module from Thermotoga maritima. J. Mol. Biol. 365555-560. [DOI] [PubMed] [Google Scholar]
- 35.van Bueren, A. L., M. Higgins, D. Wang, R. D. Burke, and A. B. Boraston. 2007. Identification and structural basis of binding to host lung glycogen by streptococcal virulence factors. Nat. Struct. Mol. Biol. 1476-84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.