Abstract
The characterization of transcriptional start sites of 14 genes encoded by the extremely AT-rich genome of “Candidatus Blochmannia floridanus” revealed a high degree of conservation with the RpoD promoter consensus sequence of the free-living relative Escherichia coli. Moreover, in agreement with the presence of the alternative heat shock sigma factor RpoH in “Ca. Blochmannia,” typical RpoH-dependent promoters were identified. However, no heat shock response resembling that of E. coli could be detected in “Ca. Blochmannia.”
In many cases obligate intracellular pathogens or mutualists have strongly size-reduced genomes which are also characterized by a very high AT content ranging from 70 to 80% (9, 16). “Candidatus Blochmannia” species are endosymbionts of carpenter ants which are required by the host for normal development and mainly contribute to nutritional upgrading of the host during larval and pupal stages (4, 13). With only 705 kb, the genome of “Candidatus Blochmannia floridanus,” the endosymbiont of Camponotus floridanus, is about six times smaller than that of the free-living relative Escherichia coli and it has an AT content of 74%, characteristic of endosymbiotic bacteria (5). Due to the obligate intracellular location in the cytosol of specialized cells, the bacteriocytes, in the midgut or in the ovaries, the bacteria are considered to encounter little environmental change. However, very little is known about gene-regulatory phenomena in “Ca. Blochmannia.” In a recent survey of transcription profiles of the endosymbiont during different developmental stages of the holometabolous host animals, only minor transcriptional changes were noted (15). This is in line with the fact that most of the transcription regulators present in free-living relatives within the Enterobacteriaceae have been lost in “Ca. Blochmannia floridanus” and only four dedicated transcription regulators and two sigma factors, RpoD and RpoH, were annotated (5).
Similarly, virtually nothing is known about the cis-acting regulatory sequences in “Ca. Blochmannia floridanus.” In other obligate cell-associated or intracellular bacteria exhibiting an extremely high AT content and reduced genomes such as pathogenic Mycoplasma pneumoniae or symbiotic “Candidatus Carsonella ruddii,” interesting genomic features were observed. In M. pneumoniae the majority of all genes are arranged in long convergent gene clusters with only very short intergenic distances, suggesting very long transcription units (7). In “Ca. Carsonella ruddii,” almost no intergenic regions are present and very long transcripts and massive translational coupling of the genes were suggested (1, 10). In contrast, in “Ca. Blochmannia floridanus” as well as in Buchnera aphidicola, the closely related obligate endosymbiont of aphids, overall gene organization resembles that of the free-living relative E. coli (5, 14). Here, numerous intergenic sequences of considerable length are present, and in many cases genes can be assigned to specific operons or consist of single transcription units (15). However, since RpoD-dependent promoters are AT rich, their reliable bioinformatic prediction is very difficult in these strongly AT-biased genomes. Thus, the question arises whether in such AT-rich genomes defined promoter sites are still used or whether RNA polymerase initiates transcription at a multitude of initiation sites randomly scattered over the entire genome. To investigate this question, we mapped the transcriptional start sites of several genes/operons of “Ca. Blochmannia floridanus” by primer extension analysis and defined a consensus sequence for RpoD-dependent promoters. Moreover, by bioinformatic analysis based on the E. coli consensus sequence, we identified four genes in the “Ca. Blochmannia floridanus” genome with putative RpoH-dependent heat shock promoters. We mapped their transcriptional start sites and defined a consensus sequence also for these promoters. Finally, we investigated whether “Ca. Blochmannia floridanus” is still able to mount a heat shock response like B. aphidicola (3, 17).
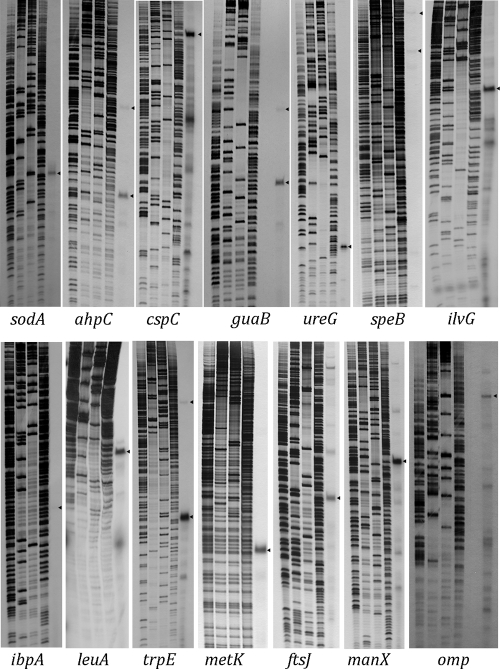
The results of the primer extension experiments with a series of genes/operons (ahpC, cspC, ftsJ, guaB, ibpA, ilvG, leuA, manX, metK, omp, sodA, speB, trpE, and ureG) are shown in Fig. 1. These factors were selected either because of the presence of relatively long nontranslated intergenic regions in front of the genes or because they are transcribed in the opposite direction from the upstream gene, suggesting the presence of specific promoters. Bacteria were isolated from their host animals as described recently by Stoll et al. (15), and primer extensions with bacterial whole-cell RNA were performed as described previously using the primers shown in Table 1 (6). In most cases one or a few defined transcription start points are found, usually with one transcript being most abundant (Fig. 1). In general, highly conserved −10 and −35 sequences are located at appropriate positions upstream of the dominant transcription start sites (Table 2). The alignment of these putative promoter sequences revealed a very good conservation with the RpoD consensus sequence of E. coli (Fig. 2). This indicates that, despite the extraordinary high AT content and many predictable transcriptional start sites, defined promoters are used by RpoD-containing RNA polymerase in “Ca. Blochmannia floridanus.” A similar result was obtained with four “Ca. Blochmannia floridanus” genes (groES, dnaK, lon, and grpE) known to be RpoH dependent in E. coli. These genes were chosen due to the presence of significant similarities in their upstream regions with the RpoH consensus sequence of E. coli. It is interesting that most of these genes belong to the most strongly expressed factors in “Ca. Blochmannia floridanus” (15). Again, well-defined transcriptional start sites were found in front of each gene which in all cases corresponded to the predicted promoters (Fig. 2 and 3; Table 2). This indicates that, despite the extreme difference in the AT contents of these genomes, the promoter structure of the RpoD- and RpoH-dependent promoters has been well conserved between “Ca. Blochmannia floridanus” and E. coli and that RNA polymerase quite selectively recognizes these promoters. However, of particular interest is the ibpA gene encoding a small heat shock protein which was found to be RpoH dependent and heat inducible in B. aphidicola strains from Acyrthosiphon pisum and Schizaphis graminum, where it contributes to thermotolerance in vivo (3, 17). In contrast, in “Ca. Blochmannia floridanus” this gene has experienced a change in its control elements since it is not preceded by any sequences resembling RpoH-dependent promoters but by a typical RpoD promoter, while in the case of its close relative “Candidatus Blochmannia pennsylvanicus,” the endosymbiont of the ant Camponotus pennsylvanicus, a possible RpoH-dependent promoter (5′-AGTGAAA-13 bp-AACCTTAT-50 bp-ATG-3′) may be used (2).
FIG. 1.
Mapping of putative RpoD-dependent promoters of “Ca. Blochmannia floridanus” by primer extension analysis. Briefly, 0.5 pmol of γ-32P-end-labeled oligonucleotides specific for the indicated genes (Table 1) was hybridized to 15 μg of “Ca. Blochmannia floridanus” RNA in reverse transcriptase buffer containing 0.4 mM deoxynucleoside triphosphates. cDNA synthesis was performed as described previously using avian myeloblastosis virus reverse transcriptase (6). cDNAs were subjected to gel electrophoresis on 6% urea-polyacrylamide gels. Sequencing ladders used as length standards were obtained by annealing the respective oligonucleotide to a plasmid containing the upstream region of the indicated genes. The arrowheads indicate transcriptional start sites preceded by sequences resembling the RpoD consensus sequence of E. coli (Table 2).
TABLE 1.
Primer sequences used for primer extension and qRT-PCR experiments
| Primer use and name | Primer sequence (5′→ 3′) |
|---|---|
| Primer extension | |
| ahpC_PE | CACCATTACCTAGTATTGTAGGAGCA |
| cspC_PE | TGCGGGTGTGATAAAACCAAACC |
| dnaK_PE | TCAATAATCGCAACACAAGAATTG |
| ftsJ_PE | CCTTAATTTTTGATTCTGTGCTTGTA |
| groES_PE | CCTCCAGAAGATTTCGATTCA |
| grpE_PE | CGTATTTGCAGAAGACGCATTA |
| guaB_PE | TCAAAAGTCAACGCCTCTTTTA |
| ibpA_PE | AACCTATAACAGAACCATACAAAGG |
| ilvG_PE | TAATCGCTCCCCCTGGAT |
| leuA_PE | AAGCTAGTTTGTAAGGATTGTTCTCC |
| lon_PE | CACATCACGCAGGGGTAAA |
| manX_PE | GTAATTGTTCAGATACTGTTCCATGAG |
| metK_PE | ATCTGGATGCCCTGCTGATA |
| speB_PE | CGGAAGTCTTAAAAATCCAAACG |
| omp_PE | AGCACCCGCAGTGCTTAT |
| sodA_PE | AAAAAGGTTCTAGTGAATCGTAAG |
| trpE_PE | GTTGGATTGGAATGATATAGTACTGG |
| ureG_PE | TTTCATAGGGTGTTTCGCTTT |
| qRT-PCR | |
| qRT-groES-F | GCGTAAAGAGGTTGAATCGAA |
| qRT-groES-R | CCAACGCGTACATCTAAAGC |
| qRT-groEL-F | TTTCTGCAAATTCCGATGAAACGG |
| qRT-groEL-R | TAACCACGATCAAACTGCATACC |
| qRT-dnaK-F | CAAGGAGAGCGTAAAAGAGCA |
| qRT-dnaK-R | TCCTTTGCGGAAACATGTAA |
| qRT-dnaJ-F | CAAGAAGGGGTTCTGATTTACG |
| qRT-dnaJ-R | TGACACAATTGATGCACTCG |
| qRT-lon-F | TTTAGTAGGTCCGCCTGGAG |
| qRT-lon-R | CCAGGCATAGAACCAATGTAAG |
| qRT-grpE-F | AGAGCGTACATTGGGTATTATCG |
| qRT-grpE-R | TTTCAGGATTAAATGGGATATGAA |
| qRT-ibpA-F | TCCTCCTTATAACGTTGAATTGG |
| qRT-ibpA-R | AATCTTGATACGAATCAGCATGAG |
| qRT-clpP-F | CCAGGAGGGGAAGTGACAG |
| qRT-clpP-R | GAATTGGGCAAACAAAATCG |
| qRT-clpB-F | TCGAATGCGATTCGTAGAAG |
| qRT-clpB-R | ACGCACCATAGCATTGTCAG |
| qRT-clpX-F | ATTGATAGAGCACAAAGCGGTA |
| qRT-clpX-R | AAAAGCTACGGTTCCCTCAA |
| qRT-rpoH-F | CATTCGCAGTACACTGGATTAAA |
| qRT-rpoH-R | TCTCCTTCGTTGAACCAACC |
| qRT-rpoB-F | CACGAACCTCAAATCCTGCT |
| qRT-rpoB-R | GCGGCTGTAAGGGAATTTTT |
TABLE 2.
Putative promoter sequences recognized by RpoD- or RpoH-containing RNA polymerase in “Ca. Blochmannia floridanus” as deduced from primer extension experiments
| Promoter type and gene | Sequencea | Distance (bp) from 5′ end of transcript to ORFb |
|---|---|---|
| RpoD dependent | ||
| grpE | TTGAAT 16 bp TATAAT | 35 |
| TTGAAT 19 bp AATAAC | 32 | |
| sodA | TTTACA 17 bp TATAAT | 65 |
| guaB | TTGTAA 17 bp TATGAT | 102 |
| TTGTAT 17 bp TATTAT | 166 | |
| ibpA | TTTAAA 16 bp TATAAT | 71 |
| ureG | TTGATT 16 bp TAGAAT | 47 |
| ilvG | TATTTA 16 bp TATTAT | 81 |
| omp | TTTTAA 18 bp TATGAT | 264 |
| leuA | TTGACA 17 bp TTTAAT | 114 |
| ahpC | TTGCAA 17 bp TACAAT | 64 |
| TAGCAA 17 bp TATAAT | 134 | |
| cspC | TTGATA 16 bp TATTAT | 235 |
| trpE | CTAACA 18 bp TATAAT | 201 |
| TTAACA 23 bp TATAAT | 53 | |
| metK | TTCACA 17 bp TATGAT | 76 |
| ftsJ | TTGAAA 17 bp TAACAT | 61 |
| manX | ATATCA 17 bp TATAAT | 112 |
| speB | TCGATA 16 bp TATAAT | 372 |
| TTTATT 17 bp TATAAT | 255 | |
| RpoH dependent | ||
| groES | CTTGAAG 13 bp CCCCACTT | 111 |
| dnaK | GTTGAAA 13 bp CCGCATAT | 37 |
| lon | GTTGAAT 13 bp CCCCATAT | 75 |
| grpE | CTTGAAT 13 bp CCTTATAA | 37 |
Bold letters indicate deviations from the respective E. coli consensus sequences.
Distance between the transcriptional start point and the start codon of the first open reading frame (ORF).
FIG. 2.
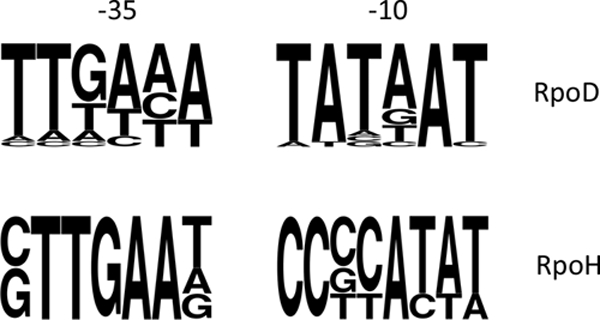
Consensus sequences of RpoD- and RpoH-dependent promoters of “Ca. Blochmannia floridanus” calculated using the WebLogo program available at http://weblogo.berkeley.edu/logo.cgi.
FIG. 3.
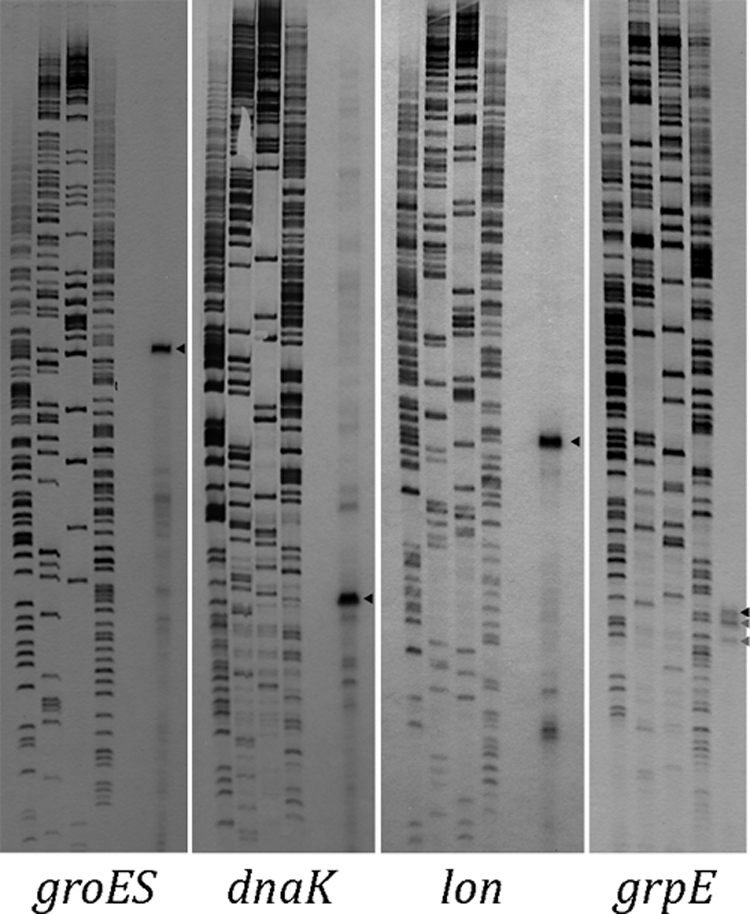
Mapping of putative RpoH-dependent promoters of “Ca. Blochmannia floridanus” by primer extension. The experiments were carried out as described for Fig. 1. Black arrowheads indicate transcriptional start sites preceded by sequences resembling the RpoH consensus sequence of E. coli, while gray arrowheads indicate possible RpoD-dependent transcripts (Table 2).
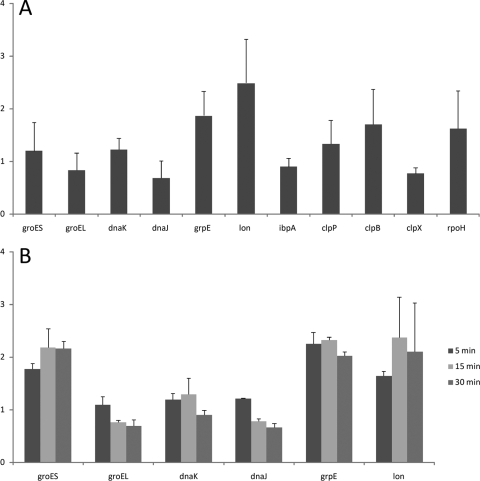
The presence of RpoH in “Ca. Blochmannia floridanus” raised the question of whether a heat shock response can be mounted by this bacterium. To investigate this matter, we quantified transcripts of 10 genes known to be heat induced on the transcriptional level in E. coli (groES, groEL, dnaK, dnaJ, lon, grpE, ibpA, clpP, clpB, and clpX) and rpoH in untreated pupae and after incubation of the whole animals for 30 min at 42°C by quantitative real-time PCR (qRT-PCR). No significant heat-dependent regulation of expression of the investigated factors was noted under these experimental conditions (Wilcoxon test for matched pairs: control versus heat shocked, n = 11, P = 0.7; two-sided t tests per gene were all nonsignificant) (Fig. 4A). To further corroborate and refine these data, we isolated the bacteria from the animals as described previously (15). Then we subjected them to a heat shock at 42°C, whereas controls were kept at 25°C, and followed expression of the groEL, groES, dnaK, dnaJ, grpE, and lon genes during a time course of up to 30 min using E. coli DH5α as a control. While, in agreement with the literature for E. coli, a strong heat-induced expression of heat shock factors was evident already 5 min after heat shock (data not shown), only mild transcriptional changes at a level comparable to those observed after heat shock of the entire animals were observed for “Ca. Blochmannia floridanus” (Fig. 4B) (18). This may indicate that in “Ca. Blochmannia floridanus” expression of the RpoH-dependent genes is quite constitutive and that RpoH is being transformed into an alternative vegetative sigma factor in this organism, dedicated to the initiation of transcription at promoters mainly controlling strongly expressed genes. However, we cannot exclude the possibility that so-far-unknown stress signals other than heat may influence expression of the RpoH-dependent genes in this endosymbiont.
FIG. 4.
(A) Changes in expression levels of “Ca. Blochmannia floridanus” genes upon heat shock exposure of late pupae of C. floridanus as detected by qRT-PCR. The genes selected are part of the heat shock regulon in E. coli (groES, groEL, dnaK, dnaJ, grpE, lon, ibpA, clpP, clpB, clpX, and rpoH). Late pupae were collected from C. floridanus colonies. Then they were either incubated further under standard conditions at 25°C or subjected to a heat shock at 42°C for 30 min. After incubation the animals were quickly chilled on ice. Isolation of the endosymbionts was performed in ice-cold isolation buffer, and RNA extraction, cDNA synthesis, and qRT-PCR were performed as described recently (15). The columns show the ratio of transcript amounts of the respective genes detected after heat shock compared to the transcript amount without heat shock. In each case, three independent experiments were performed in duplicate, including two different ant colonies. Standard deviations are indicated by vertical bars above the columns, and values were normalized to expression of the internal control rpoB. (B) Time course of change of expression of the groES, groEL, dnaK, dnaJ, grpE, and lon genes after heat shock of bacteria isolated from the animals before heat treatment. Columns show the ratio of transcript amounts of the respective genes detected by qRT-PCR after temperature shift to 42°C and different time intervals (5, 15, and 30 min) compared to the transcript amounts of the untreated controls. Two independent experiments were performed with bacteria from different ant colonies each in replicate qRT-PCRs. Standard deviations are indicated by vertical bars above the columns, and values were normalized to expression of the internal control rpoB.
Currently there is no explanation for the lack of a significant heat shock response in “Ca. Blochmannia floridanus,” since (i) amino acids of “Ca. Blochmannia floridanus” RpoH known to be involved either in binding of the RNA polymerase core subunit or in promoter recognition are conserved with those of the E. coli RpoH protein (data not shown) and (ii) the predicted secondary structure of the “Ca. Blochmannia floridanus” rpoH mRNA is very similar to the secondary structure of the E. coli counterpart, which is known to be involved in heat shock induction by translational control of RpoH expression (data not shown) (18). However, “Ca. Blochmannia floridanus” lacks genes encoding the ATP-dependent protease HslVU, which is known to play a role in the control of RpoH protein levels in E. coli (8). This may lead to expression of large amounts of RpoH in “Ca. Blochmannia floridanus” already under standard conditions. In contrast, although in a previous study no heat shock response could be detected in B. aphidicola, application of a different protocol in more recent studies revealed a moderate but significant heat shock response which contributes to survival of the animals under heat stress conditions (3, 12, 17). In this respect it is interesting that the B. aphidicola genome encodes all proteases known to affect the half-life of the RpoH protein including HslVU. Differences in the heat shock responses of “Ca. Blochmannia floridanus” and B. aphidicola may reflect behavioral differences of the host animals in their natural habitat. Extreme heat conditions may in fact not be a major problem for ants since these social animals live in large communities and are able to regulate the temperature in their nests. The developing brood is translocated toward areas within the nest with optimal conditions when temperature varies (11). Workers are mobile and able to restrict their activities outside their nest to periods of optimal temperature. In contrast, aphids are much less mobile, and once they have inserted their mouthparts into the plant tissue to suck phloem sap, they cannot easily escape and may suffer from significant thermal variation.
Acknowledgments
We thank Dagmar Beier for critically reading the manuscript.
Financial support was provided by the priority program SFB567 of the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Clark, M. A., L. Baumann, M. L. Thao, N. A. Moran, and P. Baumann. 2001. Degenerative minimalism in the genome of a psyllid endosymbiont. J. Bacteriol. 1831853-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Degnan, P. H., A. B. Lazarus, and J. J. Wernegreen. 2005. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 151023-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunbar, H. E., A. C. Wilson, N. R. Ferguson, and N. A. Moran. 2007. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 5e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldhaar, H., J. Straka, M. Krischke, K. Berthold, S. Stoll, M. J. Mueller, and R. Gross. 2007. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil, R., F. J. Silva, E. Zientz, F. Delmotte, F. González-Candelas, A. Latorre, C. Rausell, J. Kamerbeek, J. Gadau, B. Hölldobler, R. C. van Ham, R. Gross, and A. Moya. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. USA 1009388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross, R., and R. Rappuoli. 1989. Pertussis toxin promoter sequences involved in modulation. J. Bacteriol. 1714026-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Himmelreich, R., H. Plagens, H. Hilbert, B. Reiner, and R. Herrmann. 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanemori, M., K. Nishihara, H. Yanagi, and T. Yura. 1999. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J. Bacteriol. 1797219-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran, N. A., J. P. McCutcheon, and A. Nakabachi. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42165-190. [DOI] [PubMed] [Google Scholar]
- 10.Nakabachi, A., A. Yamashita, H. Toh, H. Ishikawa, H. E. Dunbar, N. A. Moran, and M. Hattori. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314267. [DOI] [PubMed] [Google Scholar]
- 11.Roces, F., and J. A. Nunez. 1995. Thermal sensitivity during brood care in workers of two Camponotus ant species: circadian variation and its ecological correlates. J. Insect Physiol. 41659-669. [Google Scholar]
- 12.Sato, S., and H. Ishikawa. 1997. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J. Bacteriol. 1792300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer, C., E. Stackebrandt, J. Gadau, B. Hölldobler, and R. Gross. 2000. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: proposal of the new taxon Candidatus Blochmannia gen. nov. Int. J. Syst. Evol. Microbiol. 501877-1886. [DOI] [PubMed] [Google Scholar]
- 14.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 40781-86. [DOI] [PubMed] [Google Scholar]
- 15.Stoll, S., H. Feldhaar, and R. Gross. 2008. Transcriptional profiling of the endosymbiont Blochmannia floridanus during developmental stages of its holometabolous ant host. Environ. Microbiol. 11877-888. [DOI] [PubMed] [Google Scholar]
- 16.Wernegreen, J. J. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3850-861. [DOI] [PubMed] [Google Scholar]
- 17.Wilcox, J. L., H. E. Dunbar, R. D. Wolfinger, and N. A. Moran. 2003. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 481491-1500. [DOI] [PubMed] [Google Scholar]
- 18.Yura, T., and K. Nakahigashi. 1999. Regulation of the heat shock response. Curr. Opin. Microbiol. 2153-158. [DOI] [PubMed] [Google Scholar]