Abstract
TraR of Agrobacterium tumefaciens is a LuxR-type quorum-sensing transcription factor that regulates genes required for replication and conjugation of the tumor-inducing (Ti) plasmid. TraR requires its cognate autoinducer N-3-oxooctanoyl-homoserine lactone (OOHL) for resistance of proteolysis in wild-type bacteria and for correct protein folding and solubility when overexpressed in E. coli. In this study, we ask whether GroESL might also play a role in TraR folding, as this molecular chaperone assists many proteins in attaining their native tertiary structure. Expression of E. coli GroESL in a strain expressing TraR increases the solubility of TraR and increases transcriptional activity of a TraR-dependent promoter. Both solubility and activity still require OOHL. We also studied the folding of TraR in the closely related bacterium Sinorhizobium meliloti. A mutation in one groEL gene slightly decreased the expression of a TraR-dependent promoter, strongly decreased the accumulation of TraR in Western immunoblot assays, and also strongly influenced the fate of pulse-labeled TraR.
Many bacterial proteins acquire their native tertiary structure during or shortly after translation and require no assistance from any other molecules. However, for many other proteins, folding requires or is facilitated by cytoplasmic chaperone proteins, which appear to test a variety of possible conformations of their substrates until the native conformation is found (20). We have extensively studied the folding of the quorum-sensing regulator TraR of Agrobacterium tumefaciens, which is a member of the LuxR family of transcription factors (25, 47, 48) and regulates genes required for vegetative replication and conjugal transfer of tumor-inducing (Ti) plasmids (18, 24, 29, 30). TraR activity requires the diffusible signal molecule N-3-oxooctanoyl-homoserine lactone (OOHL), which is synthesized by the TraI protein. OOHL is completely buried within the N-terminal domain of TraR, where it contributes to the hydrophobic environment of the protein core, and has virtually no contact with bulk solvent (37, 43).
We have demonstrated that TraR requires OOHL in order to fold into a stable, soluble, dimeric, protease-resistant form (22, 41, 42). Pulse-chase experiments showed that TraR can be rescued from proteolysis by OOHL only during its synthesis on ribosomes. Once synthesis is complete, apo-TraR cannot be rescued by OOHL from proteolysis (48). This finding strongly suggests that OOHL mediates the cotranslational folding of TraR and acts as an essential scaffold for TraR maturation. In the absence of OOHL, natively expressed apo-TraR was undetectable in A. tumefaciens by Western immunoblotting, due to its rapid proteolysis (7, 47, 48). These Western blot assays would have detected as few as one TraR molecule per cell, indicating that in the absence of OOHL, TraR is degraded extremely rapidly.
Fusion proteins containing TraR and certain other proteins are far more stable to proteolysis than is native TraR and were partially active in the absence of OOHL (6). Those foreign polypeptides of these fusion proteins were proposed to function as protein solubility enhancers and intramolecular chaperones (6). In the absence of OOHL, fused polypeptides may sequester the amino terminus of TraR from proteolysis and/or promote dimerization (6).
GroES and GroEL proteins form a complex consisting of 14 GroEL subunits and 14 GroES subunits that, together, enclose an internal chamber where protein folding occurs. GroESL is one of the major ATP-dependent cytoplasmic chaperones and is highly conserved among bacteria, archaea, and eukaryotes (2, 14, 17, 39). It interacts with a large number of unfolded or misfolded proteins and assists in their folding and remodeling during or after their synthesis. It has been proposed that in Escherichia coli, between 10 and 15% of the newly translated cytoplasmic proteins are associated with GroESL, and this fraction increases to about 30% under heat stress conditions (14). When the LuxR protein of Vibrio fischeri was expressed in E. coli, its solubility and activity were strongly enhanced by the overexpression of GroESL. An E. coli groEL mutant containing functional luxR and luxICDABE genes was only weakly luminescent (13). Overexpression of GroESL also enhanced the binding of exogenous autoinducers by LuxR (13). However, those experiments were done in a heterologous host using a high-copy plasmid, so conclusions were limited for the role of GroESL in the native strain.
Proteins orthologous to TraR and TraI were described in Sinorhizobium meliloti AK631 (26). The TraR protein of that strain is only 28% identical with TraR of the octopine-type Ti plasmid but regulates orthologous tra and trb genes and is inhibited by TraM (26). A mutation in a chromosomal groEL gene (designated groELc1583::Tn5) abolished the production of a specific set of autoinducers whose synthesis is TraR and TraI dependent (26), suggesting that GroELc plays a positive role in this process, possibly by aiding in the folding of one of these proteins. In an elegant genetic analysis, suppressor mutations that would restore autoinducer production were sought. One such mutant had a null mutation in the traM gene. This protein also has an ortholog on the Ti plasmid, and both TraM proteins are TraR antiactivators (9, 10, 22, 26, 31). Apparently, the release of TraR from antiactivation somehow compensated for the loss of GroELc. One possible interpretation of these data is that TraR in this strain requires GroELc for folding and that the reduced level of TraR caused by this mutation can be compensated for by releasing it from antiactivation. It was not clear whether the original GroELc mutation completely abolished protein function, as it was caused by a transposon insertion very close to the 3′ end of the gene. It is also not known how many other groESL operons exist in this strain of S. meliloti, though S. meliloti Rm1021 has five groESL operons, two of which are on the chromosome, two of which are on pSymA, and one of which is on pSymB (1, 5, 16, 19).
The evidence that GroESL is important for quorum sensing in S. meliloti suggested that the same might be true in A. tumefaciens. Unlike S. meliloti, A. tumefaciens has only one copy of the groESL operon, located on the circular chromosome (21, 40). In the present study, we show that overexpression of E. coli GroESL enhanced TraR solubility in E. coli and enhanced the expression of a TraR-dependent promoter in A. tumefaciens. A mutation of groELc of S. meliloti impaired the accumulation and activity of A. tumefaciens TraR in this heterologous host. These findings all support a role for GroESL in TraR folding.
MATERIALS AND METHODS
Strains, media and reagents.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were cultured in LB medium at 37°C for general purposes or at 27°C for TraR overexpression. A. tumefaciens strains were cultured at 27°C in AT minimal medium (4). S. meliloti strains were cultured at 27°C in LB medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LB/MC) (26). Antibiotics were added at the following concentrations: spectinomycin at 100 μg per ml for A. tumefaciens and at 300 μg per ml for S. meliloti; gentamicin at 100 μg per ml for A. tumefaciens, at 10 μg per ml for E. coli, and at 50 μg per ml for S. meliloti; tetracycline at 2.5 μg per ml for A. tumefaciens and at 10 μg per ml for S. meliloti; ampicillin at 200 μg per ml for E. coli. Restriction enzymes and other DNA modification enzymes were purchased from New England Biolabs. The N-3-oxooctanoyl-homoserine lactone (OOHL) used in this study was generously provided by A. Eberhard and was prepared as previously described (44).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant description | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | α-Complementation | Stratagene |
| E. coli BL21(DE3) | E. coli B, Plac-gene 1 of bacteriophage T7 | 35 |
| A. tumefaciens KYC55 | Ti plasmid-less A. tumefaciens R10; Kmr | 11 |
| S. meliloti Rm11500 | Spontaneous Smr mutant of AK361 | 26 |
| S. meliloti Rm11501 | Rm11500 containing groELc1583::Tn5 | 26 |
| Plasmids | ||
| pT-groE | PT7-groESL, ColE1; Cmr | 42 |
| pJZ335 | Plac-traR in pPZP201; Spr | 44 |
| pJZ358 | PT7-traR in pRSERA; Ampr | 47 |
| pJZ372 | PtraI-lacZ lacIq; Tetr | 44 |
| pJZ384 | PT7-traR in pPZP201; Spr | 45 |
| pJZ410 | T7 RNAP gene under PL of bacteriophage λ; Gmr | 45 |
| pYC335 | Plac-traR in pPZP201; Spr | 7 |
| pYC337 | PT7-groESL in pYC335; Spr | This study |
| pYC358 | PT7-groESL in pJZ358; Ampr | This study |
DNA manipulations.
Molecular cloning and plasmid constructions were performed according to published protocols (33). Plasmid pYC358 was constructed by cutting plasmid pT7-groESL with SalI and HindIII and cloning a 2.5-kb fragment containing groESL into the same sites of plasmid pJZ358 (47). Plasmid pYC358 therefore contains both a PT7-traR fusion and a PT7-groESL fusion. Plasmid pYC337 was constructed by cloning the same 2.5-kb fragment into the corresponding sites of pYC335 (7). Plasmid pYC337 therefore contains a Plac-traR fusion and a PT7-groESL fusion. These plasmids were introduced into A. tumefaciens and S. meliloti by electroporation (4).
TraR overexpression.
Strains BL21(DE3)(pJZ358) and BL21(DE3)(pYC358) were cultured at 27°C in 100 ml of LB broth containing 400 μg of ampicillin per ml and 10 μM OOHL. When cultures reached an optical density at 600 nm of 0.4, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the cultures to a final concentration of 500 μM. Incubation was continued at 27°C until the cultures reached an optical density at 600 nm of 1.0. Cells were collected, resuspended in 2 ml of TEDG buffer (8) supplemented with 100 mM NaCl, and disrupted using a French pressure minicell (15,000 lb/in2). Total cell lysates were separated into soluble and pellet fractions by ultracentrifugation at 45,000 × g for 30 min. Protein samples from total, soluble, and pellet fractions were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Immunodetection of TraR in vivo.
To compare intracellular concentrations of TraR in the wild-type S. meliloti strain and the S. meliloti groELc mutant, plasmids pJZ335 and pJZ372 were introduced into Rm11500 (wild type) and Rm11501 (groELc1583::Tn5). The resulting strains were cultured at 28°C in LB/MC medium supplemented with appropriate antibiotics, 500 μM IPTG, and 100 nM OOHL. Cells were harvested at early log phase, resuspended in 1 ml of TEDG buffer supplemented with 100 mM NaCl, and lysed using a French pressure minicell (15,000 lb/in2). Total cell lysates were size fractionated by SDS-PAGE. Proteins were transferred by electrophoresis to a nitrocellulose membrane (Bio-Rad) and detected using preabsorbed polyclonal anti-TraR rabbit antiserum as described previously (7).
Assays of TraR-dependent activity in vivo.
Bioassays of TraR activity were conducted by culturing cells either in AT minimal medium (for A. tumefaciens strains) or in LB/MC medium (for S. meliloti strains) supplemented with 500 μM IPTG (IPTG was omitted in A. tumefaciens cultures to reduce TraR expression) and the OOHL concentrations indicated in Fig. 2. Cells were grown with vigorous aeration for 12 h (to early log phase) at 27°C and were assayed for β-galactosidase specific activities (27). All data represent averages of at least two independent assays, and error bars represent values of the standard deviations.
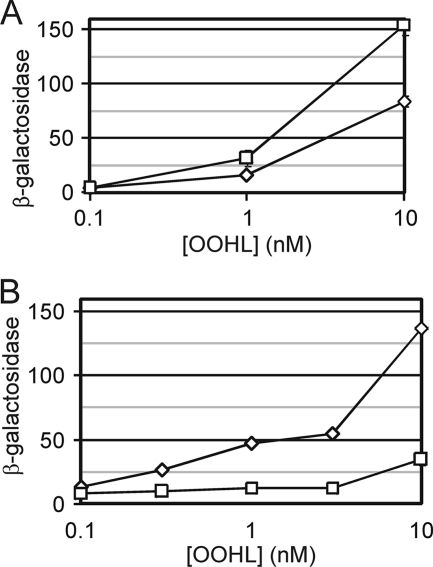
FIG. 2.
(A) Overexpression of E. coli GroESL in A. tumefaciens increases TraR-dependent gene expression. Two derivatives of strain KYC55(pJZ372)(pJZ410) are compared, one containing plasmid pYC335 (diamonds), which contains the Plac-traR fusion only, and the other containing pYC337 (squares), which contains a Plac-traR fusion and a PT7-groESL fusion. The resulting strains were treated with OOHL at the indicated concentrations and assayed for β-galactosidase specific activity. (B) Expression of the traI promoter in the presence or absence of GroELc. Plasmids pJZ335 (which contains a Plac-traR fusion) and pJZ372 (which contains a PtraI-lacZ reporter fusion) were introduced into Rm11500 (diamonds) and the groELc mutant (squares). The strains were cultured with the indicated concentrations of OOHL and assayed for β-galactosidase specific activity.
Measurements of TraR turnover.
Plasmids pJZ384 and pJZ410 were introduced into S. meliloti strains Rm11500 and Rm11501. The resulting strains were cultured at 27°C in LB/MC medium supplemented with the appropriate antibiotics and 100 nM OOHL until early log phase. Rifampin was added to the cultures to a final concentration of 200 μg per ml to block bacterial RNA polymerase activity (3). Forty minutes later, [35S]methionine was added to the cultures to a final concentration of 5 μCi per ml. After an interval of 3 min, nonlabeled methionine was added to the cultures to a final concentration of 2 mM. Cells were withdrawn at various time intervals, washed with a cold Slota buffer (50 mM Tris-HCl, 0.5 M NaCl, 20 mM EDTA [pH 8.0], 0.05% Na-Sarkosyl) and ice-cold lysis buffer (50 mM Tris-HCl [pH 7.9], 200 mM NaCl, 10 mM EDTA, 1 mM dithiothreitol, 1 mM β-mercaptoethanol, 0.1% phenylmethylsulfonyl fluoride, 1% NP-40, and 500 μg per ml freshly prepared lysozyme), and incubated on ice for 30 min. The resulting lysates were centrifuged at 12,000 × g for 3 min, and the supernatants were subjected to size fractionation by SDS-PAGE. Results were analyzed using a PhosphorImager (model 840; Molecular Dynamics).
RESULTS
Cooverexpression of TraR and the E. coli GroESL enhances accumulation of soluble TraR.
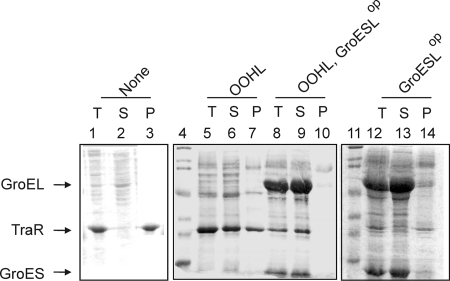
When TraR was overexpressed in E. coli strain BL21(DE3) using a T7 protein expression system (35) in the absence of OOHL, virtually all of the TraR protein was found in the particulate fraction, indicating that it accumulated as insoluble inclusion bodies, while the addition of OOHL to the culture medium dramatically increased TraR solubility, as approximately half of the total TraR protein was in the soluble fraction (47). We wanted to test whether overexpression of the E. coli GroESL proteins would further enhance the accumulation of soluble TraR. To do this, we used plasmid pYC358, which overexpresses TraR from a T7 promoter and expresses E. coli GroESL from a second T7 promoter on the same plasmid. As a control, we used plasmid pJZ358, which overexpresses TraR but not GroESL (47).
As observed previously (45), when TraR was overexpressed in the absence of OOHL, all detectable protein was insoluble (Fig. 1, lanes 1 to 3), while when it was expressed in medium containing 10 μM OOHL, approximately half of the total TraR protein was soluble and presumed to be folded (Fig. 1, lanes 5 to 7) (47). In contrast, when TraR and the E. coli GroESL were coexpressed in the presence of 10 μM OOHL, all detectable TraR protein was found in the soluble fraction (Fig. 1, lanes 8 to 10). This can best be seen by comparing lanes 7 and 10 in Fig. 1, as the former contains a large quantity of TraR, while the latter contains none. Overexpression of GroESL therefore enhanced TraR solubility, presumably by enhancing the rate at which TraR can fold. The total amount of TraR produced by these cells appears to have decreased somewhat (Fig. 1, lanes 5 and 8). It is possible that GroESL overproduction depleted the cells of the substrates for protein synthesis. Alternatively, it is possible that GroESL rescued TraR from inclusion bodies, but not from proteolysis, especially if the rescued TraR lacks OOHL and therefore is unfolded.
FIG. 1.
Cooverexpression of TraR and E. coli GroESL. Strain BL21(DE3)(pJZ358) was used to detect soluble TraR in the absence of GroESL overexpression, while BL21(DE3)(pYC358) was used to detect soluble TraR in the presence of GroESL overexpression. Cells were cultured in LB medium in the presence (lanes 5 to 10) or absence (lanes 1 to 3 and lanes 12 to 14) of 10 μM OOHL. Lanes 4 and 11 contain molecular-weight standards. Letters T, S, and P represent total, soluble, and pellet fractions of the cell lysates, respectively. The gel was stained with Coomassie brilliant blue dye. GroESLop refers to strains that overproduce GroESL from pYC358. This gel is representative of an experiment carried out three times with independent biological samples.
In the absence of OOHL, overexpression of GroESL did not cause the accumulation of soluble, stable TraR (Fig. 1, lanes 12 to 14). Furthermore, GroESL caused a drastic decrease in the total abundance of TraR (Fig. 1, lanes 1 and 12). One possible contribution to this effect could be that overproduction of GroESL may divert protein synthetic resources from TraR. We favor the alternative possibility that GroESL overproduction may rescue apo-TraR from protease-resistant inclusion bodies but that it cannot fold this protein into a mature form in the absence of OOHL. If so, then the soluble protein would be degraded by cellular proteases. We conclude, first, that GroEL enhances TraR folding and, second, that even when GroESL is overproduced, OOHL is still essential for TraR folding and protease resistance.
Overexpression of GroESL increases transcription of a TraR-dependent promoter.
We also expressed TraR and E. coli GroESL in A. tumefaciens to see whether overexpression of GroESL would have any effect on TraR activity. We introduced into strain KYC55(pJZ410)(pJZ372) plasmid pYC337, which contains a PT7-groESL fusion and a Plac-traR fusion on the same plasmid, and pYC335, which contains the Plac-traR fusion but lacks groESL. Plasmid pJZ410 expresses T7 RNA polymerase while pJZ372 contains a PtraI-lacZ fusion and lacIq (45). In these experiments, IPTG was omitted from the broth to help limit TraR expression. Overexpression of the GroESL caused a modest, reproducible increase in the expression of the traI promoter (Fig. 2A). The fact that GroESL overexpression caused only a modest enhancement may have been due to the endogenous expression of GroESL. Similarly, when GroESL was overexpressed from the weaker lac promoter, it did not detectably enhance expression of the reporter (data not shown). When IPTG (0.5 mM) was added into the medium to induce TraR expression from the lac promoter, overexpression of GroESL did not enhance expression of the target promoter (data not shown). We conclude that GroESL enhances TraR activity only when the two proteins are overexpressed at a particular ratio, as too little GroESL or too much TraR abolishes this effect.
TraR activity is defective in an S. meliloti groELc mutant.
A previous study of S. meliloti (strain RM11500) has shown that mutation of the groELc gene blocked the activity of a TraR ortholog encoded by a conjugal plasmid of that bacterium (26). The full sequence of groELc is not available, and we do not know how many other homologous genes are found in that strain. However, the sequenced S. meliloti strain, Rm1021, has five groESL operons, two borne on the chromosome, two on pSymA, and one on pSymB (19). In contrast, A. tumefaciens has only one copy of the groEL genes, and despite repeated attempts, we were unable to disrupt this gene, suggesting that it may be essential. Inasmuch as A. tumefaciens and S. meliloti are closely related members of the Rhizobiaceae (15, 41), we decided to use the S. meliloti groELc mutant (Rm11501) and its isogenic parent strain (RM11500) in our study. Both strains carry an endogenous copy of traR on plasmid pRme41a, although it is apparently not significantly expressed under these conditions, as control strains lacking the A. tumefaciens traR gene did not express a TraR-dependent reporter. We introduced two plasmids into these strains: pJZ372, which has a PtraI-lacZ fusion and lacIq, and pJZ335, which has a Plac-traR fusion and expresses the native TraR protein. Both of the resulting strains grew at similar rates in LB/MC medium supplemented with the appropriate antibiotics and OOHL (data not shown). The TraR-dependent expression of the traI promoter was reproducibly higher in the wild-type strain than in the groELc mutant (Fig. 2B). We conclude that GroELc is required for maximal activity of the A. tumefaciens TraR and believe that residual TraR activity in the mutant may be due at least in part to GroESL proteins encoded by other groESL operons.
TraR accumulation in the S. meliloti groELc mutant.
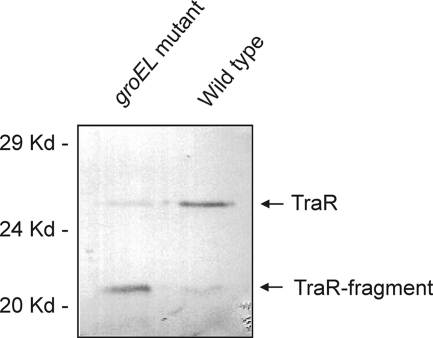
The hypothesis that GroESLc promotes TraR folding predicts that the groELc mutation should cause a decrease in the accumulation of TraR. Western immunoblot assays comparing TraR accumulation in Rm11500 and that in Rm11501 showed abundant levels of full-length TraR in the wild-type strain, but they also showed that only trace levels accumulated in the groELc mutant (Fig. 3). The mutant instead accumulated a smaller protein, the size of which is very close to the size of the N-terminal domain of TraR (residues 1 to 170). This protein was present only in trace amounts in the wild-type strain and could represent a proteolytic breakdown product of TraR. If this fragment consists of the TraR N-terminal domain, accumulation of such a protein would be predicted to inhibit the activity of the wild-type protein by the formation of inactive heterodimers (8, 28, 46).
FIG. 3.
Western immunoblot assays detecting TraR accumulation in the wild-type S. meliloti strain Rm11500 and in the groELc mutant Rm11501. TraR was expressed in strains Rm11500(pJZ372)(pJZ335) and Rm11501(pJZ372)(pJZ335). Accumulation of TraR was assayed as described in Materials and Methods.
TraR folds more effectively in the wild-type S. meliloti strain than in the groELc mutant.
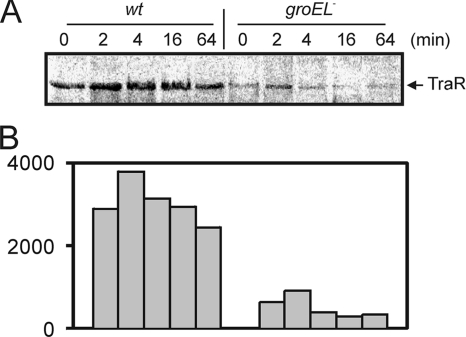
We performed pulse-chase experiments to directly compare TraR stability in a groELc mutant (Rm11501) with that in a congenic wild-type strain (Rm11500). Two plasmids were introduced into both strains, one of which (pJZ384) expresses TraR from a T7 promoter, while the other (pJZ410) contains the T7 RNA polymerase gene under the control of a PL promoter of the bacteriophage lambda and also contains the thermosensitive cI857 allele of the lambda repressor. We used these two plasmids to specifically radiolabel TraR under conditions where the host RNA polymerase was inactivated using rifampin (3). Although expression of the T7 RNA polymerase was designed to be heat inducible, we previously found that transcription activity levels from the T7 promoter were similar at either 27°C or 42°C in this system (45). Therefore, we conducted all assays at 27°C. Radiolabeling of TraR was far stronger in the wild-type strain than in the groELc mutant (Fig. 4A), both in the presence of 100 nM OOHL. However, the TraR radiolabeled in the mutant strain had a half-life similar to that of TraR expressed in the wild-type strain. We conclude, first, that the groELc mutant is starved for GroEL and, second, that other copies of this gene provide low levels of functional GroEL protein. Under these conditions, the majority of TraR fails to be folded by GroESL and is rapidly degraded. A fraction of the total TraR pool is able to interact with the remaining pool of GroESL, and this TraR fraction is resistant to proteolysis, just as it is in the wild-type strain.
FIG. 4.
Pulse-chase experiment comparing TraR stabilities in the wild-type S. meliloti strain Rm11500 (wt) and in the groELc mutant strain Rm11501 (groEL). (A) Strains Rm11500(pJZ384)(pJZ410) and Rm11501(pJZ384)(pJZ410) were used to overexpress TraR from a T7 promoter in the presence of 100 nM OOHL. [35S]methionine-labeled TraR proteins were collected at various time intervals as indicated and were size fractionated by SDS-PAGE. Results were analyzed using a PhosphorImager. (B) Relative signal strength of labeled TraR shown in panel A.
DISCUSSION
One of the most intriguing and puzzling properties of TraR may be its inability to accumulate and resist proteolysis in the absence of OOHL (47, 48). Most well-studied ligand binding proteins are equally stable in their ligand-free form and their ligand-bound form (12), and for such proteins, binding of ligands often causes protein conformational changes that lead to altered activity. Like TraR, several other LuxR-type proteins require acyl-homoserine lactone-type autoinducers for correct folding and protease resistance (34, 36, 38).
GroESL has been proposed to facilitate the folding of 10 to 30% of all nascent cytoplasmic proteins in E. coli and can also facilitate the refolding of purified proteins that have been denatured in vitro. However, at least some of these same proteins can also fold cotranslationally in vitro in the absence of GroESL. In fact, cotranslational folding of a protein in the absence of GroESL often occurs more rapidly than posttranslational folding of the same protein by GroESL. For most protein substrates, the cotranslational folding event happens within 10 to 30 s, although in other cases folding may take a much longer time (14). In general, GroESL binds to proteins that have exposed hydrophobic residues. Our data are consistent with models in which GroESL helps to protect apo-TraR from misfolding and maintains it in a state that is competent to bind OOHL, which is also essential for folding. If OOHL is absent, the protein is eventually degraded by the major cellular proteases Clp and Lon (47, 48).
The genomes of several bacterial genera encode multiple copies of GroESL (23, 32), leading to speculation that the different copies of these genes may encode proteins with unique functions (39). S. meliloti RM1021 bears five copies of groESL (1, 16, 19), and it has been proposed that each copy may have a unique biological role (26). If the same is true of strain RM15000, it might help to explain the finding that the mutation of just one of these genes causes a quorum-sensing phenotype (26). The residual level of TraR activity in the groESLc mutant could well be due to the other copies of groESL.
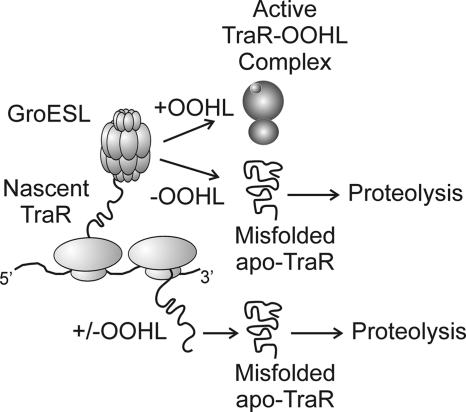
Our data suggest that GroESL may be just as essential for TraR maturation as is OOHL. We therefore propose a model for GroESL-dependent, OOHL-dependent cotranslational TraR folding. In this model, we consider TraR folding under two different conditions—one in the absence of OOHL (Fig. 5A) and one in the presence of OOHL (Fig. 5B). In the absence of OOHL, during translation or immediately afterwards, most or all apo-TraR proteins are bound by GroESL. However, even this chaperone cannot impart a protease-resistant conformation, and when TraR is released from the folding chamber of the chaperone, it is quickly degraded by the cytoplasmic proteases Clp and Lon (48). TraR must interact directly with OOHL and with GroESL in order to fold. It remains unclear whether GroESL and OOHL act upon TraR simultaneously or sequentially and whether OOHL binds TraR within the folding chamber of this chaperone.
FIG. 5.
A proposed model of nascent TraR folding in the absence or presence of GroESL and OOHL. TraR protein interacts with GroESL complexes during or directly after synthesis (top). In the presence of OOHL, GroESL can fold TraR monomers into soluble forms capable of dimerization and DNA binding. In the absence of OOHL, GroESL is unable to fold TraR into an active form and releases it for rapid proteolysis (middle). TraR protein that fails to interact with GroESL is targeted for proteolysis, even in the presence of OOHL (bottom).
Acknowledgments
We thank Juan E. Gonzalez at University of Texas, Dallas, for generously providing S. meliloti strains Rm11500 and Rm11501, and we thank members of S.C.W.'s lab for helpful discussions and critical reviews of the manuscript during its preparation.
This work was supported by grant GM42893 from the NIH.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K.-C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 989883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92351-366. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104901-912. [DOI] [PubMed] [Google Scholar]
- 4.Cangelosi, G. A., E. A. Best, G. Martinetti, and E. W. Nester. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204384-397. [DOI] [PubMed] [Google Scholar]
- 5.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 989877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai, Y., and S. C. Winans. 2005. Amino-terminal protein fusions to the TraR quorum-sensing transcription factor enhance protein stability and autoinducer-independent activity. J. Bacteriol. 1871219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai, Y., and S. C. Winans. 2004. Site-directed mutagenesis of a LuxR-type quorum-sensing transcription factor: alteration of autoinducer specificity. Mol. Microbiol. 51765-776. [DOI] [PubMed] [Google Scholar]
- 8.Chai, Y., J. Zhu, and S. C. Winans. 2001. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40414-421. [DOI] [PubMed] [Google Scholar]
- 9.Chen, G., P. D. Jeffrey, C. Fuqua, Y. Shi, and L. Chen. 2007. Structural basis for antiactivation in bacterial quorum sensing. Proc. Natl. Acad. Sci. USA 10416474-16479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, G., J. W. Malenkos, M. R. Cha, C. Fuqua, and L. Chen. 2004. Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 521641-1651. [DOI] [PubMed] [Google Scholar]
- 11.Cho, K., C. Fuqua, B. S. Martin, and S. C. Winans. 1996. Identification of Agrobacterium tumefaciens genes that direct the complete catabolism of octopine. J. Bacteriol. 1781872-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wolf, F. A., and G. M. Brett. 2000. Ligand-binding proteins: their potential for application in systems for controlled delivery and uptake of ligands. Pharmacol. Rev. 52207-236. [PubMed] [Google Scholar]
- 13.Dolan, K. M., and E. P. Greenberg. 1992. Evidence that GroEL, not σ32, is involved in transcriptional regulation of the Vibrio fischeri luminescence genes in Escherichia coli. J. Bacteriol. 1745132-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewalt, K. L., J. P. Hendrick, W. A. Houry, and H. F. Ulrich. 1997. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90491-500. [DOI] [PubMed] [Google Scholar]
- 15.Farrand, S. K., P. B. van Berkum, and P. Oger. 2003. Agrobacterium is a definable genus of the family Rhizobiaceae. Int. J. Syst. Evol. Microbiol. 531681-1687. [DOI] [PubMed] [Google Scholar]
- 16.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Puhler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 989889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frydman, J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70603-647. [DOI] [PubMed] [Google Scholar]
- 18.Fuqua, C., and S. C. Winans. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 1762796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F.-J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K.-C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293668-672. [DOI] [PubMed] [Google Scholar]
- 20.Georgopoulos, C. 2006. Toothpicks, serendipity and the emergence of the Escherichia coli DnaK (Hsp70) and GroEL (Hsp60) chaperone machines. Genetics 1741699-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 2942323-2328. [DOI] [PubMed] [Google Scholar]
- 22.Hwang, I., D. M. Cook, and S. K. Farrand. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karunakaran, K. P., Y. Noguchi, T. D. Read, A. Cherkasov, J. Kwee, C. Shen, C. C. Nelson, and R. C. Brunham. 2003. Molecular analysis of the multiple GroEL proteins of chlamydiae. J. Bacteriol. 1851958-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, P., and S. K. Farrand. 2000. The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J. Bacteriol. 182179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, Z. Q., and S. K. Farrand. 1999. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. USA 969009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marketon, M. M., and J. E. Gonzalez. 2002. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 1843466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Oger, P., K. S. Kim, R. L. Sackett, K. R. Piper, and S. K. Farrand. 1998. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27277-288. [DOI] [PubMed] [Google Scholar]
- 29.Pappas, K. M., and S. C. Winans. 2003. A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol. Microbiol. 481059-1073. [DOI] [PubMed] [Google Scholar]
- 30.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature (London) 362448-450. [DOI] [PubMed] [Google Scholar]
- 31.Qin, Y., S. Su, and S. K. Farrand. 2007. Molecular basis of transcriptional antiactivation. TraM disrupts the TraR-DNA complex through stepwise interactions. J. Biol. Chem. 28219979-19991. [DOI] [PubMed] [Google Scholar]
- 32.Rusanganwa, E., and R. S. Gupta. 1993. Cloning and characterization of multiple groEL chaperonin-encoding genes in Rhizobium meliloti. Gene 12667-75. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 10115833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polemerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 36.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vannini, A., C. Volpari, C. Gargioli, E. Muraglia, R. Cortese, R. De Francesco, P. Neddermann, and S. D. Marco. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 214393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C. S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57452-467. [DOI] [PubMed] [Google Scholar]
- 39.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 2861888-1893. [DOI] [PubMed] [Google Scholar]
- 40.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M.-J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z.-Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J.-F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2942317-2323. [DOI] [PubMed] [Google Scholar]
- 41.Yanagi, M., and K. Yamasato. 1993. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol. Lett. 107115-120. [DOI] [PubMed] [Google Scholar]
- 42.Yasukawa, T., C. Kanei-Ishii, T. Maekawa, J. Fujimoto, T. Yamamoto, and S. Ishii. 1995. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 27025328-25331. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, R., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417971-974. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 1805398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, J., Y. Chai, Z. Zhong, S. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 696949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, J., and S. C. Winans. 1998. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol. Microbiol. 27289-297. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 964832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 981507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]