Abstract
RpoS is a bacterial sigma factor of RNA polymerase which is involved in the expression of a large number of genes to facilitate survival under starvation conditions and other stresses. The results of our study demonstrate that the frequency of emergence of base substitution mutants is significantly increased in long-term-starved populations of rpoS-deficient Pseudomonas putida cells. The increasing effect of the lack of RpoS on the mutation frequency became apparent in both a plasmid-based test system measuring Phe+ reversion and a chromosomal rpoB system detecting rifampin-resistant mutants. The elevated mutation frequency coincided with the death of about 95% of the cells in a population of rpoS-deficient P. putida. Artificial overexpression of superoxide dismutase or catalase in the rpoS-deficient strain restored the survival of cells and resulted in a decline in the mutation frequency. This indicated that, compared to wild-type bacteria, rpoS-deficient cells are less protected against damage caused by reactive oxygen species. 7,8-Dihydro-8-oxoguanine (GO) is known to be one of the most stable and frequent base modifications caused by oxygen radical attack on DNA. However, the spectrum of base substitution mutations characterized in rpoS-deficient P. putida was different from that in bacteria lacking the GO repair system: it was broader and more similar to that identified in the wild-type strain. Interestingly, the formation of large deletions was also accompanied by a lack of RpoS. Thus, the accumulation of DNA damage other than GO elevates the frequency of mutation in these bacteria. It is known that oxidative damage of proteins and membrane components, but not that of DNA, is a major reason for the death of cells. Since the increased mutation frequency was associated with a decline in the viability of bacteria, we suppose that the elevation of the mutation frequency in the surviving population of carbon-starved rpoS-deficient P. putida may be caused both by oxidative damage of DNA and enzymes involved in DNA replication and repair fidelity.
Accumulation of reactive oxygen species (ROS) such as superoxide anion, hydrogen peroxide, and hydroxyl radicals leads to nucleic acid, protein, and cell membrane damage. ROS have been implicated in cancer, aging, and various diseases in humans but also in the death of microorganisms (53). Many microorganisms are continuously faced with ROS derived from different sources. For example, during infection, pathogenic bacteria are exposed to the exogenous oxidative stress that phagocytes use as a host defense mechanism (25, 47). Additionally, ROS are constantly generated as by-products of aerobic metabolism. To counteract oxidative stress, both prokaryotic and eukaryotic cells maintain inducible defense systems to detoxify oxidants and repair damage (14, 32). Gram-negative bacteria commonly synthesize both cytoplasmic and periplasmic isozymes of superoxide dismutases (SOD) to eliminate superoxide anions (40). Hydrogen peroxide is scavenged in most organisms by peroxidases and catalases (13, 32). Bacteria use distinct sensing mechanisms for the detection of discrete forms of oxidative stress. For instance, the SoxRS system is a regulator of superoxide stress whereas the regulators OxyR and PerR respond to hydrogen peroxide stress (32).
RpoS is a sigma subunit of RNA polymerase that is involved in the induction of a large number of genes when the environment becomes unable to sustain the growth of bacteria. The rpoS gene (also named katF) was initially characterized as a locus which affects the synthesis of catalase (38). Later it was shown that RpoS is a central regulator of the general stress response in Escherichia coli, where it controls the expression of at least 100 genes and is involved in cross-protection against osmotic, acidic, and oxidative stress (27, 78). RpoS-deficient cells of E. coli are less protected against oxidative or osmotic stress and are less viable in the stationary phase (35, 42).
Several lines of evidence indicate that RpoS positively regulates many mutagenic processes in growth-limiting environments of bacteria (reviewed in references 20 and 22). RpoS enhances spontaneous mutations by upregulating specialized DNA polymerase Pol IV in E. coli (36). Pol IV is responsible for 50 to 80% of the Lac+ revertants that occur because of 1-bp deletions in runs of iterated bases in the F′ episome in E. coli strain FC40 (19, 43). The emergence of Lac+ revertants is reduced about 10-fold in the absence of RpoS (36, 39). Additionally, RpoS controls a switch that changes normally error-free double-strand break repair into an error-prone process under stress (60). RpoS is also required for base substitution mutations in aging E. coli colonies (8). Some evidence indicates that downregulation of mismatch repair proteins MutS and MutH by RpoS (17, 75) increases mutation rates in starving E. coli (8, 21, 24, 79). Phase variation in Pseudomonas sp. strain PCL1171 is also dependent on RpoS (76).
We have studied the mechanisms of mutagenic processes in bacteria by using Pseudomonas putida as a model organism. The genus Pseudomonas represents one of the largest groups of bacteria including both pathogenic and nonpathogenic species. Bacteria of this genus are known for their abilities to colonize multiple habitats and to adapt rapidly to new environments. P. putida is a fast-growing bacterium found in most temperate soil and water habitats where oxygen is present. It is also able to colonize surfaces of living organisms. We have found that RpoS can act as a positive regulator in the transposition of Tn3 family transposon Tn4652 in P. putida (31). Additionally, the occurrence of 2- to 3-bp deletions, characteristic of long-term starvation, was not detected in an RpoS− background (66). RpoS also promotes genome instability in other Pseudomonas species (76).
In the present study, we focused on the role of RpoS in the occurrence of point mutations in starving P. putida. The results presented herein demonstrate a negative role for RpoS in the emergence of base substitutions in this organism. We found that the formation of base substitutions, estimated with both plasmid-based and chromosomal test systems, was significantly elevated in rpoS-deficient P. putida after 5 to 7 days of carbon starvation. The increased mutation frequency in a starving population coincided with the rapid death of the majority of the cells. Suppression of the effects of the lack of RpoS by overexpression of SOD or catalase indicated that damage caused by ROS reduces survival and elevates the mutation frequency in a starving rpoS-deficient strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Complete medium was Luria-Bertani medium (46), and minimal medium was M9 (1). Solid medium contained 1.5% Difco agar. Casamino Acids and glucose were added to the minimal medium at final concentrations of 0.4 and 0.2%, respectively. Phenol minimal plates contained 2.5 mM phenol as the sole carbon and energy source. Antibiotics were added at the following concentrations: ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, tetracycline at 50 μg/ml, chloramphenicol at 1,500 to 3,000 μg/ml, streptomycin at 500 μg/ml, rifampin at 100 μg/ml, and carbenicillin at 1,500 to 3,000 μg/ml. Potassium tellurite was added at a final concentration of 25 μg/ml. E. coli was incubated at 37°C, and P. putida was incubated at 30°C. E. coli and P. putida were electrotransformed as described by Sharma and Schimke (68). E. coli strains DH5α (Invitrogen) and CC118 λpir (28) were used for DNA cloning procedures, and HB101 (10) was used as a host for helper plasmid pRK2013 (18), which was necessary for the mobilization of nonconjugative plasmids. CC118 λpir was also used as a host for the R6K replicon-based helper plasmid providing the Tn7 transposase proteins (6).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or construction | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80dlacZΔM15) recA1 endA1 hsdR17 thi-1 gyrA96 relA1 | Invitrogen |
| HB101 | subE44 subF58 hsdS3(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 10 |
| CC118 λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 λpir phage lysogen | 28 |
| P. putida | ||
| PaW85 | WT | 7 |
| PaWRpoS | PaW85, rpoS::km (the same as PKS54) | 55 |
| PaWRpoSLacI | PaWRpoS, lacIq gene in chromosome | This study |
| PaWRpoSSodAB | PaWRpoS, lacIq-Ptac-sodAB expression cassette in chromosome | This study |
| PaWRpoSKatA | PaWRpoS, Ptac-katA transcriptional fusion and lacIq gene in chromosome | This study |
| PaWRpoSSodABKatA | PaWRpoS, lacIq-Ptac-sodAB and Ptac-katA expression cassettes in chromosome | This study |
| Plasmids | ||
| pBluescript KS(+) | Cloning vector (Apr) | Stratagene |
| pUC18Not | Cloning vector (Apr) | 28 |
| pUTmini-Tn5 Cm | Delivery plasmid for mini-Tn5 Cm (Apr Cmr) | 65 |
| pJMT6 | Delivery plasmid for mini-Tn5 Tel (Apr Telr) | 65 |
| pRK2013 | Helper plasmid for conjugal transfer of mini-Tn-carrying plasmids (Kmr) | 18 |
| pBK-miniTn7-ΩSm1 | pUC19-based delivery plasmid for mini-Tn7-ΩSm1 (Smr Apr) | 34 |
| pUX-BF13 | R6K replicon-based helper plasmid providing Tn7 transposase proteins (Apr Mob+) | 6 |
| pAM10.6 | Biodegradative plasmid carrying catalase gene katA | 57 |
| pBRlacItac | pBR322 carrying Ptac promoter and lacIq gene for LacI repressor | 55 |
| pKSsodB | pBluescript KS (+) containing PCR-amplified sodB gene region inserted into EcoRV-opened vector plasmid | This study |
| pKSsodAB | PCR-amplified sodA gene in SmaI site of pKSsodB | This study |
| pBRlacItacsodAB | pBRlacItac with sodAB genes cloned from pKSsodAB within XbaI- and SalI-generated DNA fragment | This study |
| pUC18NotlacItacsodAB | pUC18Not containing lacIq-Ptac-sodAB expression cassette cloned within BamHI-generated DNA fragment from pBRlacItacsodAB | This study |
| pUTtellacItacsodAB | mini-Tn5 delivery plasmid pJMT6 containing lacIq-Ptac-sodAB expression cassette cloned from pUC18NotlacItacsodAB in NotI site | This study |
| pACtac | pACYC184 containing Ptac promoter | 71 |
| pKStac | pBluescript KS(+) containing Ptac promoter cloned from plasmid pACtac within BamHI- and HindIII-generated DNA fragment | This study |
| pKStackatA | PCR-amplified katA gene from plasmid pAM10.6 inserted into XbaI- and NotI-cleaved pKStac | This study |
| pAKNtackatA | pBK-miniTn7-ΩSm1 containing Ptac-katA transcriptional fusion inserted into ApaI- and NotI-cleaved vector plasmid | This study |
| pREP4 | LacI repressor-expressing plasmid | Qiagen |
| pUC18NotlacI | pUC18Not containing lacIq gene from pREP4 cloned within Eco47III- and SmaI-generated fragment into SmaI site | This study |
| pUTCmlacI | lacIq gene from pUC18NotlacI inserted within NotI fragment into mini-Tn5 delivery plasmid pUTmini-Tn5 Cm | This study |
| pKTpheA56+A | Test system for detection of Phe+ revertants occurring due to 1-bp deletions | 74 |
| pKTpheA22TAG | Test system for detection of Phe+ revertants occurring due to base substitutions | 74 |
Measurement of intracellular amounts of superoxide and SOD activities and detection of catalase expression in bacteria.
In order to measure the amount of superoxide radicals (SO) in carbon-starved P. putida cells, wild-type (WT) strain PaW85 and its rpoS-deficient derivative were grown overnight in M9 minimal medium supplemented with glucose and Casamino Acids. Approximately 1 × 109 cells were spread onto minimal plates and incubated under carbon starvation conditions. Cells from three agar plates were resuspended in 1.5 ml of M9 buffer at the beginning of starvation (on day 2) and after prolonged starvation (on day 8). Cell suspensions were divided into two equal amounts (each part was 750 μl). In the first half of the suspension, the cells were disrupted by sonication and the cell lysate (lysate I) cleared by centrifugation at 16,000 × g for 25 min at 4°C was used for measurement of the total amount of SO. The second half of the cell suspension was centrifuged at 12,000 × g for 3 min, and the supernatant (supernatant I) was collected for the measurement of SO. Cells from the bottom of an Eppendorf tube were resuspended in 750 μl of M9 buffer and harvested by centrifugation to obtain supernatant II for the measurement of SO. Finally, after the removal of supernatant II, the cells were resuspended in 750 μl of M9 buffer, disrupted by sonication, and cleared by centrifugation at 16,000 × g for 25 min at 4°C, yielding lysate II for the measurement of SO. Hydroethidine was added to cell lysates I and II or collected supernatants I and II at a final concentration of 63.5 μM. The reaction mixtures were incubated for 30 min at 30°C in the dark. The amount of superoxide in the reaction mixtures was expressed as hydroethidine dehydrogenation to ethidium bromide (EB) by superoxide anion. The quantity of EB generated was determined by the intensity of EB fluorescence measured with a computerized fluorometer Tecan Genios-Plus at 485-nm excitation and 595-nm emission wavelengths. The amount of SO was calculated either per plate or as the relative amount of SO per 1 × 106 CFU in a 1-ml reaction mixture. All measurements were repeated at least four times in triplicate.
To measure SOD activity in stationary-phase P. putida cells, cells starved for 8 days were resuspended and sonicated in phosphate buffer (100 mM Na2HPO4/KH2PO4 [pH 7]). SOD activity was measured in cell lysates by using the SOD Assay Kit-WST (Fluka) according to the protocol provided by the manufacturer. Still, the reaction was performed at 30°C, which is the optimal growth temperature of P. putida. SOD activity was expressed as the percent inhibition of tetrazolium salt reduction to formazan dye (absorbance at 440 nm) by superoxide anion. The percent formazan formation per 1.0 μg of total protein was measured.
The expression of catalase was monitored by determining the formation of O2 bubbles in a cell suspension after the addition of an H2O2 solution. A 100-μl volume of overnight-grown bacterial cultures was diluted into 2 ml of M9 buffer, and 20 μl of 30% H2O2 was added to detect the catalase reaction. The reaction tubes were photographed after 1 min of incubation at room temperature.
Construction of P. putida strains overexpressing SOD or catalase.
In order to overexpress SOD in P. putida (strain PaWRpoSSodAB), we cloned the sodA and sodB genes of P. putida under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptac promoter and the LacI repressor and introduced the lacIq-Ptac-sodAB expression cassette into the bacterial chromosome by using mini-Tn5-based plasmid pJMT6 (65). The genomic DNA of P. putida PaW85 was amplified with primers sodA1 and sodA2 to clone the sodA gene and with primers sodB1 and sodB2 to clone the sodB gene. The primers used for amplification are shown in Table 2, and the constructed plasmids are described in Table 1. First, the PCR product of the sodB gene was cloned into the EcoRV site of pBluescript KS(+) to obtain pKSsodB. Subsequently, the PCR product of the sodA gene was inserted into the SmaI site of pKSsodB, yielding plasmid pKSsodAB. To obtain the lacIq-Ptac-sodAB expression cassette, the XbaI- and SalI-generated DNA fragment carrying the sodAB genes was cloned into the pBRlacItac vector, resulting in plasmid pBRlacItacsodAB. The expression cassette was then subcloned within a BamHI-cleaved DNA fragment into pUC18Not (resulting in plasmid pUC18NotlacItacsodAB) to gain NotI sites for the cloning of this cluster into pJMT6. The resulting plasmid, pUTtellacItacsodAB, which does not replicate in hosts other than E. coli strain CC118 λpir, was conjugatively transferred into the P. putida WT strain and its rpoS-deficient derivative by using helper plasmid pRK2013. Transconjugants carrying random insertions of mini-Tn5 containing the lacIq-Ptac-sodAB expression cassette in the P. putida chromosome were isolated.
TABLE 2.
Oligonucleotides used in this study
| Purpose | Designation | Sequence and positiona |
|---|---|---|
| Cloning of P. putida sodA gene | sodA1 | 5′-CGCTGCCAAGCCGGATGT-3′, complementary to positions −32 to −15 upstream of ATG start codon of sodA |
| sodA2 | 5′-TTACTTCAGGGCTTCAAGGTA-3′, complementary to positions 610 to 627 downstream of ATG start codon of sodA | |
| Cloning of P. putida sodB gene | sodB1 | 5′-CGGCCTTGCGCAAACCGC-3′, complementary to positions −28 to −45 upstream of ATG start codon of sodB |
| sodB2 | 5′-TTAGGCCTTGAAGGTCTTGCC-3′, complementary to positions 547 to 591 downstream of ATG start codon of sodB | |
| Cloning of katA gene from pAM10.6 | katA1 | 5′-AATTTCTAGAAACGGGAGTTAATAGTATGAG-3′, complementary to positions −16 to + 5 relative to ATG start codon of katA |
| katA2 | 5′-TTAAGCGGCCGCTTAAAGAAAACTTGGTAAACCT-3′, complementary to positions 1499 to 1520 downstream of ATG start codon of katA |
Restriction enzyme sites are underlined (XbaI site for katA1 and NotI site for katA2).
To overexpress the catalase gene katA in P. putida (strain PaWRpoSKatA), the Ptac-katA transcriptional fusion was constructed and introduced into the attTn7 site of the P. putida chromosome by using a mini-Tn7-based integration system (34). To insert the gene for the LacI repressor into the P. putida chromosome, the Eco47III- and SmaI-generated DNA fragment containing the lacIq gene from plasmid pREP4 was inserted into pUC18Not to obtain pUC18NotlacI. The latter construct was cleaved with NotI to subclone this gene into mini-Tn5 delivery plasmid pUTmini-Tn5 Cm, yielding pUTCmlacI. Finally, the lacIq gene was introduced into the P. putida chromosome as a result of random insertions of mini-Tn5 as described above. The katA gene-containing DNA region of plasmid pAM10.6 (57) was amplified with primers katA1 and katA2 (Table 2). The PCR product was cut with XbaI and NotI and inserted into XbaI- and NotI-cleaved pBluescript (KS) derivative pKStac containing the Ptac promoter, resulting in plasmid pKStackatA. Plasmid pKStac was obtained by cloning the Ptac promoter from plasmid pACtac (71) within the BamHI- and HindIII-cleaved DNA fragment into pBluescript KS(+) opened with the same restriction endonucleases. The Ptac-katA transcriptional fusion was cut from pKStackatA with restriction endonucleases ApaI and NotI and inserted into Tn7-based mini-Tn-carrying vector pBK-miniTn7-ΩSm1 (34) opened with the same enzymes, yielding plasmid pAKNtackatA. A strain overexpressing both SodAB and KatA was constructed by using strain PaWRpoSKatA as a recipient.
Transconjugants were tested for resistance to carbenicillin. Only clones that were sensitive to carbenicillin represent true transposition event and not integration of the whole plasmid. The desired strains were confirmed by PCR analysis. To study the phenotypic effects of the constructed strains, we examined several clones carrying the same transcriptional fusions which gave the reproducible effects. P. putida carrying only the lacIq gene insertion into the chromosome was used as a reference when we compared the effects of overexpression of SOD or catalase on the survival and mutagenesis of rpoS-deficient bacteria.
Isolation and analysis of Phe+ revertants and Rifr mutants.
The assay system we used to record the frequencies of different types of point mutations in P. putida was based on activation of the phenol monooxygenase gene pheA, enabling bacteria to utilize phenol as a growth substrate and to form colonies on selective plates. The reporter gene pheA was altered in RSF1010-derived tester plasmids either by +1 frameshift mutation or by introducing a TAG translational stop codon into the pheA gene to monitor the occurrence of 1-bp deletions or base substitutions, respectively (74). Conditions for the isolation of phenol-degrading Phe+ revertants were the same as in our previous studies (67, 74). The viability of bacteria was determined on the same plates that were used for the isolation of Phe+ revertants. An approximately 350-bp DNA region covering the area of the pheA gene containing potential reversion mutations was analyzed by DNA sequencing in the Phe+ revertants as described previously (74). The frequency of spontaneous Rifr mutations in independent growing cultures of the P. putida WT strain and its rpoS-deficient derivative was calculated per 1 × 109 cells by using the Lea-Coulson method of the median (37, 63). The same cultures were plated onto phenol minimal plates, and after 10 days of starvation, the cells were collected by resuspension from the agar surface and plated onto selective Luria-Bertani medium plates containing 100 μg/ml rifampin. The number of Rifr colonies was determined after 24 h of incubation, and the median value of the frequency of mutations was determined in cells that survived the long-term starvation conditions. Data were analyzed by a program for statistical analyses (Statgraphics Centurion XV; Statpoint Inc.). Differences between average accumulation rates of Phe+ mutants were analyzed with the Student t test. A chi-square test of independence was used to compare DNA sequencing results.
RESULTS
RpoS is involved in the avoidance of base substitution mutations in stationary-phase P. putida populations.
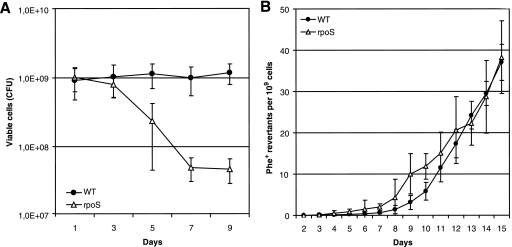
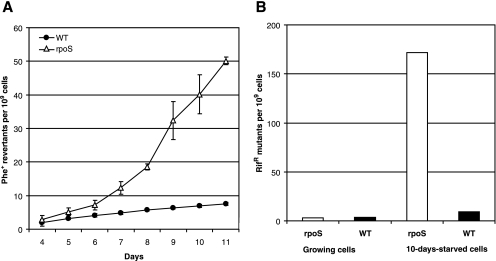
In order to study the involvement of RpoS in the occurrence of point mutations in long-term-starved populations of P. putida, we used two tester plasmids, pKTpheA56+A for the detection of restoration of the reading frame of the phenol monooxygenase gene pheA by −1 frameshift and pKTpheA22TAG to estimate various base substitutions eliminating the TAG stop codon from the pheA gene (74). Phenol minimal plates were inspected for the emergence of phenol-utilizing (Phe+) revertants each day, and the frequency of accumulation of Phe+ revertants per CFU determined from the same phenol minimal plates was calculated (Fig. 1A). The frequency of accumulation of 1-bp deletion mutants did not differ between the WT strain and its rpoS-deficient derivative (Fig. 1B). At the same time, as shown in Fig. 2A, the emergence of base substitution mutants was approximately 1.5-fold higher in the rpoS-deficient P. putida strain than in the WT strain during the initial period of starvation (days 4 to 6). After the first week of starvation, the frequency of accumulation of the Phe+ revertants increased in the rpoS-deficient P. putida strain and thereafter remained approximately up to seven times higher than that measured in the WT strain.
FIG. 1.
Effects of RpoS on P. putida survival under carbon starvation conditions and frequency of accumulation of 1-bp deletions. (A) Viability of P. putida WT strain PaW85 and its rpoS-deficient derivative on phenol minimal plates. (B) Accumulation of Phe+ revertants on phenol minimal plates in P. putida WT strain PaW85 and its rpoS-deficient derivative carrying plasmid pKTpheA56+A. About 5 × 108 P. putida cells from independent cultures grown overnight in liquid M9 medium were plated onto phenol minimal plates. Data for at least five parallel experiments are presented. In all cases, means ± standard deviations (error bars) for 10 plates calculated per 1 × 109 cells are shown.
FIG. 2.
Effect of RpoS on the frequency of base substitution mutations in P. putida under starvation conditions. (A) Accumulation of Phe+ revertants on phenol minimal plates in P. putida WT strain PaW85 and its rpoS-deficient derivative (rpoS) carrying plasmid pKTpheA22TAG. About 5 × 108 P. putida cells from independent cultures grown overnight in liquid M9 medium were plated onto phenol minimal plates. Data for at least five parallel experiments are presented. Means ± standard deviations (error bars) for 10 plates calculated per 1 × 109 cells are shown. (B) Frequency of Rifr mutants in growing cells and cells of P. putida WT strain PaW85 and its rpoS-deficient derivative starved for 10 days. The frequency of spontaneous Rifr mutations in 50 independent growing cultures of the P. putida WT strain and its rpoS-deficient derivative was calculated per 1 × 109 cells by using the Lea-Coulson method of the median (37, 63). The same cultures were plated onto phenol minimal plates, and after 10 days of starvation, the frequency of Rifr mutations was determined in the cells that survived the long-term starvation conditions. Median values for 10 plates calculated per 1 × 109 cells are shown. In total, 100 and 200 plates were examined for Rifr mutants in populations of the WT and its rpoS-deficient derivative, respectively.
To examine how general the elevated rate of the generation of base substitutions in starving cells of the rpoS-deficient P. putida strain is, we compared the mutation frequencies in starving WT and rpoS-deficient P. putida by using a chromosomal rpoB gene encoding the β subunit of the RNA polymerase as a reporter. The results presented in Fig. 2B show that the median frequencies of Rifr mutants in growing cultures of the WT strain and its rpoS-deficient strain were similar at 3.7 × 10−9 and 3.0 × 10−9, respectively. After 10 days of starvation, the frequency of Rifr mutants in the rpoS-deficient P. putida strain increased more than 50-fold. Thus, the use of both plasmid-based and chromosomal test systems revealed that the absence of RpoS caused an elevated frequency of base substitutions in long-term-starved P. putida.
Elevated mutation frequency in starving rpoS-deficient P. putida is suppressed by artificial overexpression of SOD or catalase.
ROS such as superoxide, hydrogen peroxide, and hydroxyl radicals are constantly generated as by-products of aerobic metabolism. ROS can damage proteins, lipids, and nucleic acids and may lead to the death of cells. It is shown that expression of SOD is required for the survival of E. coli during the stationary phase (2). Importantly, the time period when the frequency of accumulation of base substitution mutants increased in rpoS-deficient P. putida coincided with the rapid death of about 95% of the cells in the population (Fig. 1A and 2A). Hence, we hypothesized that starving rpoS-deficient P. putida could be less protected against damage caused by ROS than the WT strain.
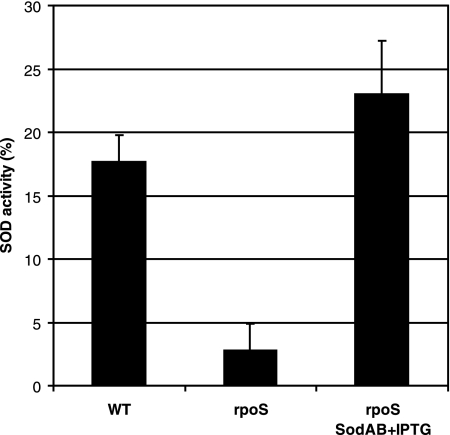
The results obtained by Park et al. (56) indicated that the response to superoxide stress in P. putida may be quite different from the SoxR mechanism in E. coli. However, the control mechanism has remained unknown. We compared SOD expression in stationary-phase cells of P. putida WT and rpoS-deficient strains. The results presented in Fig. 3 show an approximately fivefold decline in the level of SOD activity in the rpoS-deficient strain compared to that observed in the WT. These data suggest that SOD expression is under the positive control of RpoS in P. putida. Thus, it is possible that the reduced level of expression of SOD in starving rpoS-deficient P. putida cells is one of the mechanisms responsible for the accumulation of ROS and therefore causes a drop in viability and the elevation of mutations.
FIG. 3.
Effects of RpoS on the expression of SOD activity in carbon-starved P. putida. SOD activity was measured in cells of the WT strain, the rpoS-deficient strain (rpoS), and the rpoS-deficient strain overexpressing SodAB (rpoS SodAB plus IPTG) that were starved for 8 days. SOD activity is expressed as the percent inhibition of tetrazolium salt reduction to formazan dye (absorbance at 440 nm) by superoxide anion.
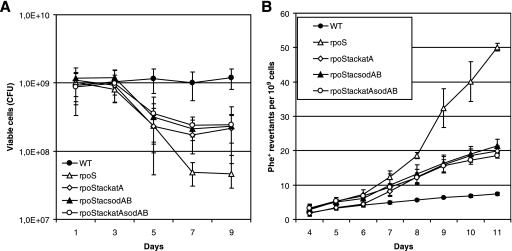
In order to examine the possibility that limitation of SOD in rpoS-deficient bacteria can elevate the accumulation of base substitutions under the conditions of long-term-starvation of bacteria, we studied the effect of overexpression of this enzyme on the mutation frequency in starving bacteria. It is known that SOD is functional in P. putida stationary-phase cells as a SodAB heterodimer (26). Therefore, we constructed a chromosomal sodA (PP0946) and sodB (PP0915) overexpression cassette where these genes are expressed as a single transcriptional unit under the control of the IPTG-inducible Ptac promoter. The rpoS-deficient P. putida strain carrying the Ptac-sodAsodB expression cassette exhibited a level of SOD activity comparable to that measured in the WT strain (Fig. 3). Notably, the overexpression of SOD increased the survival of the rpoS-deficient P. putida strain about 10-fold during long-term-starvation (Fig. 4A). Also, the frequency of accumulation of Phe+ revertants was approximately 2.5-fold lower in this strain than in the rpoS-deficient strain lacking the SOD overexpression cassette (Fig. 4B). At the same time, SOD overexpression did not affect the mutation frequency in WT cells (data not shown). Thus, our results indicate that RpoS is important in the regulation of the sodA and sodB genes, protecting cells against oxidative damage, and that the increased amount of superoxide promotes mutagenesis in starving populations of P. putida. However, the limitation of SOD in the rpoS-deficient strain was not the only factor responsible for cell death and elevation of the mutation frequency.
FIG. 4.
Effects of overexpression of SOD and catalase on the survival of rpoS-deficient P. putida cells under carbon starvation conditions and on the frequency of accumulation of base substitution mutations. (A) Viability of the P. putida rpoS-deficient strain overexpressing catalase (rpoStackatA), SodAB (rpoStacsodAB), or both catalase and SodAB (rpoStackatAsodAB) on phenol minimal plates. The viability of the WT strain and its rpoS-deficient derivative without SOD or catalase overexpression is shown as well. (B) Accumulation of Phe+ revertants on phenol minimal plates in starving populations of the same strains shown in panel A. All strains carried the tester plasmid pKTpheA22TAG. Data for at least five parallel experiments are presented. In all cases, means ± standard deviations (error bars) for 10 plates calculated per 1 × 109 cells are shown.
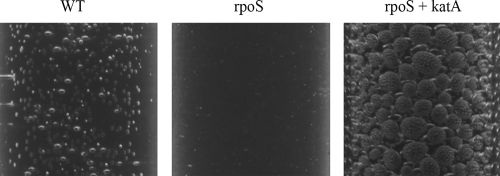
The expression of two catalases, KatA and KatB, is under the control of the OxyR regulator in P. putida (29). It is also known that RpoS positively regulates the expression of KatB (29, 48). Additionally, Ramos-González and Molin (62) have shown that the rpoS-deficient P. putida strain has an increased sensitivity to hydrogen peroxide in the stationary phase. We observed significant differences between the native levels of expression of catalase in starving WT and rpoS-deficient P. putida cells (Fig. 5). This indicated that limitation of catalase in starving rpoS-deficient P. putida cells may also affect the viability and mutagenic processes of bacteria. Therefore, we studied the effect of artificial overexpression of catalase on the survival of and the frequency of base substitutions in starving rpoS-deficient P. putida cells. For that purpose, we constructed the P. putida strain overexpressing katA-encoded catalase derived from P. fluorescens plasmid pAM10.6 (57). The gene was cloned under the control of the IPTG-inducible Ptac promoter and introduced into the P. putida chromosome. The overexpression of KatA was verified by the production of oxygen bubbles in a cell suspension after the addition of hydrogen peroxide. Remarkably, O2 production was significantly enhanced under the conditions of overexpression of P. fluorescens catalase KatA in a culture of rpoS-deficient bacteria grown overnight (Fig. 5), thereby confirming the finding of the previously published studies (57) that P. fluorescens KatA is a catalase which is functional in P. putida. We also confirmed that long-term-starved cells of the rpoS-deficient strain containing the P. fluorescens katA expression construct exhibit higher catalase activity than the same strain without this construct (data not shown). Similar to the effect of the overexpression of SOD, the IPTG-induced expression of this catalase increased the survival of starving cells lacking RpoS and reduced the frequency of accumulation of base substitution mutations (Fig. 4). Taken together, our results indicated that the limitation of SOD and catalase in starving rpoS-deficient P. putida is one of the most important factors resulting in the death of the majority of the cells and elevation of the mutation frequency in the surviving population under long-term carbon starvation conditions. Nevertheless, simultaneous overexpression of SOD and KatA catalase in the rpoS-deficient strain did not further improve cell survival and the frequency of mutations, thereby indicating that the defense mechanisms provided by SOD and catalase overexpression cannot fully prevent the death of cells under the conditions studied (Fig. 4A and B).
FIG. 5.
Detection of catalase expression in the P. putida in WT strain, its rpoS-deficient derivative (rpoS), and the rpoS-deficient strain overexpressing the catalase gene katA. Amounts of oxygen bubble formation were compared in cell suspensions of overnight cultures of these strains 1 min after the addition of a hydrogen peroxide solution.
The total amount of SO per plate is increased in rpoS-deficient bacteria.
As our results clearly demonstrated that overexpression of SOD or catalase in the P. putida rpoS-deficient strain increased the viability of cells and reduced the mutation frequency under conditions of long-term starvation, we decided to measure the cellular amount of SO in the WT strain and its rpoS-deficient derivative at the beginning of starvation (on day 2) and after prolonged starvation (on day 8). Cells from phenol minimal plates were resuspended in equal amounts of M9 buffer, and SO was measured in cleared lysates prepared from cell suspensions obtained directly from the plates (lysate I) and those prepared from cells washed with M9 buffer to get rid of lysed cells (lysate II). The results presented in Table 3 demonstrate that the amount of SO calculated per 106 CFU in the reaction mixtures was significantly increased in the case of rpoS-deficient bacteria. We detected a two- to threefold greater SO amount in this strain than in the WT strain when the cells of rpoS-deficient bacteria were starved for 2 days on minimal plates and a more-than-100-fold increase when the rpoS-deficient strain was starved for 8 days.
TABLE 3.
Amounts of superoxide in lysates and supernatants of cells of the P. putida WT strain and its rpoS-deficient derivative
| Strain (day of sampling) | Amt of superoxide/ plate | Amt of superoxide/106 CFUc |
|---|---|---|
| WT strain | ||
| Lysate Ia (2) | 35,956 ± 5,740 | 32.0 ± 10.1 |
| Supernatant I (2) | 11,509 ± 1,501 | 11.2 ± 2.1 |
| Supernatant II (2) | 561 ± 251 | 0.8 ± 0.07 |
| Lysate IIb (2) | 28,938 ± 4,772 | 33.0 ± 22.8 |
| Lysate Ia (8) | 32,677 ± 4,649 | 42.9 ± 9.8 |
| Supernatant I (8) | 14,910 ± 2,722 | 21.0 ± 6.2 |
| Supernatant II (8) | 1,829 ± 452 | 3.7 ± 1.0 |
| Lysate IIb (8) | 20,101 ± 1,888 | 39.7 ± 13.8 |
| RpoS-deficient strain | ||
| Lysate Ia (2) | 82,677 ± 11,407 | 111 ± 13.3 |
| Supernatant I (2) | 27,033 ± 2,780 | 43.8 ± 8.3 |
| Supernatant II (2) | 3,165 ± 1,324 | 7.4 ± 0.4 |
| Lysate IIb (2) | 36,667 ± 4,598 | 62.9 ± 12.1 |
| Lysate Ia (8) | 73,189 ± 11,972 | 8,832 ± 1,599 |
| Supernatant I (8) | 32,812 ± 6,370 | 4,302 ± 803 |
| Supernatant II (8) | 11,177 ± 5,446 | 1,861 ± 994 |
| Lysate IIb (8) | 28,103 ± 4,324 | 4,481 ± 1,062 |
Cells from agar plates sonicated within their resuspension buffer (see preparation of lysate I in Materials and Methods).
Cells sonicated after washing with M9 (see preparation of lysate II in Materials and Methods).
Relative amount of SO per 1 × 106 CFU in 1 ml reaction mixture; values for supernatant I and supernatant II are given per 1 × 106 CFU in 1 ml of cell suspension before and after washing of cells, respectively.
As already mentioned above, the number of CFU was reduced drastically during the prolonged starvation of rpoS-deficient bacteria (Fig. 1A). Therefore, it is possible that other cells (dead ones with cell wall integrity and those staying in a viable but nonculturable state) were sources of superoxide as well. Indeed, the total amounts of this oxygen radical measured per plate differed only twofold between the WT strain and its rpoS-deficient derivative. Interestingly, a large amount of SO was already detectable in the supernatant of cells that were centrifuged without sonication (supernatant I). Supernatants derived from the additional washing of the cells (supernatant II) also contained SO. The presence of large amounts of SO in supernatants I and II indicated that many cells derived from the surface of agar plates lysed during the initial resuspension and subsequent washing, thereby releasing SO into the environment. Additionally, there is a possibility that at least a part of the SO molecules measured in supernatants I and II diffused from surviving cells. This idea is supported by the results of measurement of SO in the WT strain grown overnight in liquid M9 medium plus glucose. When cells from the liquid culture were collected by centrifugation and washed with M9 buffer, about 20% of the total amount of SO measured in cells plus their washing solution was detectable in the washing solution alone (data not shown). Taken together, the results of our experiments indicate that the total amount of SO per plate is increased if bacteria lack RpoS whereas a large amount of SO is derived from cells which are either dead or not able to form colonies. It is difficult to say how much the intracellular amount of SO is increased in the surviving cells under the conditions studied. However, since the overexpression of SOD significantly increased the viability of rpoS-deficient cells during long-term carbon starvation (Fig. 4A), an inability to detoxify ROS seems to be a major reason for the death of the cells.
The spectrum of mutations characterized in starving RpoS-deficient bacteria is different from that identified in bacteria lacking the GO repair system.
Based on the results demonstrating that the decreased survival of rpoS-deficient P. putida under carbon starvation conditions is caused by limitation of enzymes responsible for detoxification of ROS, we supposed that the elevation of the mutation frequency under such conditions is caused by the incomplete removal of DNA lesions resulting from ROS attack. It is known that oxidative damage of DNA is an important source of mutation (9) and that the guanine oxidation product 7,8-dihydro-8-oxoguanine (GO) can give rise to mutations in starving E. coli (11, 12). Also, our previous results demonstrated that the absence of GO repair enzymes in starving P. putida leads to an increase in the mutation frequency and the characteristic spectrum of base substitution mutations (67). Therefore, in order to examine the possibility of whether the GO repair system could became saturated in rpoS-deficient P. putida under starvation conditions, we analyzed the pheA gene sequence of Phe+ colonies that were picked up from selective plates on days 7 to 10, when the mutation frequency was significantly elevated. We expected to see a pattern of mutations in the rpoS-deficient background similar to that observed in strains lacking the GO repair enzymes. Surprisingly, the analysis of the nucleotide sequence of the Phe+ revertants (Table 4) revealed that the spectrum of mutations identified in the rpoS-deficient strain was distinct from that identified by us previously in strains lacking either GO repair enzyme MutY or MutT (67). The proportions of G-to-T and A-to-C transversions, which were strictly increased in mutY- and mutT-deficient bacteria, respectively, were not significantly increased in the rpoS mutant strain (Table 4). These data indicate that the higher frequency of mutations in long-term-starved rpoS-deficient P. putida was not caused by saturation of the GO repair system due to accumulation of the guanine oxidation product GO.
TABLE 4.
Reversion of nonsense mutation (TAG) in Phe+ mutants accumulating in the P. putida WT strain and its RpoS-deficient derivative
| Change in TAG DNA sequence | No. of occurrencesa (% of total)
|
|||
|---|---|---|---|---|
| WT | mutT | mutY | rpoSb | |
| T → C | 164 (77) | 14 (8.3) | 24 (12) | 75 (40)c |
| T → G | 19 (9) | 34 (20) | 5 (2) | 25 (13) |
| T → A | 1 (0.5) | 0 | 0 | 4 (2) |
| G → T | 12 (5.6) | 1 (0.6) | 176 (85) | 18 (10) |
| A → C | 0 | 118 (70) | 0 | 1 (1) |
| A → G | 13 (6) | 2 (1.1) | 3 (1) | 48 (26)c |
| A → T | 4 (1.9) | 0 | 0 | 4 (2) |
| Deletions | 0 | 0 | 0 | 11 (6)c |
The spectra of mutations in the WT and GO repair-deficient strains are from reference 67.
Phe+ mutant colonies used for identification of stationary-phase mutations were picked up on days 7 to 15. We did not notice remarkable changes in the spectra of mutations in revertants derived from either the early or the late starvation period in the WT and GO repair-deficient derivative strains (67).
Statistically significantly different (P < 0.05) from WT strain value.
At the same time, the spectrum of mutations identified in the rpoS mutant background differed from that characterized in the WT strain. For example, the proportion of A-to-G transitions increased from 6% in the WT strain to 26% in the rpoS-deficient strain, whereas the proportion of T-to-C transitions decreased from 77% in the WT strain to 40% in the strain lacking RpoS. Interestingly, the absence of RpoS in starving P. putida stimulated the appearance of large in-frame deletions from the pheA sequence encompassing the introduced stop codon. Importantly, no deletions have been recorded with this test system in any of the other genetic backgrounds studied by us so far. In the case of rpoS-deficient bacteria, we identified one 69-nucleotide (nt)-long deletion, two identical 63-nt-long deletions, six identical 45-nt-long deletions, and one 30-nt-long deletion. One CTG deletion was found as well. Thus, as the absence of RpoS in starved P. putida affected the spectrum of mutations, certain types of DNA damage distinct from GO accumulate in starving RpoS-deficient P. putida.
DISCUSSION
The present study demonstrates that the absence of RpoS results in a significant elevation of base substitution mutations in starving P. putida. Additionally, the formation of some larger deletions was also accompanied by a lack of RpoS function in starving bacteria. The rise in the mutation frequency coincided with the death of the majority of rpoS-deficient cells after 5 to 7 days of starvation on minimal agar plates. Our results indicate that the decreased survival of carbon-starved rpoS-deficient P. putida on agar plates, i.e., in a structured environment, is caused by ROS. (i) SOD and catalase activities were reduced in starving P. putida lacking RpoS (Fig. 3 and 5). (ii) Overexpression of SOD or catalase KatA increased the survival of the rpoS-deficient strain by 10-fold and lowered the mutation frequency (Fig. 4A and B). Additionally, measurement of the amounts of SO indicated that rpoS-deficient cells have a reduced ability to detoxify ROS compared to that of WT cells (Table 3). The increase in the amount of SO in the case of the rpoS-deficient strain already became evident at the beginning of starvation (on day 2). Surprisingly, the total amounts of SO per plate measured in the rpoS-deficient strain on day 2 exceeded those measured on day 8. One possible explanation for this is that the number of CFU in populations of the rpoS-deficient strain starved for 8 days was reduced about 50-fold compared to that at the beginning of starvation and therefore the samples prepared on day 2 contained more SO-producing cells. At the same time, dead cells and those unable to form colonies could be a source of SO in our assays as well. Indeed, supernatants of the cells resuspended from agar plates (supernatant I in Table 3) contained a large amount of SO, whereas the SO values were the highest in the supernatants of the rpoS-deficient cells sampled on day 8. Unfortunately, direct measurement of SO in a fraction of viable (and colony-forming) cells in starved populations of rpoS-deficient bacteria was complicated due to the uncertainty of cell sorting by flow cytometry. The majority of rpoS-deficient cells derived from agar plates already stained with propidium iodide (PI) at the beginning of starvation, and the proportion of PI-stained cells even increased if the cells were washed before PI staining (data not shown). Although PI is frequently used to distinguish dead cells from viable ones because PI penetrates only bacteria with damaged membranes, bacteria having compromised membranes are not always dead (see, e.g., the instructions for LIVE/DEAD BacLight Bacterial Viability Kits; Molecular Probes, Invitrogen). Some of our unpublished results indicate that the cell membrane of rpoS-deficient P. putida is more permeable than that of the WT strain when the bacteria are incubated on agar plates. Additionally, many cells in long-term-starved populations of bacteria could be in a viable but nonculturable state, which complicates the separation of surviving and potentially mutating cells from the rest of the cells on selective plates even more.
Although the overexpression of SOD or catalase in the P. putida rpoS-deficient strain increased the survival of bacteria under carbon starvation conditions and reduced the mutation frequency in the surviving cells, the frequency of accumulation of Phe+ revertants was still about twofold higher than that estimated in the WT populations. Thus, it is possible that, in addition to the accumulation of superoxide and hydrogen peroxide, some other hazardous components accumulate in starving rpoS-deficient P. putida as well. One may imagine that the death of rpoS-deficient cells could be explained by changes in pH during the prolonged incubation of bacteria on selective plates. Since the growth medium of the bacteria was buffered with M9 solution, we did not expect to see pH changes. Indeed, direct measurement of pH on minimal agar plates after prolonged incubation of bacteria did not reveal any changes (data not shown).
The increase in the frequency of base substitutions in the cells that survived was observed in the cases of both the plasmid-based test system measuring Phe+ reversion and the chromosomal Rifr-based test system. This indicates that long- term-starved rpoS-deficient P. putida cells, or at least a subpopulation of these cells, have a genome-wide elevated mutability. GO is known to be one of the most stable and frequent base modifications resulting from oxygen radical attack on DNA (9). Bacteria have evolved a GO repair system to avoid mutations occurring due to GO (45). Analysis of the mutation spectrum of the Phe+ revertants in the P. putida rpoS-deficient strain did not reveal any similarity to the spectra identified by us previously in P. putida derivatives lacking GO repair enzyme MutT or MutY (67). While the lack of particular DNA repair functions specifically elevated certain types of mutations, A to C or G to T in strains lacking MutT or MutY, respectively (67), the mutation spectrum characterized in the rpoS-deficient strain was broader and more similar to that of the WT. These data imply that the GO repair system is functional in the rpoS-deficient strain.
The proportion of A-to-G transitions was increased in the absence of RpoS. Another significant dissimilarity from the WT strain was the occurrence of large deletions from the pheA sequence. Thus, it is possible that the occurrence of certain types of DNA damage not recognized by the GO repair system is responsible for these mutations in the rpoS mutant background. It is known that GO is highly susceptible to further oxidation products which are highly mutagenic (51). 2-Hydroxyadenine also induces mutations (33). Indeed, the results of our recent study imply that adenine oxidation products might also be an important source of mutation in starving bacteria (67). Additionally, aging cells are particularly subject to alkylation damage (73) and loss of enzymes that remove alkyl groups from damaged bases increases the mutation frequency in nongrowing bacteria (reviewed in reference 20). Our preliminary results indicate that long-term-starved rpoS-deficient bacteria accumulate DNA damage which is repaired by the nucleotide excision repair pathway (data not shown). The substrate repertoire of nucleotide excision repair extends from bulky lesions to other damage, such as certain oxidative base lesions, alkyl lesions, abasic (AP) sites, etc. (49). These results also support the idea that the accumulation of certain types of DNA damage produced by ROS and/or by methylation agents may facilitate the generation of mutations in rpoS-deficient P. putida. Furthermore, damage of DNA bases, if not repaired, and generation of AP sites due to limitation of AP endonuclease may cause accumulation of DNA strand breaks, thereby inducing RecA and facilitating recombination processes in starving cells. DNA synthesis occurring during recombinational repair may be error prone due to the involvement of DNA damage-induced specialized DNA polymerases. One may also hypothesize that the appearance of large deletions, especially in the rpoS-deficient background of P. putida, might be caused by ROS-stimulated recombination. Additionally, nonhomologous end joining (NHEJ), an essential pathway responsible for the repair of double-strand breaks and composed of Ku and a multifunctional DNA ligase (LigD), has recently been identified in many prokaryotes, including Pseudomonas species (58, 69). The DNA repair provided by the bacterial NHEJ system has been shown to be inaccurate, resulting in single nucleotide additions or deletions with various lengths at the break site (23, 41, 72). Recent studies also suggest that NHEJ acts under conditions where DNA replication is reduced or absent, such as in spores or in the stationary phase (59, 72, 77). Hence, it is possible that the deletions identified by us in this study were due to NHEJ taking place in starving cells of rpoS-deficient P. putida.
On the other hand, we suppose that the elevation of the mutation frequency in starved rpoS-defective P. putida cannot be caused by DNA damage only. This assumption is based on the findings that the rise in mutation frequency in the starving population of the rpoS-deficient strain coincided with the death of the majority of the cells and that the overexpression of SOD or catalase had stronger effects on the survival of the cells than on their mutation frequency. It is argued that the increased mutation frequency in stationary-phase cells does not limit the cellular life span and DNA does not seem to be the primary target of oxidative damage in the cells (53). Instead, the oxidative damage of proteins and membrane lipids is the major reason for aging and mortality (53). Bacterial stasis results in an increase in the oxidation and differential oxidation of target proteins (15). Oxidative modifications may inhibit or alter the activity of proteins, whereas carbonylated proteins have increased susceptibility to proteolysis (54). Studies by Dukan and Nyström (15) have demonstrated that protein carbonylation is increased in E. coli rpoS mutant cells starved for 1 to 2 days and that stasis-induced oxidation targets specific proteins. Importantly, the results presented by Dukan and Nyström (15) revealed that protein carbonylation can be mitigated by overproducing SOD. Also, some earlier studies have revealed that the lack of SOD or catalase activity in E. coli elevates the mutation frequency at doses of various oxidants that were nonmutagenic for WT cells (61, 64). Similar to the results of our studies, the contribution of GO to mutagenesis seemed to be negligible in the studies by Ruiz-Laguna et al. (64).
Oxidative damage of proteins is connected to errors in the translational machinery: damage to components of protein synthesis increases mistranslation, and vice versa, mistranslated proteins are more susceptible to oxidative damage (16). Mistranslation can be promoted by various factors in aging cells. For example, amino acid starvation can lead to a 10- to 100-fold increase in mistranslation (70). As first proposed in 1991 by Ninio (52) and later confirmed by others (3, 4, 5, 30), mistranslation of DNA repair and replication proteins can create a transient mutator phenotype. Hence, taking these results together, we suppose that, in addition to oxidative DNA lesions, mistranslation and oxidative damage of proteins may also affect DNA replication fidelity and thereby increase the mutation frequency in rpoS-deficient bacteria under the conditions studied. So far, direct evidence supporting this hypothesis is lacking. However, oxidative modification of replication proteins has recently been reported in eukaryotic systems (44, 50). Additionally, hypermutagenesis in E. coli mutA cells mistranslating aspartate as glycine due to a mutation in the glycine tRNA anticodon was mediated by modifications of DNA polymerase III due to elevated mistranslation (3).
The results of the present study (Fig. 1B) did not reveal any differences in the frequency of accumulation Pol IV-dependent 1-bp deletion mutants in starving P. putida WT and rpoS-deficient cells. In the light of the idea that genome-wide elevation of base substitutions occurs in long-term-starved rpoS-deficient P. putida as a result of an increased amount of ROS attacking both DNA and proteins, it is difficult to explain why the frequency of 1-bp deletions was not affected. DNA polymerase Pol IV has been shown to be upregulated by RpoS in starving E. coli cells (36). On the contrary, we found that the transcription of the Pol IV dinB gene in P. putida was not dependent on RpoS (74). However, we cannot exclude the possibility that RpoS indirectly controls the stability of Pol IV and/or the activity and access of Pol IV to the DNA replication machinery. If this is true, then two opposite effects, the reduced amount of Pol IV-provided error-prone DNA synthesis and increased mutagenesis due to ROS-induced damage may balance each other in starving rpoS-deficient cells. Further studies are needed to investigate this possibility.
To sum up, the results presented in this study indicate that the stationary-phase sigma factor RpoS is important for the survival of P. putida in the stationary phase. RpoS-deficient cells with limiting amounts of SOD and catalase activities (due to their positive control by RpoS) are more exposed to ROS on a solid surface under conditions of carbon starvation, and this may be lethal to bacteria because of the oxidative damage to proteins and the cell membrane. So far, the role of oxidative damage to proteins in DNA integrity has been underestimated. It certainly needs more thorough investigation. We suggest that the surviving population of carbon-starved rpoS-deficient P. putida has an elevated mutation frequency as a result of the accumulation of both DNA damage and a decline in DNA replication and repair fidelity due to oxidative damage of enzymes and/or errors occurring during the translation of proteins.
Acknowledgments
We thank Rita Hõrak and Marta Putrinš for their comments on the manuscript and Niilo Kaldalu and Dmitri Lubenets for their technical advice on characterizing populations of starving bacteria.
This work was supported by grants 5757 and 7376 from the Estonian Science Foundation, by funding of Targeted Financing Project TLOMR0031 from the Estonian Ministry of Research and Education, and by grant 55005614 from the Howard Hughes Medical Institute International Research Scholars Program.
Footnotes
Published ahead of print on 3 April 2009.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY.
- 2.Al-Maghrebi, M. A., and L. T. Benov. 2001. Polyphosphate accumulation and oxidative DNA damage in superoxide dismutase-deficient Escherichia coli. Free Radic. Biol. Med. 311352-1359. [DOI] [PubMed] [Google Scholar]
- 3.Al Mamun, A. A. M., S. Gautam, and M. Z. Humayun. 2006. Hypermutagenesis in mutA cells is mediated by mistranslational corruption of polymerase, and is accompanied by replication fork collapse. Mol. Microbiol. 621752-1763. [DOI] [PubMed] [Google Scholar]
- 4.Balashov, S., and M. Z. Humayun. 2002. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J. Mol. Biol. 315513-527. [DOI] [PubMed] [Google Scholar]
- 5.Balashov, S., and M. Z. Humayun. 2003. Escherichia coli cells bearing a ribosomal ambiguity mutation in rpsD have a mutator phenotype that correlates with increased mistranslation. J. Bacteriol. 1855015-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109167-168. [DOI] [PubMed] [Google Scholar]
- 7.Bayley, S. A., C. J. Duggleby, M. J. Worsey, P. A. Williams, K. G. Hardy, and P. Broda. 1977. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol. Gen. Genet. 154203-204. [DOI] [PubMed] [Google Scholar]
- 8.Bjedov, I., O. Tenaillon, B. Gérard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 3001404-1409. [DOI] [PubMed] [Google Scholar]
- 9.Bjelland, S., and E. Seeberg. 2003. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 53137-80. [DOI] [PubMed] [Google Scholar]
- 10.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41459-472. [DOI] [PubMed] [Google Scholar]
- 11.Bridges, B. A. 1993. Spontaneous mutation in stationary-phase Escherichia coli WP2 carrying various DNA repair alleles. Mutat. Res. 302173-176. [DOI] [PubMed] [Google Scholar]
- 12.Bridges, B. A., M. Sekiguchi, and T. Tajiri. 1996. Effect of mutY and mutM/fpg-1 mutations on starvation-associated mutation in Escherichia coli: implications for the role of 7,8-dihydro-8-oxoguanine. Mol. Gen. Genet. 251352-357. [DOI] [PubMed] [Google Scholar]
- 13.Chelikani, P., I. Fita, and P. C. Loewen. 2004. Diversity of structures and properties among catalases. Cell Mol. Life Sci. 61192-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Autréaux, B., and M. B. Toledano. 2007. ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. 8813-824. [DOI] [PubMed] [Google Scholar]
- 15.Dukan, S., and T. Nyström. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 123431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dukan, S., A. Farewell, M. Ballesteros, F. Taddei, M. Radman, and T. Nyström. 2000. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 975746-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, G., H.-C. T. Tsui, and M. E. Winkler. 1996. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J. Bacteriol. 1782388-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, P. L. 2000. Adaptive mutation: implications for evolution. Bioessays 221067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, P. L. 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42373-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, P. L., G. Gudmundsson, J. M. Trimarchi, H. Cai, and M. Goodman. 1995. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 927951-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galhardo, R. S., P. J. Hastings, and S. M. Rosenberg. 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42399-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong, C., P. Bongiorno, A. Martins, N. C. Stephanou, H. Zhu, S. Shuman, and M. S. Glickman. 2005. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 12304-312. [DOI] [PubMed] [Google Scholar]
- 24.Harris, R. S., G. Feng, K. J. Ross, R. Sidhu, C. Thulin, S. Longerich, S. K. Szigety, P. J. Hastings, M. E. Winkler, and S. M. Rosenberg. 1999. Mismatch repair is diminished during stationary-phase mutation. Mutat. Res. 43751-60. [PubMed] [Google Scholar]
- 25.Hassett, D. J., and M. S. Cohen. 1989. Bacterial adaptation to oxidative stress: implications for pathogenesis and interaction with phagocytic cells. FASEB J. 32574-2582. [DOI] [PubMed] [Google Scholar]
- 26.Heim, S., M. Ferrer, H. Heuer, D. Regenhardt, M. Nimtz, and K. N. Timmis. 2003. Proteome reference map of Pseudomonas putida strain KT2440 for genome expression profiling: distinct responses of KT2440 and Pseudomonas aeruginosa strain PAO1 to iron deprivation and a new form of superoxide dismutase. Mol. Microbiol. 51257-1269. [DOI] [PubMed] [Google Scholar]
- 27.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 161-178. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 28.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1726557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hishinuma, S., M. Yuki, M. Fujimura, and F. Fukumori. 2006. OxyR regulated the expression of two major catalases, KatA and KatB, along with peroxiredoxin, AhpC in Pseudomonas putida. Environ. Microbiol. 82115-2124. [DOI] [PubMed] [Google Scholar]
- 30.Humayun, M. Z. 1998. SOS and Mayday: multiple inducible mutagenic pathways in Escherichia coli. Mol. Microbiol. 30905-910. [DOI] [PubMed] [Google Scholar]
- 31.Ilves, H., R. Hõrak, and M. Kivisaar. 2001. Involvement of σS in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J. Bacteriol. 1835445-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imlay, J. A. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77755-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamiya, H. 2003. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 31517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch, B., L. E. Jensen, and O. Nybroe. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45187-195. [DOI] [PubMed] [Google Scholar]
- 35.Lange, R., and R. Hengge-Aronis. 1991. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 549-59. [DOI] [PubMed] [Google Scholar]
- 36.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea, D. E., and C. A. Coulson. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49264-265. [DOI] [PubMed] [Google Scholar]
- 38.Loewen, P. C., and B. L. Triggs. 1984. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J. Bacteriol. 160668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombardo, M. J., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch, M., and H. Kuramitsu. 2000. Expression and role of superoxide dismutases (SOD) in pathogenic bacteria. Microbes Infect. 21254-1255. [DOI] [PubMed] [Google Scholar]
- 41.Malyarchuk, S., D. Wright, R. Castore, E. Klepper, B. Weiss, A. J. Doherty, and L. Harrison. 2007. Expression of Mycobacterium tuberculosis Ku and ligase D in Escherichia coli results in RecA and RecB-independent DNA end-joining at regions of microhomology. DNA Repair 61413-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCann, M. P., J. P. Kidwell, and A. Matin. 1991. The putative σ factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 1734188-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenzie, G. J., P. L. Lee, M.-J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7571-579. [DOI] [PubMed] [Google Scholar]
- 44.Men, L., M. Roginskaya, Y. Zou, and Y. Wang. 2007. Redox-dependent formation of disulfide bonds in human replication protein A. Rapid Commun. Mass Spectrom. 212743-2749. [DOI] [PubMed] [Google Scholar]
- 45.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 1746321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 47.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 101-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura, K., S. Inouye, and A. Nakazawa. 1998. The rpoS gene regulates OP2, an operon for the lower pathway of xylene catabolism on the TOL plasmid, and the stress responses in Pseudomonas putida mt-2. Mol Gen. Genet. 25972-78. [DOI] [PubMed] [Google Scholar]
- 49.Møller, P., and H. Wallin. 1998. Adduct formation, mutagenesis and nucleotide excision repair of DNA damage produced by reactive oxygen species and lipid peroxidation product. Mutat. Res. 410271-290. [DOI] [PubMed] [Google Scholar]
- 50.Montaner, B., P. O'Donovan, O. Reelfs, C. M. Perrett, X. Zhang, Y. Z. Xu, X. Ren, P. Macpherson, D. Frith, and P. Karran. 2007. Reactive oxygen-mediated damage to a human DNA replication and repair protein. EMBO Rep. 81074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neeley, W. L., and J. M. Essigmann. 2006. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 19491-505. [DOI] [PubMed] [Google Scholar]
- 52.Ninio, J. 1991. Transient mutators: a semiquantitative analysis of the influence of translation and transcription errors on mutation rates. Genetics 129957-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nyström, T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58161-181. [DOI] [PubMed] [Google Scholar]
- 54.Nyström, T. 2005. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 241311-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ojangu, E.-L., A. Tover, R. Teras, and M. Kivisaar. 2000. Effect of combination of different −10 hexamers and downstream sequences on stationary-phase-specific sigma factor σS-dependent transcription in Pseudomonas putida. J. Bacteriol. 1826707-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park, W., S. Peña-Llopis, Y. Lee, and B. Demple. 2006. Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem. Biophys. Res. Commun. 34151-56. [DOI] [PubMed] [Google Scholar]
- 57.Peters, M., A. Heinaru, and A. Nurk. 2001. Plasmid-encoded catalase KatA, the main catalase of Pseudomonas fluorescens strain Cb36. FEMS Microbiol. Lett. 200235-240. [DOI] [PubMed] [Google Scholar]
- 58.Pitcher, R. S., A. J. Green, A. Brzostek, M. Korycka-Machala, J. Dziadek, and A. J. Doherty. 2007. NHEJ protects mycobacteria in stationary phase against the harmful effects of desiccation. DNA Repair 61271-1276. [DOI] [PubMed] [Google Scholar]
- 59.Pitcher, R. S., N. C. Brissett, and A. J. Doherty. 2007. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 61259-282. [DOI] [PubMed] [Google Scholar]
- 60.Ponder, R. G., N. C. Fonville, and S. M. Rosenberg. 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19791-804. [DOI] [PubMed] [Google Scholar]
- 61.Prieto-Álamo, M. J., N. Abril, and C. Pueyo. 1993. Mutagenesis in Escherichia coli K-12 mutants defective in superoxide dismutase or catalase. Carcinogenesis 14237-244. [DOI] [PubMed] [Google Scholar]
- 62.Ramos-González, M. I., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 1803421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 204-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruiz-Laguna, J., M.-J. Prieto-Álamo, and C. Pueyo. 2000. Oxidative mutagenesis in Escherichia coli strains lacking ROS-scavenging enzymes and/or 8-oxoguanine defenses. Environ. Mol. Mutagen. 3522-30. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Romero, J. M., R. Diaz-Orejas, and V. de Lorenzo. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 644040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saumaa, S., A. Tover, L. Kasak, and M. Kivisaar. 2002. Different spectra of stationary-phase mutations in early-arising versus late-arising mutants of Pseudomonas putida: involvement of the DNA repair enzyme MutY and the stationary-phase sigma factor RpoS. J. Bacteriol. 1846957-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saumaa, S., A. Tover, M. Tark, R. Tegova, and M. Kivisaar. 2007. Oxidative DNA damage defense systems in avoidance of stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 1895504-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma, R. C., and R. T. Schimke. 1996. Preparation of electrocompetent E. coli using salt-free growth medium. BioTechniques 2042-44. [DOI] [PubMed] [Google Scholar]
- 69.Shuman, S., and M. S. Glickman. 2007. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 5852-861. [DOI] [PubMed] [Google Scholar]
- 70.Sørensen, M. A. 2001. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 307785-798. [DOI] [PubMed] [Google Scholar]
- 71.Sørensen, M. A., J. Elf, E. Bouakaz, T. Tenson, S. Sanyal, G. R. Björk, and M. Ehrenberg. 2005. Over expression of a tRNALeu isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J. Mol. Biol. 35416-24. [DOI] [PubMed] [Google Scholar]
- 72.Stephanou, N. C., F. Gao, P. Bongiorno, S. Ehrt, D. Schnappinger, S. Shuman, and M. S. Glickman. 2007. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 1895237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taverna, P., and B. Sedgwick. 1996. Generation of an endogenous DNA-methylating agent by nitrosation in Escherichia coli. J. Bacteriol. 1785105-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tegova, R., A. Tover, K. Tarassova, M. Tark, and M. Kivisaar. 2004. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 1862735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsui, H.-C. T., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 1797476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van den Broek, D., T. F. C. Chin-A-Woeng, G. V. Bloemberg, and B. J. J. Lugtenberg. 2005. Role of RpoS and MutS in phase variation of Pseudomonas sp. PCL1171. Microbiology 1511403-1408. [DOI] [PubMed] [Google Scholar]
- 77.Wang, S. T., B. Setlow, E. M. Conlon, J. L. Lyon, D. Imamura, T. Sato, P. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 35816-37. [DOI] [PubMed] [Google Scholar]
- 78.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao, J., and M. E. Winkler. 2000. Reduction of GC → TA transversion mutation by overexpression of MutS in Escherichia coli K-12. J. Bacteriol. 1825025-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]