Abstract
Measles virus remains a substantial cause of morbidity and mortality, producing acute infection with a potential for development of viral persistence. To study the events underlying acute and persistent measles virus infection, we performed a global transcriptional analysis on murine neuroblastoma cells that were acutely or persistently infected with measles virus. In general, we found that acute infection induced significantly more gene expression changes than did persistent infection. A functional enrichment analysis to identify which host pathways were perturbed during each of these infections identified several pathways related to cholesterol biosynthesis, including cholesterol metabolic processes, hydroxymethylglutaryl-coenzyme A (CoA) reductase activity, and acetyl-CoA C-acetyltransferase activity. We also found that measles virus colocalized to lipid rafts in both acute and persistent infection models and that the majority of genes associated with cholesterol synthesis were downregulated in persistent infection relative to acute infection, suggesting a possible link with the defective viral budding in persistent infection. Further, we found that pharmacological inhibition of cholesterol synthesis resulted in the inhibition of viral budding during acute infection. In summary, persistent measles viral infection was associated with decreased cholesterol synthesis, a lower abundance of cholesterol and lipid rafts in the cell membrane, and inhibition of giant-cell formation and release of viral progeny.
Measles virus (MV) remains a significant health burden, claiming 450,000 lives worldwide every year, and most of the deaths occur in children (35). MV is the etiological agent of both acute measles infection and subacute sclerosing panencephalitis (SSPE), a rare and devastating persistent infection of the central nervous system (7, 8, 12). MV is typically highly cytolytic, resulting in an acute infection that confers lifelong immunity. The reasons and underlying mechanisms of transformation into a persistent infection in some individuals remain unknown, and the pathological implications of such an infection are controversial.
Although no mechanism for the transformation from acute to persistent infection has been established, partial explanations have been proposed. Defective measles matrix (M) protein has been recovered in some SSPE cases and was suggested as a possible underlying mechanism for persistence (11, 15, 20). Our previous research also demonstrated that there is significantly less viral budding in persistent versus acute MV infection of murine neuroblastoma cell lines and that this is not due to decreased viral protein synthesis, which is unimpaired (22, 25). Viral budding in MV infection involves the incorporation of envelope proteins into the host cell membrane (5). This has been recently shown to occur in lipid rafts—cholesterol-rich domains in the cellular membrane (17, 19, 21, 33).
In this study, we used microarray analysis to compare gene expression during acute and persistent infection with MV and to identify pathological mechanisms unique to persistent infection that could be targeted for further analyses. We then used a gene set enrichment analysis (GSEA) (29) to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways (13) and Gene Ontology (GO) (2) functions that were more perturbed during persistent infection than acute infection. Since several of the most perturbed pathways involved cholesterol biosynthesis, we chose to focus on the cholesterol biosynthesis pathway in general. We then verified the proposed mechanisms using various experimental protocols. Our results suggest an unexpected role for cholesterol biosynthesis in the regulation of MV infection persistence. These data represent not only the first study utilizing microarrays to examine gene expression in cells that are persistently infected with MV but also the first global transcriptional comparison of persistent and acute viral infection.
MATERIALS AND METHODS
Cell lines, viruses, and plaque assays.
In this study, we used mouse neuroblastoma C1300 cells, clone NS20Y, of the A/J mouse strain. We propagated the cells as loosely adherent monolayers and then passaged them when confluent by trypsinization or by shaking the culture vigorously. A detailed description of the persistently infected NS20Y cell clone (NS20Y/MS) is given in Rager-Zisman et al. (22). Mouse neuroblastoma cells that were mock infected (NS20Y), persistently infected (NS20Y/MS), and acutely infected (NS20Y+MV) with MV were grown in Dulbecco's modified Eagle's medium (DMEM; Beit Haemek, Israel) supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% glutamine, and 1% antibiotics. We incubated all cultures in a humidified atmosphere of 5% CO2 in air at 37°C. For the acute infection, we infected 24-h-old NS20Y cultures with the Edmonston strain of MV at a multiplicity of infection (MOI) of 0.01. We harvested cells at 48 h postinfection for cholesterol quantification studies and microarray analysis. At this time point the expression level of MV nucleoprotein (NP) was similar to that in NS20Y/MS cells. Again, we followed the same procedure described by Rager-Zisman (22) in performing and expressing virus plaque assays as PFU per ml (PFU/ml).
Antibodies and reagents.
We were kindly provided with anti-MV hemagglutinin (HA) monoclonal antibodies (anti MV HA MAbs) by S. Schneider-Schaulies and V. ter Meulen (Institute for Virology and Immunology, Wurzburg, Germany). We purchased Cy3-conjugated goat anti-mouse secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA). We used anti-mouse Alexa 647-conjugated cholera toxin B subunit (CTXB) (Molecular Probes, Eugene, OR) for fluorescent visualization of the lipid rafts. We purchased filipin for the staining of cellular cholesterol and simvastatin for the cholesterol depletion analysis from Sigma (S6196; Rehovot, Israel).
Total RNA isolation and mRNA amplification.
We disrupted cultured cells using Trizol reagent (Invitrogen, Carlsbad, CA) and isolated total RNA according to the Trizol protocol. We double-amplified all total RNA samples using a RiboAmp RNA Amplification kit (Arcturus, Mountain View, CA). We evaluated the quality of amplified RNA by capillary electrophoresis using an Agilent 2100 Bioanalyzer.
Expression microarray format and data analysis.
We used mouse oligonucleotide microarrays, carrying approximately 22,000 oligonucleotides representing approximately 21,000 genes, from Agilent Technologies, Santa Clara, CA. For each infection type, we competitively hybridized four replicate arrays using the dye label reverse technique and scanned as previously described by Geiss et al. (9), using two samples, one from an infected cell plate and one from a plate containing an uninfected NS20Y control. We uploaded the resulting data into Rosetta Resolver System, version 6.0 (Rosetta Biosoftware, Seattle, WA). We used this system to compute mean ratios between the expression levels of each gene in the analyzed sample pair, standard deviations, and P values (10, 27) and for subsequent analyses. Resolver generates P values and error measurements, and the Agilent system performs error modeling before data are loaded into the Resolver system. Resolver performs a squeeze operation that creates ratio profiles by combining replicates while applying error weighting. The error weighting consists of adjusting for additive and multiplicative noise. A P value is generated that represents the probability that a gene is differentially expressed. Resolver allows users to set thresholds below which genes of a P value are considered to be significantly differentially expressed. Resolver then combines ratio profiles to create ratio experiments using an error-weighted average, as described in R. Stoughton and H. Dai (28).Gene expression data from this study are available at http://viromics.washington.edu.
Identifying perturbed metabolic pathways.
We used a GSEA (29) to identify KEGG metabolic pathways (13) and GO (2) functions that were perturbed differentially during acute and persistent infection. In preparation for this analysis, we used the Rosetta Resolver System to compute a new ratio for each gene that was a measure of the gene's expression during a persistent infection compared to acute infection. In short, given a list of genes ranked in decreasing order by their recomputed ratios, GSEA allowed us to identify which pathways and functions contain genes concentrated at the top of the list (signifying a greater perturbation in a persistent infection) and those at the bottom (signifying a greater perturbation in an acute infection). We used a dynamic program method to determine the exact P value for each pathway (23). We retained only pathways with P values of ≤0.05. We tested the significance only of GO functions with a depth of at least four in the GO hierarchy to avoid nonspecific terms. Given the hierarchical structure of GO, it is possible that several related functions can appear as enriched. In these cases, we removed any function if a child function was more enriched (i.e., lowest P value).
qRT-PCR.
We used quantitative real-time reverse transcription-PCR (qRT-PCR) to verify expression levels of selected genes and to determine the intracellular viral load. We treated total RNA samples with DNase, using a DNA-free DNase kit (Ambion, Inc., Austin, TX). We generated cDNA using reverse transcription reagents and random hexamers (Applied Biosystems, Foster City, CA). We performed qRT-PCR on an ABI 7500 real-time PCR system using TaqMan chemistry (Applied Biosystems, Foster City, CA). We performed two independent biological replicates on each target gene, each in quadruplicate in 20-μl reaction volumes with TaqMan 2× PCR Universal Master Mix (Applied Biosystems, Foster City, CA). We chose 18S rRNA as an endogenous control to normalize quantification of the target. We calculated the quantification of each gene relative to the calibrator on the instrument by using the 2−ΔΔCT (16) equation (where CT is threshold cycle) within the Applied Biosystems Sequence Detections software, version 1.3.
Immunofluorescence staining and confocal microscopy.
We fixed NS20Y/MS and NS20Y cells (1 × 105) grown on poly-l-lysine-treated glass coverslips with 2% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS) for 30 min at 40°C and then washed the cells three times with PBS. We then permeabilized cells using 0.01% Triton X-100 in PBS containing 3% bovine serum albumin (BSA) and 4% FCS and incubated them for 1 h at 40°C. Following permeabilization, we stained the cells at 40°C for 1 h with anti MV HA antibodies (1:1,000) in PBS containing 3% BSA and 4% FCS. After three washes with PBS, we diluted Cy3-conjugated secondary antibodies 1:200 in PBS-3% BSA-4% FCS and added CTXB (1 μg/ml) at 40°C for 1 h in the dark. Finally, we washed the samples three times with PBS and analyzed the samples by fluorescent confocal microscopy (LSM510, Carl Zeiss, Jena, Germany).
Flow cytometry.
For quantitative evaluation of lipid rafts, cells were suspended in 20 μl of PBS containing CTXB (1 μg/ml), incubated at 40°C for 1 h, washed three times with PBS, and analyzed with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). For cholesterol measurements, we fixed 0.3 × 106 cells in 4% paraformaldehyde in 0.1 M PBS at 40°C for 10 min and then at room temperature for 30 min. After four washes with PBS, cells were suspended in 100 μl of PBS containing filipin (20 μg/ml), incubated at 40°C for 1 h in the dark, and analyzed using a FACSCanto II flow cytometer. We recorded fluorescent signal emitted by filipin with a 450/30 band-pass filter and excitation using a 405-nm laser.
Simvastatin treatment of acute MV infection.
For immunofluorescence studies, we grew 105 NS20Y cells on poly-l-lysine-treated glass coverslips. For giant-cell evaluation, we seeded 105 NS20Y cells in six-well plates and removed the growth medium 24 h later. Then, we washed the monolayers and infected them with MV at an MOI of 0.01 (this MOI was selected since in our experience using higher virus titers resulted in extremely rapid development of cytopathic effect [CPE] and loss of the cell culture, prohibiting analysis) for 1 h at room temperature. We then washed the infected monolayers and added various amounts (5 and 20 μM) of simvastatin in DMEM without FCS and incubated the cells for 3 h at 37°C. After the incubation, we washed the cells and incubated them for an additional 24 or 48 h with DMEM containing 2% FCS. We evaluated cell viability to be no less then 90% using the trypan blue exclusion test. We counted multinucleated cells (containing at least six nuclei) 48 h later under light microscopy (average of three fields per sample) and collected culture supernatants for viral titration on Vero cells as previously described (22).
RESULTS
Global gene expression patterns distinguish acute and persistent infection.
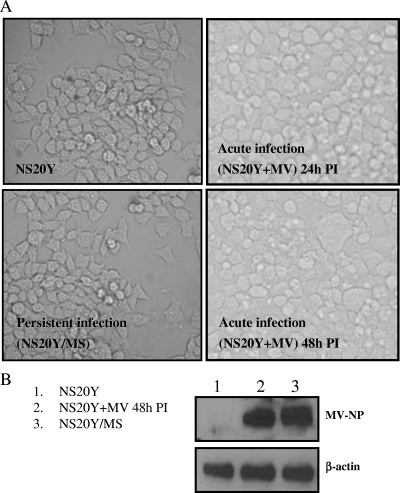
Acute infection of neuroblastoma cells (NS20Y) with MV is characterized by a CPE, formation of giant multinucleated cells (Fig. 1A), viral budding, and the production of infectious viral progeny, whereas the persistently infected cell line (NS20Y/MS) is morphologically indistinguishable from the uninfected NS20Y cell line. The NS20Y/MS cell line contains a large number of MV NP matrices, with very few budding virus particles (22). To screen for host cell pathways that could mediate differences between acute and persistent infection, we used acutely infected cells (NS20Y+MV) 48 h postinfection and NS20Y/MS cells after 48 h in culture. At this time point, the expression of MV NP protein was similar in acutely and persistently infected cells (Fig. 1B). We performed two microarray experiments, one for the acute infection and one for the persistent infection. Data were collected from four technical replicates for each infection type and combined using the Rosetta Resolver System software (see Materials and Methods). Every individual experiment independently compared gene expression of NS20Y+MV cells or NS20Y/MS cells to uninfected NS20Y cells.
FIG. 1.
Acute and persistent infection with MV of NS20Y cells. NS20Y cells were acutely infected with MV as described in Materials and Methods. (A) Photomicrographs (magnification, ×20), of parental cell line NS20Y, acutely infected NS20Y+MV cells at 24 and 48 h postinfection with MV, and persistently infected NS20Y/MS cells. CPE in the acutely infected cells is noticeable at 24 h postinfection. At 48 h postinfection, abundant giant cells and a significant CPE are observed. Persistently infected NS20Y/MS cells are morphologically indistinguishable from the NS20Y cells. (B) MV NP expression in NS20Y+MV cells (48 h postinfection) and in NS20Y/MS cells. PI, postinfection.
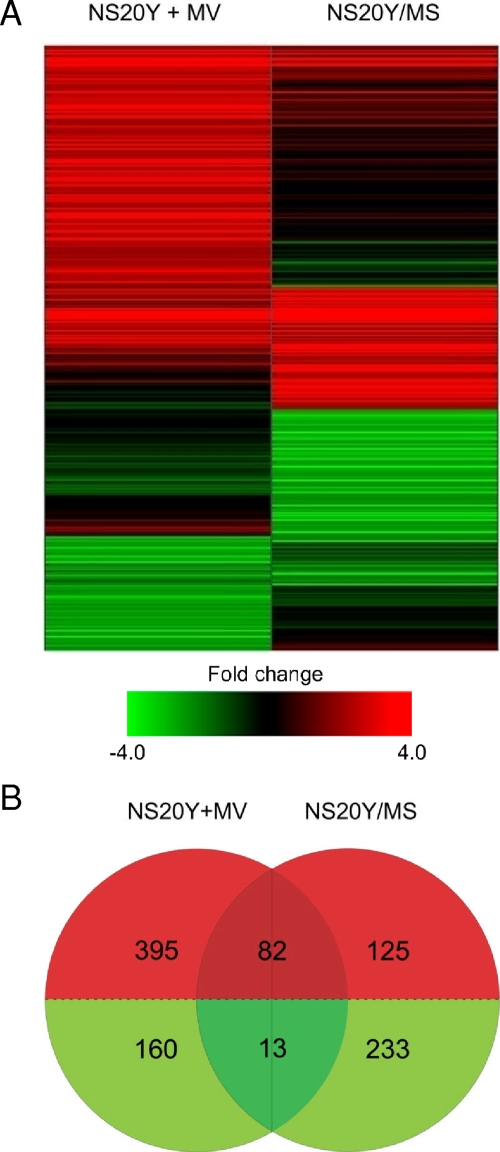
Of the approximately 21,000 transcripts measured, 1,008 genes (approximately 5%) were differentially expressed (≥2.0-fold change; P ≤ 0.01) in either persistent or acute infection, or both. The distinct global gene expression profiles of cells acutely and persistently infected with MV enabled infection types to be easily distinguished by the two-dimensional clustering algorithm (Fig. 2A). More gene expression changes (650 genes) were associated with acute infection than with persistent infection (453 genes) (Fig. 2B). Genes induced during acute infection outnumbered genes induced during persistent infection by more than a 2:1 ratio. Differential gene expression during persistent infection was characterized predominantly by gene downregulation, and only 95 genes were coregulated in both infection types. Coregulation during acute and persistent infection was almost solely associated with gene induction as there were very few genes downregulated in both infection modes (Fig. 2B). These results indicate that acute and persistent infections result in a different cascade of transcriptional events.
FIG. 2.
Global gene expression profile of NS20Y+MV cells (48 h postinfection) and NS20Y/MS cells. (A) Shown are 1,076 differentially expressed genes (≥2.0-fold change; P ≤ 0.01). Genes were sorted using hierarchical clustering, performed by the Rosetta Resolver System software with a divisive algorithm, average link heuristic criteria, and cosine correlation metric. Green and red bands represent decreased or increased levels of mRNA expression relative to uninfected neuroblastoma cells, respectively. (B) A Venn diagram depicting the differentially expressed genes (≥2.0-fold change; P ≤ 0.01) common or unique to acute and persistent MV infection.
Functional enrichment of KEGG metabolic pathways and GO functions in acute and persistent MV infections.
We used a GSEA (29) to determine which KEGG metabolic pathways (13) and GO (2) functions were differentially perturbed during acute and persistent infection. This analysis identified 35 KEGG pathways and 85 GO functions that were statistically enriched (P ≤ 0.05). The full list of the enriched pathways and functions can be found in File S1 in the supplemental material. Interestingly, several of these pathways were linked through common pathway metabolites to the cholesterol biosynthesis pathway, and we identified several enriched functions related to cholesterol biosynthesis, including hexosaminidase activity (P = 0.002; ranked 11 of 85 total), cholesterol metabolic processes (P = 0.004; rank, 15/85), steroid hormone receptor binding (P = 0.004; rank, 18/85), alkaline phosphatase activity (P = 0.013; rank, 31/85), hydroxymethylglutaryl-coenzyme A (CoA) reductase (HMGCR) activity (P = 0.035; rank, 65/85), acetyl-CoA C-acetyltransferase activity (P = 0.045; rank, 77/85), and several others. Alkaline phosphatase is selectively targeted and incorporated into lipid rafts, and HMGCR is the rate-limiting step in cholesterol synthesis. Thus, it is possible that the transcriptional events in these functional gene groups all stem from one regulatory cascade.
Cholesterol biosynthesis is downregulated in persistent MV infection.
Of the 15 genes associated with cholesterol biosynthesis that were measured in our microarray experiments, 10 were differentially expressed in either persistent or acute infection compared with the uninfected NS20Y parental cell line (Table 1). Expression levels of nearly all the differentially expressed genes in the cholesterol biosynthesis pathway were significantly lower during persistent infection than during acute infection. On the basis of this observation, we proceeded to validate the microarray findings and determine whether a link exists between such differential gene regulation and differences in MV infection modes in this murine neuroblastoma model.
TABLE 1.
Genes in the cholesterol biosynthesis pathway that are differentially regulated in acute or persistent infection with MV
| Gene symbol | Gene description | Protein action | Differential expression ina:
|
|||
|---|---|---|---|---|---|---|
| NS20Y+MV cells
|
NS20Y/MS cells
|
|||||
| Fold change | P value | Fold change | P value | |||
| CYP51A1 | Cytochrome P450 51A1 | Catalyzes the demethylation of lanosterol to 4,4′-dimethyl cholesta-8,14,24-triene-3-beta-ol | 0.5 | 0.003 | ||
| FDPS | Farnesyl pyrophosphate synthetase | Catalyzes condensation of isopentenyl pyrophosphate with allylic pyrophosphates, dimethylallyl pyrophosphate, and the reaction's product geranylpyrophosphate, to form farnesyl pyrophosphate | 0.5 | 0.00249 | ||
| HMGCR | Hydroxy-methylglutaryl-CoA reductase | Involved in the control of cholesterol biosynthesis; it is the rate-limiting enzyme of sterol biosynthesis | 1.3 | 0.00032 | 0.4 | 2.22E-08 |
| HMGCS1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | This enzyme condenses acetyl-CoA with acetoacetyl-CoA to form HMG-CoA, which is the substrate for HMGCR | 2.2 | 3.46E-20 | 0.5 | 2.04E-21 |
| IDI1 | Isopentenyl-diphosphate delta-isomerase 1 | Catalyzes allelic rearrangement of IPP to its allylic isomer DMAPP | 2.4 | 1.26E-19 | 0.6 | 0.00002 |
| MVD | Diphosphomevalonate decarboxylase | Performs the first committed step in the biosynthesis of isoprenes | 0.4 | 1.37E-10 | ||
| NSDHL | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | Steroid dehydrogenase, NAD(P) dependent | 0.5 | 0.00011 | ||
| ERG25 | C-4 methylsterol oxidase | Catalyzes the first of three steps required to remove two methyl groups from an intermediate in sterol biosynthesis | 2 | 2.68E-12 | 0.3 | 0 |
| SQLE | Squalene monooxygenase | Catalyzes the first oxygenation step in sterol biosynthesis and is suggested to be one of the rate-limiting enzymes in this pathway | 2.4 | 1.19E-10 | 0.4 | 1.04E-25 |
| ERG24 | Delta(14)-sterol reductase | Involved in the conversion of lanosterol to cholesterol | 2.3 | 1.11E-15 | 0.4 | 0.00355 |
Changes in expression are relative to the uninfected control cell line, as measured by the expression arrays, and P values were derived using the Rosetta Resolver error model, as described in Materials and Methods. We discarded expression data for genes not meeting our P value cutoff of 0.01.
qRT-PCR verification of HMGCR and SQLE.
We next used TaqMan qRT-PCR to verify expression levels of the HMGCR and SQLE genes, which encode the rate-limiting steps of cholesterol biosynthesis. Measurements were obtained using two independent probe sets, each in quadruplicate. Using the 2−ΔΔCT method, HMGCR expression in NS20Y/MS cells was shown to be 38.5% of HMGCR expression levels in NS20Y+MV cells (ΔΔCT, 1.376 ± 0.406). For SQLE, the expression levels in NS20Y/MS cells were 25.5% of those found in NS20Y+MV cells (ΔΔCT, 1.94 ± 0.849). The significance of the difference between the means was determined using a homoscedastic t test with a one-tailed distribution. The associated probabilities were 7.736E-8 for HMGCR and 1.399E-4 for SQLE.
Cholesterol downregulation in NS20Y/MS cells.
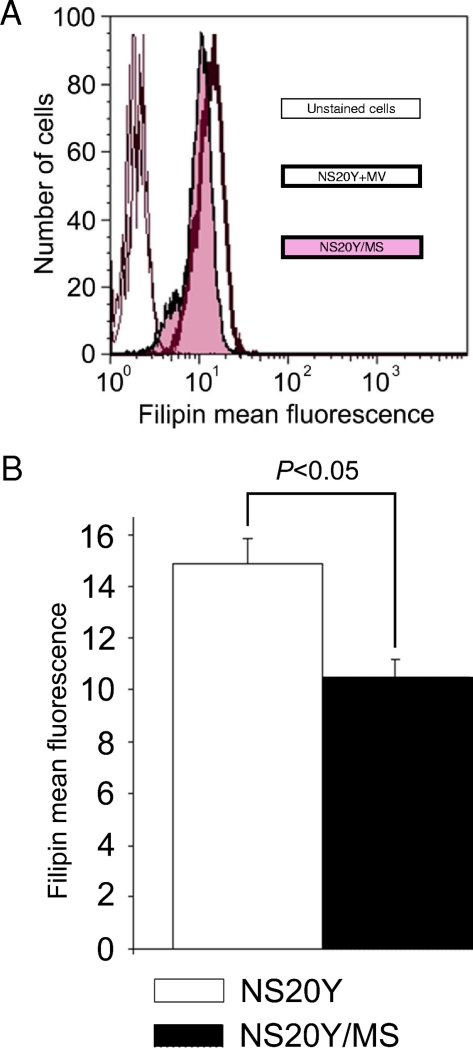
To examine whether transcriptional downregulation of cholesterol biosynthesis in NS20Y/MS cells was being manifested as lower cellular cholesterol levels, we used filipin staining to measure nonesterified cholesterol, the end product of the cholesterol biosynthesis pathway. For each infection mode, we calculated the mean and standard deviation of filipin binding to 3 × 105 cells and compared the values using a Student's t test. We observed that cellular cholesterol levels were lower in persistently infected cells than in uninfected NS20Y cells to a modest but reproducible degree, which was statistically significant (P < 0.05) (Fig. 3).
FIG. 3.
Decreased cellular content of cholesterol in NS20Y/MS cells. NS20Y and NS20Y/MS cells were probed for cellular cholesterol using filipin staining and analyzed by flow cytometry as described in Materials and Methods. (A) A histogram overlay representing a typical experiment. Overlapping thin curves located in the first logarithmic decade represent unstained NS20Y and NS20Y/MS cells. Bold and tinted curves represent filipin staining of NS20Y and NS20Y/MS cells, respectively. (B) Average ± standard deviation of mean fluorescence intensities of filipin-stained NS20Y and NS20Y/MS cells (six replicates of two independent experiments).
Downregulation of lipid rafts in NS20Y/MS cells.
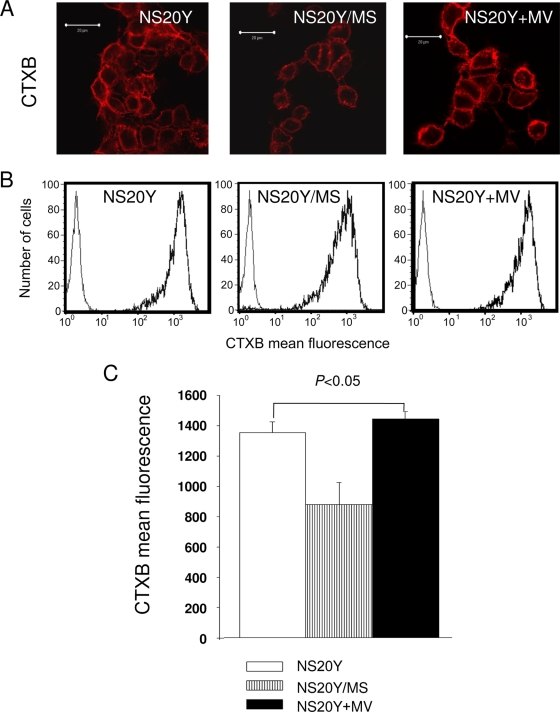
It has been previously shown that MV assembly and budding take place at cholesterol-rich domains on the cellular membrane known as lipid rafts (17). Thus, we investigated whether there was a link between transcriptional downregulation of cholesterol biosynthesis and the inhibition of viral budding observed in cells persistently infected with MV (22). For this purpose, we treated cells with CTXB, a fluorochrome-conjugated probe that interacts with GM1, a glycolipid marker associated with lipid rafts (3). Using confocal microscopy, we found that in persistently infected NS20Y/MS cells, GM1 was markedly underexpressed at the plasma membrane relative to control NS20Y cells or to acutely infected (NS20Y+MV) cells (Fig. 4A). Mean standard deviation and P values were calculated as described earlier for the filipin-binding assay. We also found a small but reproducible and statistically significant (P < 0.05) decrease in the CTXB mean fluorescence intensity of NS20Y/MS cells compared to that of control or acutely infected cells using a quantitative analysis by flow cytometry (Fig. 4B and C). These observations indicate that in comparison to acute MV infection of NS20Y cells, persistently infected NS20Y/MS cells expressed significantly lower amounts of lipid rafts at their plasma membranes.
FIG. 4.
Persistent infection with MV decreases the expression of GM1. NS20Y, NS20Y/MS, and NS20Y+MV cells were stained with Alexa 647-conjugated CTXB and analyzed by confocal microscopy and flow cytometry. (A) Fluorescent images of CTXB-stained cells (magnification, ×100). Scale bar corresponds to 20 μm. (B) Flow cytometry showing CTXB binding to the cells (bold). Thin curves in the first logarithmic decade of the x axis show nonstained control cells. The x axis depicts fluorescence intensity while the y axis depicts cell numbers. (C) Quantitative analysis of CTXB binding performed by flow cytometry. Average ± standard deviation of mean fluorescence intensities of CTXB-stained cells (nine replicates of three independent experiments).
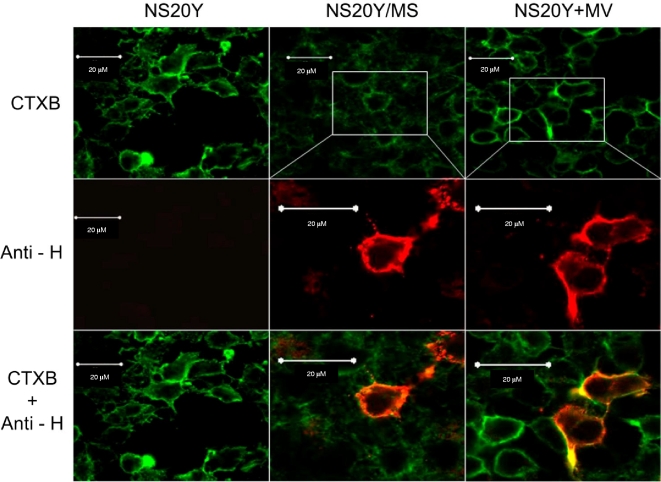
MV colocalizes to a depleted pool of lipid rafts in NS20Y/MS cells.
To verify the tropism of MV proteins to lipid rafts, we double-labeled NS20Y/MS cells and NS20Y+MV cells with anti MV HA antibodies and CTXB. Confocal microscopy revealed that in acutely infected cells, MV HA protein colocalized with the lipid raft marker GM1. Although NS20Y/MS cells displayed a significant depletion in membrane lipid raft content, the distribution of MV HA was exclusive to the plasma membrane (Fig. 5). These observations establish the tropism of MV HA to lipid rafts during both persistent and acute MV infection. The absence of a significant effect on MV HA presence in the lipid raft-depleted membrane indicated that the amounts of viral protein produced during persistent and acute MV infections were comparable; thus, the lack of infective particles in NS20Y/MS supernatant must stem from a defect in viral budding or in the assembly stage. To verify this, we used qRT-PCR to quantify the amount of intracellular RNA coding the highly conserved MV L protein. Using two biological replicates and four technical replicates each, we were not able to detect any significant difference in RNA levels between persistently and acutely infected cells (data not shown). These findings strengthen our premise that the very limited amount of infectious viral particles found in the medium of persistently infected cells is the result of a defect in viral assembly or budding rather than of a decrease in viral replication.
FIG. 5.
MV HA protein colocalizes with the lipid raft marker GM1. Confocal microscopy of NS20Y, NS20Y/MS, and NS20Y+MV cells stained with CTXB (green) or anti-HA MAb (anti-H) and Cy3-conjugated secondary antibodies (red; second row). Merged images are shown in the third row, and yellow regions denote overlaps between red- and green-stained membrane areas.
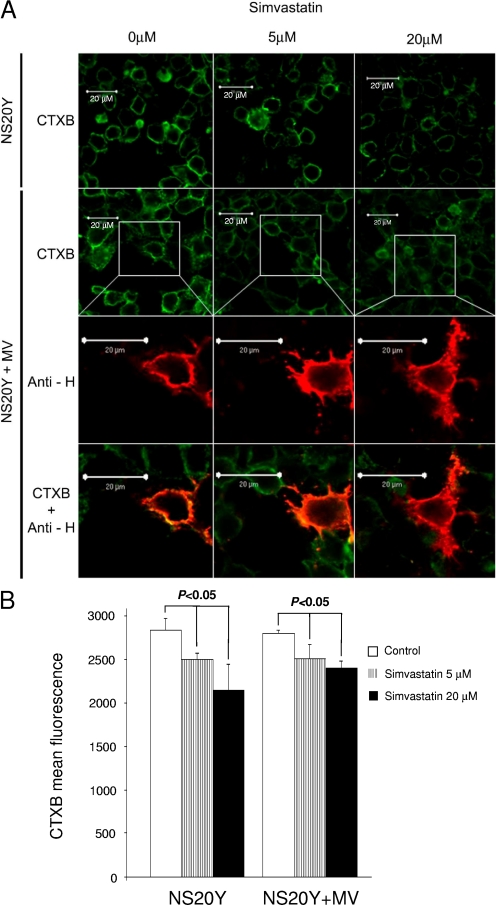
Simvastatin treatment impairs lipid raft integrity and reduces giant-cell formation and virus yields during acute infection.
We speculated that cholesterol downregulation, which impairs lipid raft integrity, may result in the inhibition of viral budding and infectivity in NS20Y/MS cells. If so, then pharmacological inhibition of cholesterol biosynthesis should have a similar effect on the productivity of acute MV infection. To test this hypothesis, we acutely infected NS20Y cell cultures with MV at an MOI of 0.01 and then treated the cultures for 3 h at 37°C with various concentrations of simvastatin (0 to 20 μM), an HMGCR inhibitor routinely used for inhibition of cholesterol synthesis. We examined the effect of this treatment on lipid rafts 24 and 48 h later by looking for the formation of giant cells, which are characteristic of acute MV infection. Using immunofluorescence staining, the effect on CTXB binding to uninfected or NS20Y+MV cells was already apparent with 5 μM simvastatin, and a more profound effect was obtained after treatment with 20 μM simvastatin (Fig. 6A, top two rows). This treatment did not have any significant effect on cell viability or morphology at any of the treatment concentrations. Figure 6A (rows 3 and 4) demonstrates that simvastatin treatment impairs raft integrity but does not effect the expression of MV HA protein at the cell membrane; thus, intracellular viral protein synthesis is apparently unaffected by simvastatin. We performed a fluorescence-activated cell sorting analysis to quantify the effect of simvastatin on the amount of lipid rafts in NS20Y and NS20Y+MV cells (Fig. 6B), which demonstrated a significant (P < 0.05) decrease in CTXB binding in response to simvastatin treatment.
FIG. 6.
The effect of simvastatin on the expression of lipid rafts in NS20Y and NS20Y+MV cells. NS20Y cells were acutely infected with MV and treated with 5 μM or 20 μM simvastatin as described in Materials and Methods. Cells were probed with Alexa 647-conjugated CTXB. MV HA protein was stained with anti-H MAb and Cy3-conjugated secondary antibodies. (A) Confocal microscopy of NS20Y cells treated with simvastatin and probed with CTXB are shown in the first row. NS20Y+MV cells treated with simvastatin and probed with CTXB are shown in the second row. The third row shows NS20Y+MV cells treated with simvastatin and stained with anti-HA MAb (anti-H) and Cy3-conjugated secondary antibodies. The fourth row represents merged images of NS20Y+MV cells treated with simvastatin. Scale bar, 20 μm. (B) Fluorescence-activated cell sorting analysis of CTXB binding to simvastatin-treated cells. Data are mean fluorescence intensities ± standard deviations of CTXB-stained NS20Y and NS20+MV cells (nine replicates of three independent experiments).
We assessed the effects of simvastatin treatment on the formation of giant cells and viral production 48 h after infection. The reduction in the number of giant cells and virus plaques was dose dependent, suggesting that cholesterol synthesis and raft integrity are important factors for successful budding and production of infectious MV progeny (Table 2).
TABLE 2.
A dose-dependent reduction of MV virus production in acute infection of NS20Y cells following simvastatin treatmenta
| Simvastatin treatment (μM) | No. of giant cells | PFU/ml |
|---|---|---|
| 0 | 66 | 3,300 |
| 5 | 49 | 650 |
| 20 | 20 | 0 |
In comparison, in NS20Y/MS cells in our previous study (22), the number of giant cells was zero, and virus was detected in supernatant at 1 to 10 PFU/ml.
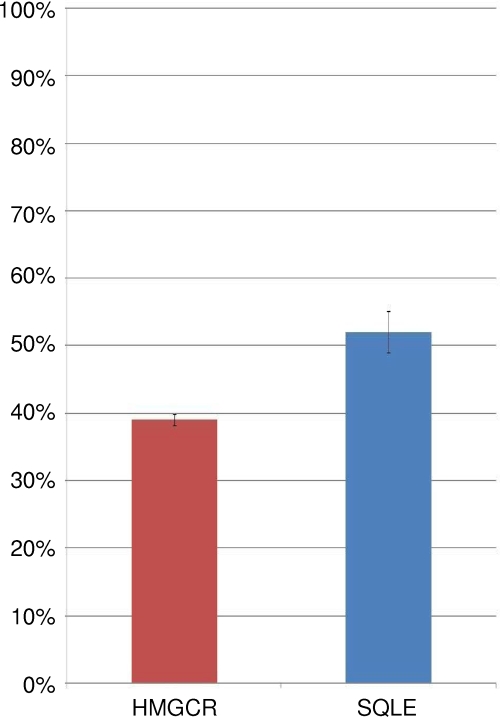
qRT-PCR quantification of human HMGCR and SQLE in MV infection.
In order to determine whether the downregulation of rate-limiting enzymes in the pathway for cholesterol biosynthesis also occurred in human cells, we performed qRT-PCR measurements on human neuroblastoma cells persistently and acutely infected with MV. The results, summarized in Fig. 7, indicate that downregulation of the cholesterol biosynthesis pathway takes place during persistent MV infection of human as well as mouse neuroblastoma cells.
FIG. 7.
TaqMan qRT-PCR quantification of HMGCR and SQLE, the rate-limiting steps of cholesterol synthesis, in human neuroblastoma cells infected with MV. Data are presented as percent downregulation of the cholesterol synthesis gene transcripts in persistent infection, relative to their expression levels during acute MV infection. Means and standard errors from four technical replicates are shown.
DISCUSSION
Persistent MV infection is characteristically distinct from acute infection. Although a persistent infection is virtually indistinguishable histologically from uninfected cells, persistently infected cells display very low or absent viral budding while indefinitely maintaining viral RNA expression and the capability of again developing into an acute infection.
Previous global transcriptional profiling studies involving MV have primarily focused on acute MV infection (4, 37, 38). These studies measured the expression of 3,000 to 13,000 genes and described the upregulation of genes associated with apoptosis, antigen presentation, interferon-mediated antiviral response, and the downregulation of genes associated with oxidative phosphorylation and protein synthesis. The only previous microarray study of persistent MV infection was done using peripheral blood mononuclear cells (which do not become persistently infected) from patients with SSPE (31).
The susceptibility of murine neuroblastoma cells and other types of murine cells to infection with MV has previously been reported by us (22) and by other groups (6, 24). In this study, we used a well-established murine neuroblastoma model of persistent and acute infection, together with microarray analysis, to construct a global profile of transcriptional events associated with acute and persistent infection. These analyses revealed that acute MV infection was associated with the upregulation of over twice as many genes as persistent infection. We then used GSEA to identify KEGG metabolic pathways and GO functions perturbed by MV infection. GSEA revealed several pathways and functions that were more perturbed in persistent infection than acute infection and identified a link to cholesterol biosynthesis, leading to our exploration of the cholesterol biosynthesis pathway.
Disruption of lipid metabolism during MV infection has been previously reported (1, 14, 30, 34) and is thought to play a role in pathology. However, the activity of the cholesterol synthesis pathway has not been previously compared between acute and persistent infections. In our experiments, most genes associated with the cholesterol biosynthesis pathway were downregulated in persistent versus acute infections. This included HMGCR and SQLE, the genes associated with the rate-limiting steps of cholesterol synthesis. Furthermore, we found that lower cholesterol synthesis in persistent infection was directly correlated with lower membrane cholesterol levels and also a decrease in the abundance of GM1, a marker for lipid rafts, the cholesterol-rich microdomains in the cell membrane where MV assembly and budding are thought to take place. This integration process into the membrane is believed to be regulated as only a fraction of virus proteins are ultimately incorporated into the rafts, and even minute differences in membrane cholesterol content have been shown to be biologically meaningful (18).
Pharmacological depletion of cellular cholesterol by simvastatin inhibition of HMGCR results in significantly impaired lipid raft formation (26, 36). However, the quantitative contribution of lipid raft abundance to virus production has not been previously assessed due to impaired cell viability following treatment with cholesterol-lowering drugs. Here, we were able to downregulate cholesterol synthesis successfully without affecting cell survival. By treating acutely infected cells with simvastatin, we were able to reduce viral titers to undetectable levels. Taken together, these findings serve to recapitulate the proposed association between cholesterol synthesis and the state of MV infection.
Establishing an association between cholesterol downregulation and inhibition of viral budding does not imply a causative relationship. Viral budding could indirectly drive cholesterol synthesis by depleting the cell membrane, and if budding was impaired for any reason, less cholesterol synthesis would be required. Alternatively, the amount of cholesterol synthesized could determine the rate of viral budding, and inhibition of cholesterol synthesis is the initiating event. To distinguish between these alternatives, we compared the levels of lipid rafts present in persistent and uninfected cells. If viral budding is driving cholesterol synthesis by depleting the cell membrane of lipids, then persistent MV infection (which is associated with little or no viral budding) would not be expected to affect lipid raft content. The reduced abundance of lipid rafts observed in persistently infected cells would therefore be unaccounted for. Our findings therefore suggest that it is the downregulation of cholesterol synthesis that drives the inhibition of viral budding.
To summarize, our findings suggest that regulation of cellular cholesterol synthesis is a primary step in the establishment of persistent MV infection. A possible explanation for the existence of a cellular mechanism for downregulation of cholesterol during viral infection is to serve as an innate antiviral mechanism aimed at limiting viral budding. While this may result in the containment and elimination of infection in cells with a life span of several days or weeks, it can result in long-term or lifelong infection if the infected cells are immortal. This cholesterol-based antiviral mechanism could also possibly interfere with the life cycle of other enveloped viruses that depend on lipid rafts for assembly. Interestingly, cholesterol synthesis was one of the few affected pathways common to both human immunodeficiency virus type 1 and influenza virus when these were compared (32). Additional studies are therefore warranted to determine whether this phenomenon can be exploited therapeutically through the utilization of widely available cholesterol-inhibiting drugs as a primary or complementary therapeutic approach to viral infection.
Supplementary Material
Acknowledgments
This study was supported in part by the Center for the Study of Emerging Diseases and the National Institute of Allergy and Infectious Disease (grant P01 AI058113).
We thank Jindrich Cinatl, Jr., at the Zentrum der Hygiene, Institut fur Medizinische Virologie, Germany, for providing the human neuroblastoma cell line. We thank S. Schneider-Schaulies and V. ter Meulen (Institute for Virology and Immunology, Wurzburg, Germany) for providing the anti-MV hemagglutinin monoclonal antibodies.
Footnotes
Published ahead of print on 18 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderton, P., T. F. Wild, and G. Zwingelstein. 1982. Phospholipids in a measles virus persistent infection: modification of fatty acid metabolism and fatty acid composition of released virus. J. Gen. Virol. 62249-258. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 2525-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blank, N., M. Schiller, S. Krienke, G. Wabnitz, A. D. Ho, and H. M. Lorenz. 2007. Cholera toxin binds to lipid rafts but has a limited specificity for ganglioside GM1. Immunol. Cell Biol. 85378-382. [DOI] [PubMed] [Google Scholar]
- 4.Bolt, G., K. Berg, and M. Blixenkrone-Moller. 2002. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 831157-1165. [DOI] [PubMed] [Google Scholar]
- 5.Cattaneo, R., and J. K. Rose. 1993. Cell fusion by the envelope glycoproteins of persistent measles viruses which caused lethal human brain disease. J. Virol. 671493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunster, L. M., J. Schneider-Schaulies, M. H. Dehoff, V. M. Holers, R. Schwartz-Albiez, and V. ter Meulen. 1995. Moesin, and not the murine functional homologue (Crry/p65) of human membrane cofactor protein (CD46), is involved in the entry of measles virus (strain Edmonston) into susceptible murine cell lines. J. Gen. Virol. 762085-2089. [DOI] [PubMed] [Google Scholar]
- 7.Freeman, J. M., R. L. Magoffin, E. H. Lennette, and R. M. Herndon. 1967. Additional evidence of the relation between subacute inclusion-body encephalitis and measles virus. Lancet 2129-131. [DOI] [PubMed] [Google Scholar]
- 8.Gascon, G. G. 1996. Subacute sclerosing panencephalitis. Semin. Pediatr. Neurol. 3260-269. [DOI] [PubMed] [Google Scholar]
- 9.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 754321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 776367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirano, A., A. H. Wang, A. F. Gombart, and T. C. Wong. 1992. The matrix proteins of neurovirulent subacute sclerosing panencephalitis virus and its acute measles virus progenitor are functionally different. Proc. Natl. Acad. Sci. USA 898745-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horta-Barbosa, L., D. A. Fuccillo, J. L. Sever, and W. Zeman. 1969. Subacute sclerosing panencephalitis: isolation of measles virus from a brain biopsy. Nature 221974. [DOI] [PubMed] [Google Scholar]
- 13.Kanehisa, M., S. Goto, S. Kawashima, Y. Okuno, and M. Hattori. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32D277-D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ledeen, R. W., C. A. Miller, J. E. Haley, and C. S. Raine. 1976. Lipids and slow viruses: comparison of measles and SSPE virions. Adv. Exp. Med. Biol. 68567-584. [DOI] [PubMed] [Google Scholar]
- 15.Lin, F. H., and H. Thormar. 1980. Absence of M protein in a cell-associated subacute sclerosing panencephalitis virus. Nature 285490-492. [DOI] [PubMed] [Google Scholar]
- 16.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 17.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh, H. Y., E. J. Lee, S. Yoon, B. H. Chung, K. S. Cho, and S. J. Hong. 2007. Cholesterol level of lipid raft microdomains regulates apoptotic cell death in prostate cancer cells through EGFR-mediated Akt and ERK signal transduction. Prostate 671061-1069. [DOI] [PubMed] [Google Scholar]
- 19.Ono, A., and E. O. Freed. 2005. Role of lipid rafts in virus replication. Adv. Virus Res. 64311-358. [DOI] [PubMed] [Google Scholar]
- 20.Patterson, J. B., T. I. Cornu, J. Redwine, S. Dales, H. Lewicki, A. Holz, D. Thomas, M. A. Billeter, and M. B. Oldstone. 2001. Evidence that the hypermutated M protein of a subacute sclerosing panencephalitis measles virus actively contributes to the chronic progressive CNS disease. Virology 291215-225. [DOI] [PubMed] [Google Scholar]
- 21.Pohl, C., W. P. Duprex, G. Krohne, B. K. Rima, and S. Schneider-Schaulies. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J. Gen. Virol. 881243-1250. [DOI] [PubMed] [Google Scholar]
- 22.Rager-Zisman, B., J. E. Egan, Y. Kress, and B. R. Bloom. 1984. Isolation of cold-sensitive mutants of measles virus from persistently infected murine neuroblastoma cells. J. Virol. 51845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riancho, J. A. 2007. Authorship of scientific articles. Acta Otorrinolaringol. Esp. 58281-283. [In Spanish.] [PubMed] [Google Scholar]
- 24.Schubert, S., K. Moller-Ehrlich, K. Singethan, S. Wiese, W. P. Duprex, B. K. Rima, S. Niewiesk, and J. Schneider-Schaulies. 2006. A mouse model of persistent brain infection with recombinant measles virus. J. Gen. Virol. 872011-2019. [DOI] [PubMed] [Google Scholar]
- 25.Segev, Y., B. Rager-Zisman, N. Isakov, S. Schneider-Schaulies, V. ter Meulen, S. Udem, S. Segal, and M. Wolfson. 1994. Reversal of the measles virus-mediated increase of phosphorylating activity in persistently infected mouse neuroblastoma cells by anti-measles virus antibodies. J. Gen. Virol. 75819-827. [DOI] [PubMed] [Google Scholar]
- 26.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 131-39. [DOI] [PubMed] [Google Scholar]
- 27.Smith, M. W., Z. N. Yue, G. K. Geiss, N. Y. Sadovnikova, V. S. Carter, L. Boix, C. A. Lazaro, G. B. Rosenberg, R. E. Bumgarner, N. Fausto, J. Bruix, and M. G. Katze. 2003. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res. 63859-864. [PubMed] [Google Scholar]
- 28.Stoughton, R., and H. Dai. February 2002. Statistical combining of cell expression profiles. U.S. patent 6,351,712.
- 29.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 10215545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi, M., E. Watari, E. Shinya, T. Shimizu, and H. Takahashi. 2007. Suppression of virus replication via down-modulation of mitochondrial short chain enoyl-CoA hydratase in human glioblastoma cells. Antivir. Res. 75152-158. [DOI] [PubMed] [Google Scholar]
- 31.Takemoto, M., R. Kira, K. Kusuhara, H. Torisu, Y. Sakai, and T. Hara. 2005. Gene expression profiles in peripheral blood mononuclear cells from patients with subacute sclerosing panencephalitis using oligonucleotide microarrays. J. Neurovirol. 11299-305. [DOI] [PubMed] [Google Scholar]
- 32.van't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 771392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent, S., D. Gerlier, and S. N. Manie. 2000. Measles virus assembly within membrane rafts. J. Virol. 749911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wild, T. F., A. Bernard, N. A. Malak, G. Brichon, and G. Zwinglestein. 1986. Imprints of virus infection: can paramyxoviruses permanently modify triacylglycerol metabolism? Lipids 21608-611. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 2007. Measles. WHO fact sheet 286. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs286/en/.
- 36.Zhuang, L., J. Kim, R. M. Adam, K. R. Solomon, and M. R. Freeman. 2005. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J. Clin. Investig. 115959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zilliox, M. J., W. J. Moss, and D. E. Griffin. 2007. Gene expression changes in peripheral blood mononuclear cells during measles virus infection. Clin. Vaccine Immunol. 14918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zilliox, M. J., G. Parmigiani, and D. E. Griffin. 2006. Gene expression patterns in dendritic cells infected with measles virus compared with other pathogens. Proc. Natl. Acad. Sci. USA 1033363-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.