Abstract
A genetic screen for suppressors of bile sensitivity in DNA adenine methylase (dam) mutants of Salmonella enterica serovar Typhimurium yielded insertions in an uncharacterized locus homologous to the Escherichia coli asmA gene. Disruption of asmA suppressed bile sensitivity also in phoP and wec mutants of S. enterica and increased the MIC of sodium deoxycholate for the parental strain ATCC 14028. Increased levels of marA mRNA were found in asmA, asmA dam, asmA phoP, and asmA wec strains of S. enterica, suggesting that lack of AsmA activates expression of the marRAB operon. Hence, asmA mutations may enhance bile resistance by inducing gene expression changes in the marRAB-controlled Mar regulon. In silico analysis of AsmA structure predicted the existence of one transmembrane domain. Biochemical analysis of subcellular fractions revealed that the asmA gene of S. enterica encodes a protein of ∼70 kDa located in the outer membrane. Because AsmA is unrelated to known transport and/or efflux systems, we propose that activation of marRAB in asmA mutants may be a consequence of envelope reorganization. Competitive infection of BALB/c mice with asmA+ and asmA isogenic strains indicated that lack of AsmA attenuates Salmonella virulence by the oral route but not by the intraperitoneal route. Furthermore, asmA mutants showed a reduced ability to invade epithelial cells in vitro.
Bile is a fluid containing cholesterol, bile salts, phospholipids, proteins, bilirubin, and a variety of electrolytes (23). A fraction of the bile synthesized in the liver flows directly into the small intestine, while another fraction is stored in the gallbladder and released into the duodenum during food passage. About two-thirds (dry weight) of bile is made of bile salts, a family of molecules with steroid structure which derive from cholesterol (23). Aside from their role in digestion, bile salts have two distinct antibacterial activities, as detergents that disrupt the bacterial envelope and as DNA-damaging agents (5, 18, 39).
The mechanisms employed by Escherichia coli and Salmonella enterica to survive in the presence of bile are diverse and only partially understood (18) and involve a variety of cell functions: envelope structures that provide physical barriers to bile salts (31, 45, 55), efflux pumps that transport bile salts outside the cell (34, 38, 46, 52), and DNA repair functions that maintain genome integrity (39, 40). Resistance to bile is especially relevant in Salmonella physiology, since systemic infection leads to colonization of the hepatobiliary tract (35), where the concentration of bile is high and steady (23). Furthermore, the gallbladder is a major niche for Salmonella in chronic carriers of Salmonella enterica serovar Typhi (14). The ability of Salmonella to survive in the mammalian gallbladder reflects its ability to adapt to virtually any concentration of bile (18) and to form biofilms on the surface of gallstones (9, 42, 43).
A strategy that has proven useful to identify cellular functions required for bile resistance in S. enterica is the isolation of bile-sensitive mutants and the subsequent identification of the mutations involved. In certain cases, however, mutation identification does not permit straightforward inference of the mechanisms whose disruption causes bile sensitivity. An example of this kind is found in S. enterica mutants lacking DNA adenine methylase (Dam), which suffer pleiotropic virulence effects including extreme bile sensitivity (15, 21, 22, 44). However, a functional relationship between DNA methylation and bile resistance is by no means obvious. In such circumstances, a strategy of classical genetics that can identify genetic partners is suppressor analysis: mutations that suppress a mutant phenotype often affect genes involved in the process under study (20). In the case of Salmonella dam mutants, their extreme bile sensitivity makes suppressor analysis easy: plating of a dam-null mutant on a medium containing ox bile produces bile-resistant revertants that carry extragenic suppressor mutations (39). Using this strategy, a previous study showed that bile sensitivity in Salmonella dam mutants was suppressed by inactivation of the Dam-dependent mismatch repair system, MutHLS (39). Below we describe the characterization of a second, unsuspected class of suppressors of bile sensitivity in Salmonella dam mutants, involving loss of function in an S. enterica locus homologous to the asmA gene of Escherichia coli.
In E. coli, AsmA appears to be involved in preventing misfolding of outer membrane proteins (OMPs), but its precise role has not been established. Mutations in asmA were initially described as suppressors that permitted assembly of mutant OmpF proteins (29), a role later extended to mutant OmpC proteins (56). The idea of involvement of AsmA in the assembly of wild-type OMPs was, however, discarded (30). An interpretation was that the presence of AsmA might create an environment refractory to the assembly of mutant (misfolded) OMPs (11). In the absence of AsmA, a more permissive environment would thus permit mutant OMP assembly (11). For Salmonella enterica, the only AsmA reference in the literature is a recent study that found asmA among the genes required for survival of Salmonella enterica serovar Typhimurium in the swine gastric environment (4). We show that the asmA gene of S. enterica encodes an OMP. The absence of AsmA in the outer membrane enhances bile resistance in diverse genetic backgrounds, presumably by increasing marRAB expression. We propose that marRAB transcriptional activation in the absence of AsmA may be an indirect consequence of an envelope rearrangement associated with loss of this OMP. Another consequence of AsmA absence is attenuation of virulence by the oral route, which can be tentatively correlated with the reduced ability of S. enterica asmA mutants to invade cultured epithelial cells.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and plasmids.
The strains of Salmonella enterica used in this study (Table 1) belong to Salmonella serovar Typhimurium and derive from the mouse-virulent strain ATCC 14028. Exceptions are the LT2 derivatives TT1704 and TT10288, obtained from J. R. Roth, University of California, Davis, CA. E. coli DH5α was the standard host for recombinant plasmids (57). Plasmid pGE108 (Kmr) is a ColE1 derivative carrying a cea::lacZ fusion (47). pIZ1581 is a pBAD18 derivative expressing the S. enterica asmA gene from the arabinose-dependent pBAD promoter (see below). pIZ53 is a pUC19 derivative carrying the internal HindIII fragment of Tn5; this fragment includes the kanamycin resistance gene (26). Transductional crosses using phage P22 HT 105/1 int201 (48; G. Roberts, unpublished data) were used for strain construction operations involving chromosomal markers and for transfer of plasmids among Salmonella strains. The transduction protocol was described elsewhere (16). To obtain phage-free isolates, transductants were purified by being streaked on green plates (7). Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5.
TABLE 1.
Strains of Salmonella enterica serovar Typhimurium
| Strain | Genotype | Reference or source |
|---|---|---|
| SV4392 | dam201::Tn10dTc | 39 |
| SV4429 | wecD::MudJ | 45 |
| SV4536 | Δdam230 | 39 |
| SV4699 | phoP7953::Tn10 | Lab stock |
| SV4704 | asmA1::MudJ dam201::Tn10dTc | This study |
| SV4708 | asmA1::MudJ | This study |
| SV4742 | Δdam230/pIZ1581 | This study |
| SV4759 | wecA::Tn10dTc | This study |
| SV4813 | wecA::Tn10dTc asmA::MudJ | This study |
| SV4873 | trg::MudJ | 49 |
| SV4929 | wecD::MudJ asmA::MudQ | This study |
| SV5056 | asmA::Cmr | This study |
| SV5057 | asmA::Kmr | This study |
| SV5058 | dcd::Kmr | This study |
| SV5059 | udk::Kmr | This study |
| SV5061 | asmA::3× FLAG | This study |
| SV5062 | asmA::3× FLAG Δdam230 | This study |
| SV5084 | udk::Kmr Δdam230 | This study |
| SV5085 | dcd::Kmr Δdam230 | This study |
| SV5396 | asmA::Kmr Δdam230 | This study |
| SV5397 | asmA::Cmr Δdam230 | This study |
| SV5426 | asmA::Cmr/pGE108 | This study |
| SV5562 | ΔtolC::Cmr | This study |
| SV5577 | ΔtolC::CmrasmA::Kmr | This study |
| SV5736 | phoP7953::Tn10 asmA::Kmr | This study |
| SV5809 | ΔasmA Δdam230 | This study |
| SV5810 | ΔasmA Δdam230 marA::Kmr | This study |
| TT1704 | Δhis-9533 | J. R. Roth |
| TT10288 | hisD9953::MudJ his-9944::MudA | 24 |
Construction of pIZ1581.
The asmA gene of S. enterica ATCC 14028 was PCR amplified using primers 5′ CAT GGA GCT C CTG AGC CAT TCG GCG CAA TAC 3′ and 5′ CAT GT CTA GAG ATG TAA CGT CTG TTC GCC TG 3′. Underlined are the SacI and XbaI restriction sites used for digestion of the amplified fragment, which was then cloned onto pBAD18 (19).
Construction of asmA, dcd, udk, tolC, and marA mutants.
Targeted disruption of the asmA, dcd, udk, and marA genes was achieved by the procedure of Datsenko and Wanner (10). Antibiotic resistance cassettes introduced during construction were excised by recombination with plasmid pCP20 (10). The oligonucleotides used for disruption were as follows: for asmA, 5′ TGC CGG TCC ATT GAG GGT AGC ATG AGA CGA TTT CTG GTG TAG GCT GGA GCT GCT TC 3′ and 5′ CTC GCC ATC CGG CTC TGC CCT TTG GCC AAA AAC TAC CAT ATG AAT ATC CTC CTT AG 3′; for dcd, 5′ TAA GGG CTT GAT GCG CGA AAG GAG AAA GTG CCA TGC GTG TAG GCT GGA GCT GCT TC 3′ and 5′ GGC AGT ATT GCG CCG AAT GGC TCA GTC TTT ATC AAT CAT ATG AAT ATC CTC CTT AG 3′; for udk, 5′ TTA CAT CCA GGT TAA TCA GGT CGC TAA ATT TAT GAC GTG TAG GCT GGA GCT GCT TC 3′ and 5′ GGT TAT CAC TGA ACG GTA CAC AAT TCG CCA GAT TTA CAT ATG AAT ATC CTC CTT AG 3′; for tolC, 5′ AGC CAG GCA GAG AAC CTG ATG CAA GTT TAT CAG CAA GCA CGT GTA GGC TGG AGC TGC TTC 3′ and 5′ GGT CTG ATA AGC GCA GCG CCA GCG AAT AAC TTA TCA ATG CCA TAT GAA TAT CCT CCT TAG 3′; and for marA, 5′ CTT AAC GGC GGAC GAA GTG GCA ACG CTT GAG TAT TTG CTC TGT AGG CTG GAG CTG CTT CG 3′ and 5′ GTG CTC TTC GCG TGG CGC ATA AAC AAA CTA GTA GTT GCC CAT ATG AAT ATC CTC CTT AG 3′. The following external primers were employed for PCR amplification of the resulting alleles: for asmA::Kmr and ΔasmA, 5′ CAT GGA GCT CCT GAG CCA TTC GGC GCA ATA C 3′ and 5′ ATG AGC AAT ACG CGC CTT GAA G 3′; for dcd::Kmr, 5′ GTT CAG TGA TAA CCT GGT TAG 3′ and 5′ AGC GTC GTC AGA AAT CGT CTC 3′; for udk::Kmr, 5′ CCC TAT AAT TGC CGC GTT TG 3′ and 5′ GCC CTT AAA TCA AGC ACA TC 3′; for tolC::Cmr, 5′ TGG CGG ATT CTG CTA GAA TC 3′ and 5′ TGG CGG ATT CTG CTA GAA TC 3′; and for marA::Kmr, 5′ TAT CCC CGC TGG ATA TCA C 3′ and 5′ TCA GCG GAT GAG GCA TTA TG 3′. PCR amplification products were sequenced at the facilities of Sistemas Genómicos SL, Paterna, Valencia, Spain.
Medium and culture conditions.
Luria-Bertani broth (LB) was used in all experiments. Unless otherwise indicated, the carbon source was 0.2% glucose. Arabinose was used at the final concentration of 0.2%. Solid LB contained agar at a 1.5% final concentration. Green plates were prepared according to the method of Chan et al. (7), except that methyl blue (Sigma) was used instead of aniline blue. Antibiotics were used at the final concentrations described previously (53). Deoxycholic acid (sodium salt) was from Sigma Chemical Co., St. Louis, MO.
MICs of DOC and tetracycline.
Exponential cultures in LB were prepared as previously described. Samples containing around 3 × 102 CFU were transferred to polypropylene microtiter plates (Soria Genlab, Valdemoro, Spain) containing known amounts of sodium deoxycholate (DOC) or tetracycline. After 12 h of incubation at 37°C, growth was visually monitored. Assays were carried out in triplicate.
Mutagenesis with MudJ.
A P22 HT lysate grown on strain TT10288 was used to transduce TT1704, selecting kanamycin resistance (24). The recipient strain carried the nontransducible deletion Δhis-9533. Transductants were selected on LB supplemented with kanamycin. Kmr transductants were made phage free by replica printing (>2 times) to LB plates containing 10 mM EGTA. Pools of ∼500 colonies were prepared and lysed with P22. These pools were then used to transduce SV4392, and Kmr transductants were selected on plates containing 1% DOC (39).
Characterization of MudJ insertion sites in the Salmonella chromosome.
A genomic DNA preparation (500 ng) from the strain carrying the MudJ insert was digested with either SmaI plus SspI or SmaI plus EcoRV. The sizes of the resulting DNA fragments were determined by Southern hybridization, using the HindIII fragment of plasmid pIZ53 as a probe; this fragment contains the Tn5 kanamycin resistance gene, which is the same Kmr determinant carried by the MudJ element. The DNA fragments obtained, all 2 to 3 kb long, were treated overnight with T4 ligase at 15°C and PCR amplified with primers derived from the MudJ DNA sequence: 5′ AGC TGT GCT CGA CGT TGT CA 3′ and 5′ CGA ATA ATC CAA TGT CCT CC 3′. If unspecific amplification had occurred, its products were used as templates for a second round of PCR amplification using two additional MudJ primers: 5′ GAT CTG GAC GAA GAG CAT C 3′ and 5′ ATT GCA CTA CAG GTT GCA AG 3′. Whenever PCR amplification was successful, the amplification product was purified with the GFX PCR DNA and gel band purification kit (Amersham Biosciences) and cloned onto pGEM-T Easy (Promega Corporation, Madison, WI). Standard DNA sequencing was performed with T3 and T7 primers.
Tagging of the AsmA protein with a 3× FLAG epitope.
Addition of a 3× FLAG epitope tag at the 3′ end of the asmA gene was carried out using plasmid pSUB11 (Kmr, 3× FLAG) as template (54). An S. enterica chromosomal fragment containing the appropriate region of the asmA gene was PCR amplified using primers 5′ GGA AAT TTT CGC GGT AAC CAC AAT AAC GAG GAA GTC TAT GGT GTA GGC TGG AGC TGC TTC 3′ and 5′ CCA GAA TAG ACG CCA TGT CTT CAC TCT GGG ATT TGC GAA TCA TAT GAA TAT CCT CCT TAG 3′. The resulting PCR fragment was purified and used to electroporate an ATCC 14028 derivative carrying pKD46. Transformants were selected on LB-kanamycin. Incorporation of the 3× FLAG tag was verified by PCR amplification and DNA sequencing. The primers used for this amplification were 5′ AAT GAA GGA TGT CGG GCA TC 3′ and 5′ CGT GCC AGT AAC GTT CTT CG 3′.
Subcellular fractionation.
Bacteria were fractionated as described elsewhere (44). Briefly, bacteria were grown overnight in LB medium at 37°C with vigorous shaking (200 rpm) and spun down by centrifugation at 15,000 × g for 15 min at 4°C. These bacteria were suspended in cold phosphate-buffered saline (PBS; pH 7.4) buffer and either mixed with Laemmli buffer (total protein extract) or disrupted by sonication. Unbroken cells were further removed by low-speed centrifugation, 5,000 × g for 5 min at 4°C. The supernatant was centrifuged at high speed (200,000 × g, 15 min, 4°C), and the new supernatant was recovered as cytosol fraction. The pellet containing envelope material was suspended in PBS containing 1% Triton X-100. Upon incubation of this material for 1.5 h at 4°C, the sample was centrifuged at 15,000 × g for 30 min at 4°C. The supernatant contained mostly soluble inner membrane proteins. The insoluble fraction enriched in OMPs was prepared upon suspension of the pellet in PBS, pH 7.4. An appropriate volume of Laemmli buffer was added to all fractions, and upon heating (100°C, 5 min) and clearing by centrifugation (15,000 × g for 5 min at room temperature), samples were analyzed for protein content by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in Tris-Tricine buffer by using 8% or 10% acrylamide gels.
Western blotting.
The AsmA protein was detected using anti-FLAG M2 monoclonal antibody (1:10,000; Sigma Chemical Co.) and anti-mouse horseradish peroxidase-conjugated secondary antibody (Bio-Rad, Hercules, CA).
Quantitative reverse transcriptase PCR (RT-PCR; real-time PCR) and calculation of relative expression levels.
Salmonella RNA was extracted from exponential- and stationary-phase cultures using the SV total RNA isolation system (Promega Corporation, Madison, WI) according to the manufacturer's instructions. The quantity and quality of the extracted RNA were determined using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). To diminish genomic DNA contamination, the preparation was treated with DNase I (Turbo DNA free; Applied Biosystems/Ambion, Austin, TX). An aliquot of 0.6 μg of DNase I-treated RNA was used for cDNA synthesis using the high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA). Real-time PCRs were performed in an Applied Biosystems 7500 Fast Real-Time PCR System. Each reaction was carried out in a total volume of 15 μl on a 96-well optical reaction plate (Applied Biosystems) containing 7.5 μl Power SYBR green PCR Master Mix (Applied Biosystems), 6.9 μl cDNA (1/10 dilution), and two gene-specific primers at a final concentration of 0.2 μM each. Real-time cycling conditions were as follows: (i) 95°C for 10 min and (ii) 40 cycles at 95°C for 15 s and 60°C for 1 min. No-template controls were included for each primer set and template. Melting-curve analysis verified that each reaction contained a single PCR product. Reported gene expression levels were normalized to transcripts of ompA, a housekeeping gene that served as an internal control. Gene-specific primers, designed with Primer Express v2.0.0 software, were as follows: for ompA, 5′ TGT AAG CGT CAG AA CCG ATA CG 3′ and 5′ GAG CAA CCT GGA TCC GAA AG 3′; for marA, 5′ AGA GCA ACG AGC CCA TTC TC 3′ and 5′ GGG TCA ATG TTT GCT GTG AC 3′; and for acrA, 5′ TTT GCG CGC CAT CTT CCC 3′ and 5′ GAC GTG CGC GAA CGA AC 3′.
Virulence assays in mice.
Groups of three to four 8-week-old female BALB/c mice (Charles River Laboratories, Santa Perpetua de Mogoda, Spain) were inoculated with a 1:1 ratio of two strains. For oral inoculation, bacterial cultures were grown overnight at 37°C in LB without shaking. Each strain was grown overnight at 37°C in LB with shaking, diluted into fresh medium (1:100), and grown to an optical density at 600 nm of 0.3 to 0.6. Oral inoculation was performed by feeding the mice with 25 μl of saline containing 0.1% lactose and 108 bacterial CFU. Intraperitoneal inoculation was performed with 0.2 ml of saline containing 105 CFU. Salmonellae were recovered from spleen 48 h after inoculation, and CFU were enumerated on LB and on selective medium. A competitive index (CI) for each mutant was calculated as the ratio between the mutant and the wild-type strain in the output (bacteria recovered from the host after infection) divided by their ratio in the input (initial inoculum) (6). The “cancelled out” CI (COI) is the CI corresponding to mixed infections of double mutants with corresponding single mutant strains and is defined as the ratio between a double mutant strain and the corresponding single mutant in the output divided by their ratio in the input (6).
Invasion assay in HeLa epithelial cells with mixed bacterial strains.
HeLa cells (ATCC CCL2) were seeded with 5 × 104 to 8 × 104 CFU the day before the infection, using 24-well plates (Costar; Corning, New York, NY) and grown at 37°C and 5% CO2. Bacteria were grown overnight in LB at 37°C without shaking. A 1:1 or 10:1 mix of two bacterial strains was prepared in Dulbecco modified Eagle medium. The CFU of the two strains in the input were enumerated by plating a dilution series of the inoculum, using the appropriate antibiotic or the colony color to distinguish the strains. The bacterial mixture was added to HeLa cells to reach a multiplicity of infection of 50 bacteria per eukaryotic cell. Thirty minutes after the infection, cells were washed twice with PBS and incubated in fresh Dulbecco modified Eagle medium containing 100 μg/ml gentamicin for 90 min. Numbers of viable intracellular bacteria were obtained after lysis of infected cells with 1% Triton X-100 and plating on appropriate medium. Strain discrimination was achieved as described for the input. Infections were carried out in triplicate. The CI in invasion is defined as the ratio between the two strains in the output (intracellular bacteria recovered 2 hours after infection) divided by their ratio in the input (49).
Statistical analyses.
Each CI or COI value is the mean of at least three independent infections ± standard error. Student's t test was used to analyze every CI or COI. The null hypothesis was that mean CI was not significantly different from 1. Every COI was analyzed with two null hypotheses: (i) mean is not significantly different from 1 and (ii) mean COI is not significantly different from the CI of the corresponding single mutant. P values of 0.05 or less were considered significant. Student's t test was likewise used to analyze differences in MICs and in mRNA levels detected by quantitative RT-PCR.
β-Galactosidase assays.
Levels of β-galactosidase activity were assayed as described by Miller (27), using the CHCl3-sodium dodecyl sulfate permeabilization procedure.
Bioinformatic analysis.
Sequence alignment was carried out at http://www.ncbi.nlm.nih.gov/BLAST/. PPSearch (http://www2.ebi.ac.uk/ppsearch) was used to search for motifs in the asmA sequence against patterns in the PROSITE database. The SignalP 3.0 algorithm (http://www.cbs.dtu.dk/services/SignalP/) predicts the location of potential signal cleavage sites in amino acid sequences. Secondary structure prediction (helix, sheet, and coil) was carried out at http://www.bork.embl-heidelberg.de/SSCP/. Transmembrane segment predictions were made using both a sliding window hydropathy plot (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and the TMpred algorithm (http://www.ch.embnet.org/software/TMPRED_form.html) based on the analysis of naturally occurring transmembrane-spanning segments.
RESULTS
Trials for MudJ-induced, DOC-resistant revertants of a dam mutant.
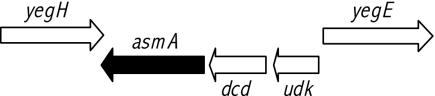
The inability of dam strains of S. enterica to grow on plates containing 1% DOC permitted the positive selection of isolates carrying extragenic suppressor mutations. MudJ insertions that suppressed the DOC-sensitive phenotype of a dam mutant (SV4392) were sought using this strategy. SV4392 was mutagenized with MudJ, and DOC-resistant Kmr mutants were selected on LB-DOC-kanamycin plates. Putative suppressor-carrying isolates were lysed with P22 HT, and the lysates were used to transduce SV4392, selecting Kmr. A 100% linkage between DOC resistance and Kmr resistance confirmed the existence of a suppressor mutation induced by MudJ. To identify the loci where the MudJ element had inserted, the boundaries of MudJ insertions were amplified by reverse PCR and sequenced. This procedure provided us with three independent MudJ insertions in asmA. In both E. coli and S. enterica, the asmA locus lies between yegH and dcd. DNA analysis in silico indicated that asmA is transcribed in the same orientation as that of the udk and dcd genes encoding uridine/cytidine kinase and dihydrouridine triphosphatase, respectively (Fig. 1). However, in silico analysis did not provide evidence that asmA, udk, and dcd might be part of the same transcriptional unit (see below).
FIG. 1.
Diagram showing known and putative genes in the 2208 to 2215 region of the chromosome of Salmonella enterica serovar Typhimurium. This region corresponds to centisome 45 on the genetic map. The diagram is based on the sequence of strain LT2, but the same gene arrangement is found in SL1344 and ATCC 14028 (data not shown).
Disruption of asmA caused a 20-fold increase in DOC resistance (Table 2). Only one asmA::MudJ insertion allele is included in Table 2 because all three showed identical suppressor abilities. Strains carrying null asmA alleles constructed by gene targeting (SV5056 and SV5057) had identical phenotypes (Table 2).
TABLE 2.
MIC of DOC in strains carrying dam, asmA, dcd, udk, phoP, wec, tolC, and marA mutations, alone or combined
| Strain | Genotype | MIC (g/100 ml) of DOC |
|---|---|---|
| ATCC 14028 | Wild type | 7 |
| SV4536 | Δdam230 | 0.2 |
| SV4708 | asmA1::MudJ | 9 |
| SV4704 | asmA1::MudJ dam201::Tn10dTc | 6 |
| SV5057 | asmA::Kmr | 9 |
| SV5396 | asmA::Kmr Δdam230 | 7 |
| SV5056 | asmA::Cmr | 9 |
| SV5397 | asmA::Cmr Δdam230 | 6 |
| SV5058 | dcd::Kmr | 4 |
| SV5085 | dcd::Kmr Δdam230 | 0.2 |
| SV5059 | udk::Kmr | 7 |
| SV5084 | udk::Kmr Δdam230 | 0.2 |
| SV4699 | phoP7953::Tn10 | 0.5 |
| SV5736 | phoP7953::Tn10 asmA::Kmr | 4 |
| SV4759 | wecA::Tn10dTc | 2 |
| SV4813 | wecA::Tn10dTc asmA::MudJ | 7 |
| SV4429 | wecD::MudJ | 1 |
| SV4929 | wecD::MudJ asmA::MudQ | 5 |
| SV5562 | ΔtolC::Cmr | 0.02 |
| SV5577 | ΔtolC::CmrasmA::Kmr | 0.02 |
| SV5809 | ΔasmA Δdam230 | 7 |
| SV5810 | ΔasmA Δdam230 marA::Kmr | 3 |
Suppression of bile sensitivity in dam mutants is specifically caused by asmA disruption.
Although no evidence exists that the clustered dcd, udk, and asmA genes might be part of the same transcriptional unit (30), the dcd and udk genes were disrupted, and the ability of dcd and udk mutations to suppress bile sensitivity in dam strains was tested. Neither mutation was able to increase resistance to DOC in a dam background (Table 2). In fact, disruption of dcd caused moderate sensitivity to DOC (Table 2). Lack of dUTPase is known to induce the SOS response as a consequence of replication impairment (37). Because bile salts may also impair DNA replication (39), it is not surprising that lack of dUTPase renders the cell bile sensitive. For the purpose of this study, however, the relevant observation was that neither udk nor dcd mutations were suppressors of bile sensitivity. A corollary was that udk and dcd knockouts were not polar on asmA, thus supporting the view that asmA may be part of an independent transcriptional unit as previously proposed for E. coli (30).
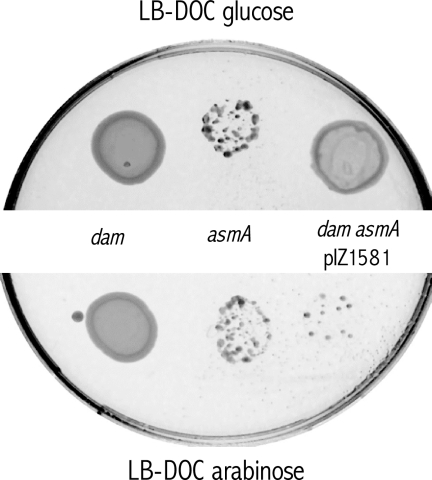
Additional support for the conclusion that the lack of AsmA suppressed bile sensitivity in a dam background was provided by complementation analysis using a previously described “drop” assay (44). A plasmid carrying the asmA gene under the control of the arabinose-inducible pBAD promoter (pIZ1581) was constructed for this purpose. Arabinose-induced asmA expression did not impair growth of the wild type (ATCC 14028/pIZ1581) in LB (data not shown). Plasmid pIZ1581 was introduced in a dam asmA strain (SV4704), and growth on LB-DOC plates containing either glucose or arabinose was tested. As shown in Fig. 2, the dam asmA derivative carrying pIZ1581 grew in the presence of glucose but not in the presence of arabinose, indicating that expression of the plasmid-borne asmA gene restored bile sensitivity.
FIG. 2.
Complementation analysis of DOC sensitivity in an asmA mutant. Ten microliters of the appropriate dilutions of exponential cultures of the wild-type and mutant strains, each containing 104 CFU, was incubated for 24 h at 37°C in an LB plate containing 1% DOC and 0.2% glucose (top) or an LB plate containing 1% DOC and 0.2% arabinose (bottom). Strains were ATCC 14028 (wild type), SV4536 (Δdam230), and SV4742 (Δdam230/pIZ1581).
Expression of the asmA gene is not under Dam methylation control.
Because the asmA gene conferred bile sensitivity when expressed from a heterologous promoter, we considered the possibility that asmA expression might be under Dam methylation control. Specifically, repression of asmA by Dam might be required for bile resistance, and uncontrolled expression might occur in dam mutants, resulting in bile sensitivity. This possibility was examined by comparing the expression of a MudJ-induced transcriptional asmA::lac fusion in dam+ and dam hosts (strains SV4708 and SV4704, respectively). The fusion showed similar β-galactosidase activities in the two backgrounds: 120 ± 18 Miller units in SV4708 and 138 ± 28 Miller units in SV4704. Hence, asmA transcription does not seem to be under Dam methylation control. This conclusion is further supported by a transcriptomic study which detected similar levels of asmA mRNA in dam+ and dam hosts (2).
Lack of AsmA suppresses bile sensitivity in phoP and wec mutants.
The ability of asmA mutations to suppress bile sensitivity in dam mutants raised the question of whether suppression was dam specific or broader. This issue was investigated by constructing asmA derivatives in other S. enterica mutants known to be bile sensitive, such as phoP (55) and wec (45) strains. Albeit it was less efficient than in dam asmA mutants, suppression was observed in both phoP asmA and wecA asmA strains (Table 2). Introduction of an asmA mutation in the wild type also caused a small but significant increase in the MIC of DOC (Table 2).
Lack of AsmA causes transcriptional activation of the marRAB operon but does not affect acrAB expression.
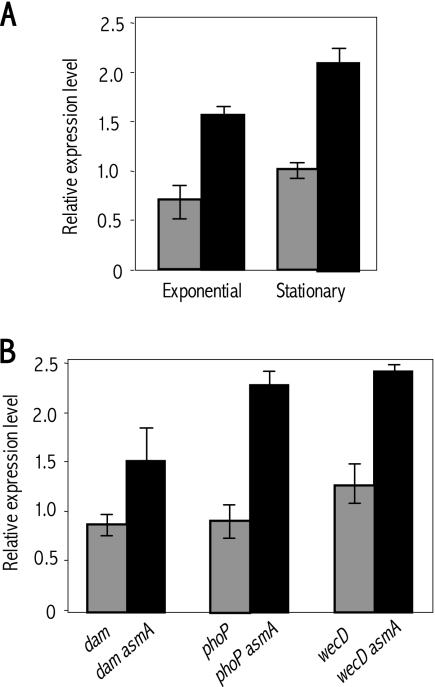
The observation that asmA mutations behaved as broad suppressors of bile sensitivity suggested the potential involvement of a cell response able to exert its effects in a variety of genetic backgrounds. One response of this kind in enteric bacteria is transcriptional activation of the marRAB operon, which in turn controls the so-called “Mar regulon” involved in resistance to multiple toxic substances including bile (1). On these grounds, we investigated whether lack of AsmA increased the level of marA mRNA. Quantitative RT-PCR data shown in Fig. 3 unambiguously indicated that asmA mutants contained higher levels of marA mRNA than did the wild type (Fig. 3A). Increased marA mRNA levels were also found in pairwise comparisons between dam, phoP, and wec mutants and isogenic asmA derivatives (Fig. 3B). These observations provided evidence that lack of AsmA activates marRAB expression. Because marRAB activation is known to enhance bile resistance in S. enterica (41), we tentatively correlated marRAB activation with the ability of asmA mutations to behave as suppressors and enhancers of bile resistance. Furthermore, the ability of asmA mutations to suppress bile sensitivity in dam mutants was greatly reduced in the absence of MarA (Table 2), thus providing additional evidence that asmA mutations confer bile resistance by activating the Mar regulon.
FIG. 3.
(A) Relative amounts of marA mRNA in exponential and stationary cultures of the wild type (ATCC 14028, gray histograms) and of an asmA mutant (SV5057, black histograms). RNA amounts were normalized using ompA mRNA as a control. (B) Relative amounts of marA mRNA, normalized to ompA mRNA, in exponential cultures of dam, phoP, and wecD mutants and in isogenic asmA derivatives. The strains used were as follows: dam (SV4536), dam asmA (SV4704), phoP (SV4699), phoP asmA (SV5736), wecD (SV4429), and wecD asmA (SV4929) strains. In both panels, bars represent averages and standard deviations from three independent experiments.
Quantitative RT-PCR experiments were likewise performed to monitor expression of the acrAB operon in asmA+ and asmA strains. AcrAB-TolC is an efflux pump regulated by MarRAB-dependent and MarRAB-independent mechanisms (41, 46) and previously shown to enhance bile resistance in S. enterica (41). The amounts of acrA mRNA in the wild type and in an asmA mutant were thus compared. Statistical analysis indicated that the small differences found were not significant. Even more clear was the absence of differences in acrA mRNA content in pairwise comparisons between dam and dam asmA, phoP and phoP asmA, and wecD and wecD asmA strains (data not shown). Altogether, these results indicated that lack of AsmA does not increase acrAB transcription and suggested that bile resistance mediated by asmA mutations may involve MarRAB-regulated genes other than acrAB. The observation that suppression of bile sensitivity by asmA mutations requires TolC (Table 2) suggests that TolC-dependent efflux pumps other than AcrAB may be involved in MarRAB-mediated bile resistance. An alternative possibility is that tolC mutations may cause an envelope defect that does not permit suppression by asmA mutations.
Lack of AsmA does not increase resistance to tetracycline.
The finding that asmA mutations activated expression of marRAB but not acrAB was at first sight perplexing, given the ability of the AcrAB efflux pump to enhance bile resistance (41). Hence, we examined whether asmA mutations affected tetracycline resistance, another well-known phenotype associated with AcrAB expression (36). The MIC of tetracycline hydrochloride was 0.5 mg/liter for ATCC 14028 and 0.4 mg/liter for an isogenic asmA mutant (SV5057). This observation provided further evidence that lack of AsmA does not activate acrAB expression.
Lack of AsmA reduces SOS induction by DOC.
Previous studies had shown that exposure of S. enterica to bile salts induces the SOS response (39, 40). To investigate whether suppression of bile sensitivity by asmA mutations was accompanied by reduced bile-induced DNA damage, induction of the SOS system was monitored using a cea::lacZ fusion carried on pGE108 (47). The β-galactosidase activities of the cea::lacZ fusion were 749 ± 45 Miller units in ATCC 14028/pGE108 versus 319 ± 20 Miller units in an isogenic asmA derivative, SV5426 (means and standard deviations of three independent experiments). Reduced SOS induction in the presence of DOC further supports the view that lack of AsmA enhances bile resistance.
In silico analysis of AsmA protein structure.
Sequence alignment using BLAST (NCBI database) detected no relatives of the AsmA protein with known function up to an E value of e−8 (data not shown). To characterize structural features of the S. enterica AsmA gene product (accession number gi29141254, NCBI), secondary structure content (percentages of alpha-helix, beta-sheet, and coil) was predicted based on both amino acid sequence and composition. The results indicated that the secondary structure of AsmA is characterized by high alpha-helix and coil content (47.0 and 41.8%, respectively) and low beta-sheet content (11.8%). In turn, in silico analysis of AsmA supersecondary structure (transmembrane segments, signal peptides, and motifs) suggested that AsmA has one transmembrane segment, from amino acid 5 to amino acid 26; an N-terminal signal peptide; and one cleavable site between amino acids 25 and 26 (VLL-VN). These results imply that AsmA may be a secreted protein. A signal typical of secreted proteins has been also described for the AsmA amino acid sequence of E. coli (30).
The Salmonella enterica AsmA protein localizes in the outer membrane.
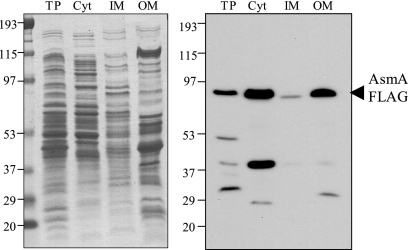
Construction of an AsmA protein derivative tagged with a 3× FLAG epitope permitted the detection of AsmA by Western immunoblot analysis. The tagged AsmA variant proved to be functional, since the MIC of DOC for strain SV5062 (asmA::3×FLAG dam) was identical to that for the dam strain SV4536 (data not shown). An electrophoretic separation of cell fractions (cytosol, cytoplasmic membrane, and outer membrane) is shown in Fig. 4 (left panel). Western analysis of the separated protein preparations was carried out with a commercial anti-FLAG antibody. The results unambiguously showed that AsmA is located in the S. enterica outer membrane (Fig. 4, right panel). This result involves a discrepancy with a previous study indicating that AsmA is a cytoplasmic membrane protein in E. coli (30).
FIG. 4.
Distribution of the AsmA protein tagged with a 3× FLAG epitope in subcellular fractions of S. enterica serovar Typhimurium (strain SV5061). Coomassie blue-stained proteins (left) and anti-FLAG Western hybridization (right) are shown for the following fractions: total protein (TP), cytosol (Cyt), inner membrane (IM), and outer membrane (OM). The volume loaded for all fractions was normalized to the same number of bacteria (5 × 107 CFU). The positions of the AsmA-FLAG protein and the prestained molecular mass standards in kDa are indicated.
Lack of AsmA causes attenuation of virulence in mice by the oral route.
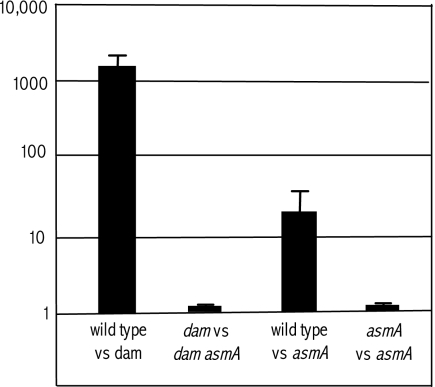
A previous study showed that mutHLS mutations, which suppress bile sensitivity in dam mutants of Salmonella, caused partial relief of virulence attenuation: dam mutHLS mutants were found to be >100 times more virulent than the parental dam strain (39). To investigate whether suppression of bile sensitivity by asmA mutations likewise resulted in attenuation relief, we compared the virulence of a dam mutant with that of a dam asmA mutant. Mixed infections were carried out in BALB/c mice upon inoculation by the oral route, and a COI was calculated. As a control, the CI of the dam mutant versus the wild type was also calculated. To facilitate colony identification after recovery of bacterial cells from the animal spleen, strain SV4873 (trg::MudJ) (49) was used as the wild type instead of ATCC 14028. Null trg mutations have been previously shown to be neutral for Salmonella virulence (49). Competitive infection experiments, summarized in Fig. 5, indicated that lack of AsmA does not relieve attenuation in a dam strain despite the fact that an asmA mutation renders dam mutants bile resistant (Table 2). This paradox suggested that lack of AsmA might impair Salmonella virulence on its own. This possibility was confirmed in mixed infections using SV4873 (trg) and SV4708 (asmA): the CI of the asmA mutant was found to be 30- to 40-fold lower than that of the virulent strain SV4873 (Fig. 5). As a control, strains carrying two different asmA alleles (SV4708 and SV5057) were also subjected to competitive infection of mice. No difference was found between the asmA::MudJ allele and the asmA::Cmr allele (Fig. 5), providing further evidence that lack of AsmA causes oral attenuation. Mixed infections by the intraperitoneal route provided small, nonsignificant differences between SV4873 (trg) and SV4708 (asmA) (data not shown), suggesting that asmA mutations may hamper S. enterica serovar Typhimurium virulence in the mouse intestine but not in further stages of infection.
FIG. 5.
CI and COI analysis of dam, asmA, and dam asmA strains after oral infection of BALB/c mice. Mixed infections were as follows: wild-type strain (SV4873)/dam strain (SV4536), dam strain (SV4536)/dam asmA strain (SV4704), wild-type strain (SV4873)/asmA strain (SV4708), and asmA strain (SV4708)/asmA strain (SV5057). The CIs represented are the means from four infections. Error bars represent the standard deviations.
Lack of AsmA causes an invasion defect.
Because lack of AsmA does not affect growth in standard media, we reasoned that attenuation of asmA mutants by the oral route might reflect impaired interaction of bacteria with animal tissues. To test this hypothesis, we compared the ability of an asmA mutant to invade epithelial (HeLa) cells with that of a fully invasive strain, SV4873. CI analysis upon mixed infection with SV4708 (asmA) and SV4873 (trg) indicated that the CI for the asmA mutant was around 0.2. Hence, lack of AsmA causes a partial impairment for invasion of nonphagocytic cells, indicating that the presence of AsmA in the S. enterica outer membrane may be required to trigger efficient bacterial uptake by nonphagocytic cells.
DISCUSSION
The ability of asmA mutations to enhance bile resistance in various genetic backgrounds suggested the involvement of a general mechanism such as mutational activation of a defense response. Note that the causes of bile sensitivity in dam, phoP, and wec mutants of S. enterica are diverse, perhaps unrelated (31, 39, 44, 45), and that enhanced bile resistance is also observed upon introduction of asmA null alleles in the wild type (Table 2). We provide evidence that the general mechanism by which asmA mutations enhance bile resistance in diverse genetic backgrounds involves transcriptional activation of the marRAB operon (Fig. 3). Initially identified in E. coli (17) and later in S. enterica (51), the marRAB operon plays a major role in the response of enteric bacteria to toxic substances (1). One of its products, MarA, is a transcriptional regulator that controls multiple genes which constitute the so-called “Mar regulon” (3). In S. enterica, transcription of marAB is activated by bile in a dose-dependent manner (41). This activation confers bile resistance, presumably via MarA-mediated activation of acrAB and other unidentified genes of the Mar regulon (41). Our observation that asmA mutants undergo increased marRAB expression can likewise explain their ability to increase bile resistance.
The mechanism of marRAB activation in asmA mutants remains to be studied. Bioinformatic analysis indicates that AsmA is unrelated to any known transport protein. A dual role of AsmA as a membrane component and a transcriptional regulator cannot be ruled out (32) but seems a priori unlikely because a putative DNA-binding domain is not found in AsmA. Hence, a tentative hypothesis is that lack of AsmA may cause an outer membrane reorganization that directly or indirectly results in marRAB activation. The possibility that AsmA absence causes changes in the outer membrane has been previously proposed to explain the phenotypes of E. coli asmA mutants (11, 56). On the other hand, the Mar regulon is known to respond to many kinds of stimuli, including intake of antibiotics, oxidative stress, and metabolic signals (3, 12, 28). Hence, it is conceivable that the absence of AsmA in the outer membrane may generate a signal that results in Mar regulon activation. An analogy may be found in the RcsBCD signaling system (25), which is activated by external stimuli but also responds to envelope perturbations caused by mdo mutations (13).
The mechanism by which marRAB activation in asmA mutants enhances bile resistance remains also to be identified. MarA regulates, either up or down, at least 60 genes (3). Hence, bile resistance in asmA mutants might result from activation or repression of genes of the Mar regulon. Although OmpF is known to be under MarRAB control (1), the possibility that bile resistance in asmA mutants might result from reduced OmpF synthesis was ruled out since a previous study had shown that lack of AsmA does not affect assembly of wild-type OMPs in E. coli (30). The list of genes under MarA control includes also the components of an efflux pump belonging to the RND family (AcrAB-TolC), which is known to transport bile salts (46). However, asmA mutations do not increase transcription of acrA, suggesting that increased bile resistance in asmA mutants is caused by altered expression of Mar-regulated genes other than acrAB. The fact that asmA mutations activate transcription of marRAB but not of acrAB is curious but not unusual: regulation of acrAB expression is known to be complex and multilayered (33, 46). A tentative hypothesis is that asmA mutations might activate a MarA-regulated efflux pump hitherto unknown to transport bile salts. A more speculative possibility is that lack of AsmA might activate one or more genes shared by the SoxRS and MarRAB regulons (28). In support of this view, the SoxRS regulon has been previously shown to be activated by bile salts (40), and asmA mutations reduce SOS induction by DOC.
A priori, any mutation that restores bile resistance in bile-sensitive mutants can be expected to favor survival of infecting Salmonella populations during intestinal passage, thus increasing their virulence by the oral route. However, asmA mutations failed to suppress oral attenuation in dam mutants (Fig. 5). This paradox was, however, solved when we observed that asmA mutations caused attenuation on their own: the CI of an asmA mutant by the oral route was found to be 30- to 40-fold lower than that of the wild type (Fig. 5). Hence, we tentatively infer that the benefits derived from increased bile resistance had been compensated (in fact, overrun) by other consequences associated with asmA mutations. In tests for invasion of epithelial cells, asmA mutants displayed a fivefold reduction in invasion capacity, suggesting that AsmA is required for optimal invasion of the intestinal epithelium.
If lack of AsmA causes indeed a major outer membrane rearrangement as proposed above and in a previous E. coli study (11), reduced invasion of epithelial cells might be a direct consequence of envelope alteration. Involvement of outer membrane components in Salmonella invasion has been previously described (8, 50). An alternative possibility is that uncontrolled activation of the Mar regulon might impair the interaction between Salmonella and epithelial cells. However, a previous study has shown that the marRAB operon is dispensable for virulence in the mouse model (51).
Acknowledgments
This work was supported by collaborative grants BIO2007-67457-CO2 and CSD2008-00013 (to F.G.-D.P. and J.C.) from the Spanish Ministry of Science and Innovation (MCINN). A.I.P. was supported by a predoctoral fellowship from the Fundación Ramón Areces. S.B.H. and I.C. are recipients of FPU fellowships from the MCINN. M.G.P. is an investigator of the Ramón y Cajal program of the MCINN. Y.O.'s sabbatical leave at the University of Seville was supported by a grant from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, Spain.
We are grateful to Javier López-Garrido for advice in the design of experiments, to Clara García-Calderón and Meritxell García-Quintanilla for helpful discussions, and to Modesto Carballo and Alberto García-Quintanilla of the Servicio de Biología (CITIUS, Universidad de Sevilla) for help in experiments performed at the facility.
Footnotes
Published ahead of print on 3 April 2009.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7410-413. [DOI] [PubMed] [Google Scholar]
- 2.Balbontín, R., G. Rowley, M. G. Pucciarelli, J. López-Garrido, Y. Wormstone, S. Lucchini, F. García-del-Portillo, J. C. D. Hinton, and J. Casadesus. 2006. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 1888160-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 1823467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearson, S. M., B. L. Bearson, and M. A. Rasmussen. 2006. Identification of Salmonella enterica serovar Typhimurium genes important for survival in the swine gastric environment. Appl. Environ. Microbiol. 722829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, C., H. Bernstein, C. M. Payne, S. E. Beard, and J. Schneider. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 3968-72. [DOI] [PubMed] [Google Scholar]
- 6.Beuzón, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 31345-1352. [DOI] [PubMed] [Google Scholar]
- 7.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology 50883-898. [DOI] [PubMed] [Google Scholar]
- 8.Cirillo, D. M., E. J. Heffernan, L. Wu, J. Harwood, J. Fierer, and D. G. Guiney. 1996. Identification of a domain in Rck, a product of the Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect. Immun. 642019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, R. W., D. L. Gibson, W. W. Kay, and J. S. Gunn. 2008. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 765341-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 906640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, M., and R. Misra. 1996. Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Mol. Microbiol. 21605-612. [DOI] [PubMed] [Google Scholar]
- 12.Domain, F., X. R. Bina, and S. B. Levy. 2007. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol. Microbiol. 66383-394. [DOI] [PubMed] [Google Scholar]
- 13.Ebel, W., G. J. Vaughn, H. K. Peters III, and J. E. Trempy. 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J. Bacteriol. 1796858-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edelman, R., and M. M. Levine. 1986. Summary of an international workshop on typhoid fever. Rev. Infect. Dis. 8329-349. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Del Portillo, F., M. G. Pucciarelli, and J. Casadesus. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 9611578-11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon, A., D. A. Cano, and J. Casadesus. 1995. Role of Erf recombinase in P22-mediated plasmid transduction. Genetics 140427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.George, A. M., and S. B. Levy. 1983. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 55541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2907-913. [DOI] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman, P. E., and J. R. Roth. 1973. Mechanisms of suppression. Adv. Genet. 171-105. [DOI] [PubMed] [Google Scholar]
- 21.Heithoff, D. M., E. I. Enioutina, R. A. Daynes, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. Salmonella DNA adenine methylase mutants confer cross-protective immunity. Infect. Immun. 696725-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284967-970. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, A. F. 1998. Bile secretion and enterohepatic circulation of bile acids, p. 937-948. In M. Feldman, B. F. Scharschmidt, and W. B. Sleisenger (ed.), Sleisenger and Fordtran's gastrointestinal disease, 6th ed. W. B. Saunders & Co., Philadelphia, PA.
- 24.Hughes, K. T., and J. R. Roth. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 1199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59379-405. [DOI] [PubMed] [Google Scholar]
- 26.Maldonado, R., A. Garzon, D. R. Dean, and J. Casadesus. 1992. Gene dosage analysis in Azotobacter vinelandii. Genetics 132869-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Miller, P. F., L. F. Gambino, M. C. Sulavik, and S. J. Gracheck. 1994. Genetic relationship between soxRS and mar loci in promoting multiple antibiotic resistance in Escherichia coli. Antimicrob. Agents Chemother. 381773-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misra, R. 1993. OmpF assembly mutants of Escherichia coli K-12: isolation, characterization, and suppressor analysis. J. Bacteriol. 1755049-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misra, R., and Y. Miao. 1995. Molecular analysis of asmA, a locus identified as the suppressor of OmpF assembly mutants of Escherichia coli K-12. Mol. Microbiol. 16779-788. [DOI] [PubMed] [Google Scholar]
- 31.Murata, T., W. Tseng, T. Guina, S. I. Miller, and H. Nikaido. 2007. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J. Bacteriol. 1897213-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muro-Pastor, A. M., P. Ostrovsky, and S. Maloy. 1997. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J. Bacteriol. 1792788-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, E., A. Yamaguchi, and K. Nishino. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 28324245-24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 1804686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52259-274. [DOI] [PubMed] [Google Scholar]
- 36.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Reilly, E. K., and K. N. Kreuzer. 2004. Isolation of SOS constitutive mutants of Escherichia coli. J. Bacteriol. 1867149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piddock, L. J. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 4629-636. [DOI] [PubMed] [Google Scholar]
- 39.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 1681787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prouty, A. M., I. E. Brodsky, S. Falkow, and J. S. Gunn. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150775-783. [DOI] [PubMed] [Google Scholar]
- 42.Prouty, A. M., and J. S. Gunn. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 717154-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 702640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pucciarelli, M. G., A. I. Prieto, J. Casadesús, and F. García-del-Portillo. 2002. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 1481171-1182. [DOI] [PubMed] [Google Scholar]
- 45.Ramos-Morales, F., A. I. Prieto, C. R. Beuzon, D. W. Holden, and J. Casadesus. 2003. Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 1855328-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 481609-1619. [DOI] [PubMed] [Google Scholar]
- 47.Salles, B., J. M. Weiseman, and G. Weinstock. 1987. Temporal control of colicin E1 induction. J. Bacteriol. 1695028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transducing abilities. Mol. Gen. Genet. 11975-88. [DOI] [PubMed] [Google Scholar]
- 49.Segura, I., J. Casadesus, and F. Ramos-Morales. 2004. Use of mixed infections to study cell invasion and intracellular proliferation of Salmonella enterica in eukaryotic cell cultures. J. Microbiol. Methods 5683-91. [DOI] [PubMed] [Google Scholar]
- 50.Shah, J., A. A. Fadl, G. R. Klimpel, D. W. Niesel, V. L. Popov, and A. K. Chopra. 2004. The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect. Immun. 723987-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sulavik, M. C., M. Dazer, and P. F. Miller. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 1791857-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 1792512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torreblanca, J., and J. Casadesus. 1996. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics 14415-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 9815264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 671614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong, X., J. N. Deeter, and R. Misra. 1996. Assembly-defective OmpC mutants of Escherichia coli K-12. J. Bacteriol. 1781213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vector. Gene 33103-119. [DOI] [PubMed] [Google Scholar]