Abstract
Human immunodeficiency virus type 1 (HIV-1) assembly occurs predominantly at the plasma membrane of infected cells. The targeting of assembly to intracellular compartments such as multivesicular bodies (MVBs) generally leads to a significant reduction in virus release efficiency, suggesting that MVBs are a nonproductive site for HIV-1 assembly. In the current study, we make use of an HIV-1 Gag-matrix mutant, 29/31KE, that is MVB targeted. We previously showed that this mutant is severely defective for virus particle production in HeLa cells but more modestly affected in primary macrophages. To more broadly examine the consequences of MVB targeting for virus production, we investigated 29/31KE particle production in a range of cell types. Surprisingly, this mutant supported highly efficient assembly and release in T cells despite its striking MVB Gag localization. Manipulation of cellular endocytic pathways revealed that unlike Vpu-defective HIV-1, which demonstrated intracellular Gag localization as a result of Gag endocytosis from the plasma membrane, 29/31KE mutant Gag was targeted directly to an MVB compartment. The 29/31KE mutant was unable to support multiple-round replication; however, this defect could be reversed by truncating the cytoplasmic tail of the transmembrane envelope glycoprotein gp41 and by the acquisition of a 16EK change in matrix. The 16EK/29/31KE matrix mutant replicated efficiently in the MT-4 T-cell line despite maintaining an MVB-targeting phenotype. These results indicate that MVB-targeted Gag can be efficiently released from T cells and primary macrophages, suggesting that under some circumstances, late endosomal compartments can serve as productive sites for HIV-1 assembly in these physiologically relevant cell types.
Human immunodeficiency virus type 1 (HIV-1) assembly is driven by the synthesis of the Gag polyprotein precursor, Pr55Gag, in the cytosol of the infected cell, followed by Gag targeting to the site of virus assembly, Gag-membrane binding, Gag multimerization, and virion release. Concomitant with particle release, the viral protease (PR) cleaves Pr55Gag into four major structural and functional domains: p17 matrix (MA), p24 capsid (CA), p7 nucleocapsid (NC), and p6 (1, 17, 26, 36, 68, 70). The N-terminal MA domain of Pr55Gag directs the binding of Gag to the plasma membrane (PM), CA and NC domains are responsible for Gag-Gag interactions leading to Gag multimerization, and p6 promotes the pinching off of virions by interacting directly with cellular endosomal sorting machinery (3, 10, 45).
Despite the significant progress that has been made in the field of HIV-1 assembly in recent years, certain aspects of the virus assembly pathway remain to be fully understood. In particular, the identity of the subcellular compartment in which assembly takes place and the itinerary that Gag follows in trafficking to the site of assembly continue to be investigated. Considerable evidence suggests that in HeLa, 293T, Cos-7, and T-cell lines, HIV-1 assembly occurs predominantly at the PM (16, 23, 38, 54, 61, 68). However, several groups have proposed that late endosomal compartments serve as major or primary sites for HIV-1 assembly (28, 31, 50, 51, 58, 64). Furthermore, several cellular proteins and protein complexes that play important roles in protein sorting to and from the endosomal pathway have been implicated in Gag trafficking and assembly. Examples include the clathrin adapter protein complexes AP-1 (5), AP-2 (2), AP-3 (13); the Golgi-localized, γ-ear-containing, Arf-binding (GGA) proteins; and ADP ribosylation factors (Arfs) (37). The site of HIV-1 assembly in monocyte-derived macrophages (MDMs), a physiologically relevant cell type for HIV-1 infection, is perhaps the least well defined. Numerous electron microscopy (EM) studies have demonstrated that particle assembly in MDMs is evident predominantly in intracellular compartments identified as late endosomes or multivesicular bodies (MVBs) (11, 24, 37a, 50, 57-59). Moreover, virions released from infected macrophages are positive for MVB markers including the tetraspanins CD63, CD81, and CD82 (7, 50, 58), further supporting the existence of an intracellular assembly site in MDMs. This hypothesis, however, was challenged by evidence that the tetraspanin-enriched, HIV-1-positive internal compartments in MDMs are connected to the PM via a thin channel, suggesting that these compartments represent specialized PM invaginations rather than bona fide MVBs (12, 71). Also consistent with the concept that the apparently internal, virus-positive compartment in MDMs is distinct from MVBs is the observation that wild-type (WT) Gag in MDMs is rapidly transferred to sites of cell-cell contact, whereas an MVB-targeted Gag mutant is not transferred to the synapse (29).
Mutational analyses have indicated that the viral determinant of Gag targeting to the PM lies in the MA domain (15, 22, 34, 56, 66, 75, 76). The membrane-binding activity of MA is conferred by a myristic acid moiety covalently attached to the N-terminal Gly and a highly basic patch of residues near the N terminus of MA (residues 17 to 31) (4, 22, 56, 76). Point mutations in the MA basic domain or between residues 84 and 88 redirect virus assembly to late endosomal or MVB compartments (22, 54-56) and significantly reduce the efficiency of particle production in HeLa cells (22, 54-56). Mistargeting of Gag and defects in virus particle production are also caused by a disruption in the levels or distribution of phosphatidylinositol(4,5)bisphosphate [PI(4,5)P2], a PM phosphoinositide (52). The latter study demonstrated that PI(4,5)P2 is a cellular cofactor for Gag trafficking to the PM, and subsequent structural and virion incorporation analyses revealed a direct binding between PI(4,5)P2 and basic residues in MA (6, 18, 62, 63, 65). Liposome binding assays confirmed the interaction between MA and PI(4,5)P2 in the context of a lipid bilayer and full-length Gag (8). HIV-1 Gag can also be targeted to endosomal compartments by replacing the MA domain with foreign, endosome-targeting signals (38). Again, this mistargeting phenotype is associated with markedly impaired virus particle production (38). The observation of severe virus release defects induced by Gag targeting to late endosomes or MVBs (22, 38, 54) led to the conclusion that assembly in late endosomal compartments represents a dead end in the particle production pathway. Interestingly, the impact of basic domain mutations on virus release was significantly less severe in primary MDMs than in HeLa cells despite the fact that the MA mutants were still MVB targeted in MDMs (54).
In the current study, we made use of a previously described MVB-targeted MA mutant, 29KE/31KE (54) (here abbreviated 29/31KE), as a tool to resolve whether late endosomal membranes can serve as productive sites for HIV-1 assembly and release. Because T cells are the major target for HIV-1 infection in vivo, we were particularly interested in the impact of MVB targeting on virus production in this cell type. Our data demonstrate that while the MVB targeting phenotype of the 29/31KE MA mutant is maintained in HeLa, 293T, MDM, and T-cell lines, virus release efficiency is severely compromised only in HeLa and 293T cells. Most importantly, the level of virus particle production for this mutant is not significantly reduced in T cells. While the 29/31KE mutant displays a marked defect in establishing a spreading infection in both MDMs and T cells, by truncating the cytoplasmic tail of the transmembrane glycoprotein gp41 and selecting for a second-site mutation in MA (16EK), we were able to achieve efficient virus replication in the MT-4 T-cell line without altering the MVB-targeting phenotype. These results demonstrate that MVB-targeted Gag can direct efficient assembly and release in some cell types including T cells and primary MDMs.
MATERIALS AND METHODS
Cells, transfections, and infections.
HeLa and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% or 10% fetal bovine serum (FBS), respectively. T cells and MDMs were cultured in RPMI 1640 medium supplemented with 10% FBS. MDMs were obtained after culturing of elutriated monocytes for 7 days. Isolation and culture of human peripheral blood lymphocytes (PBLs) and MDMs were described previously (19). Transfections of HeLa and 293T cells were performed using Lipofectamine 2000 reagent (Invitrogen). MDMs and T cells were transfected with an Amaxa electroporator using the programs Y-10 and X-01, respectively. Infection of HeLa, 293T, MDM, and T cells and PBLs was performed with virus stocks pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G). Cells were cultured for 4 to 5 h with high-titer virus stocks at 10 counts/min of reverse transcriptase (RT) activity/cell. Before infection, PBLs were activated for 48 h with 2 μg/ml of phytohemagglutinin (Roche) in the presence of 20 U/ml of interleukin-2. The pNL4-3/29/31KE molecular clone (previously referred to as pNL4-3/29KE/31KE) was described previously (53, 54). VSV-G-pseudotyped virus stocks were prepared by transfecting 293T cells with pCMVNLGagPolRRE-, pHCMV-G-, and pNL4-3-derived molecular clones (54, 73). The 16EK MA mutant and the 16EK/29/31KE triple mutant were constructed by site-directed mutagenesis by using appropriate primers; the BssHII-SphI-mutagenized fragment was cloned back into the pNL4-3 backbone. pNL4-3CTdel144-2 was described previously (48). Gp41-truncated versions of the MA mutant clones were constructed by introducing the EcoRI-BamHI fragment from pNL4-3CTdel144-2 into the MA mutant clones.
Abs and reagents.
Mouse monoclonal antibodies (Abs) that recognize HIV-1 p17 (MA) or p24 (CA) were obtained from Advanced Biotechnologies (Columbia, MD), and mouse anti-CD63 Ab was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The construct expressing dominant-negative dynamin, green fluorescent protein (GFP)-dynamin-K44A, was kindly provided by M. Caron (Duke University), and yellow fluorescent protein-tagged dominant-negative Eps15 was kindly provided by Paul Bieniasz (Aaron Diamond AIDS Research Center, New York, NY). The HIV interdomain GFP (iGFP) molecular clone was kindly provided by B. Chen (Mt. Sinai School of Medicine, New York, NY) (35). The vpu-deficient (delVpu) molecular clone pNL4-3/U35 (67) was kindly provided by K. Strebel (NIAID, NIH). HIV immunoglobulin (HIV-Ig) was provided by the NIH AIDS Research and Reference Reagent Program.
Radioimmunoprecipitation, immunofluorescence, and EM.
One day posttransfection or after infection with VSV-G-pseudotyped virus stocks, cells were metabolically labeled with [35S]Met/Cys (Perkin-Elmer) in RPMI 1640 medium supplemented with 10% FBS but devoid of Met and Cys. Procedures for the preparation of cell lysates, ultracentrifugation of virions, and immunoprecipitation of cell- and virion-associated material with HIV-Ig were described elsewhere previously (22). Virus release efficiency was calculated as the amount of virion p24 divided by total Gag (cell Pr55Gag, cell p24, and virion p24). For virus release in PBLs, culture supernatants were harvested from infected cells 24 h postinfection, and cell and virus lysates were analyzed for HIV-1 protein expression by Western blotting using HIV-Ig. For Western blotting, protein lysates resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were transferred onto polyvinylidene difluoride membranes (Millipore) and probed with the indicated Abs. Western blot signals were quantified by using an Alpha Innotech digital imager. For immunofluorescence analysis, transfected or infected cells were cultured on glass slides, fixed with 3.7% formaldehyde in 100 mM sodium phosphate buffer, and stained with appropriate Abs. The protocol for double staining with two different mouse monoclonal Abs was described in detail previously (54). The Zenon One or Alexa Fluor 488 or 594 IgG1 labeling kit (Molecular Probes) was used according to the manufacturer's protocol. Images were acquired using the DeltaVision RT deconvolution microscope, and colocalization was measured using the softWoRx colocalization tool; R is the Pearson coefficient of correlation. For EM analysis, cells were fixed at 24 to 48 h posttransfection or postinfection in buffer containing 2% glutaraldehyde and 0.1 M sodium cacodylate. Fixed cells were then analyzed by transmission EM as described previously (23).
Virus replication and isolation of viral revertants.
T cells were transfected using the DEAE-dextran transfection protocol (22). Every 2 days, culture supernatants were harvested for measurement of RT activity, and cells were split. For isolation of revertant viruses, DNA was isolated from infected cells using the QIAamp DNA blood minikit (Qiagen), and Gag and Env coding regions were PCR amplified using specific primers, followed by DNA sequencing.
Transferrin receptor internalization.
Twenty-four hours posttransfection, cells were washed three times with serum-free medium containing 20 mM HEPES and 1% bovine serum albumin and incubated for 1 h at 37°C in the same medium. Cells were then placed on ice for 5 min followed by incubation for 30 min at 4°C in serum-free medium containing 10 μg/ml fluorescently labeled transferrin (Tr-594; Molecular Probes). Cells were washed three times with phosphate-buffered saline and incubated at 37°C for 15 min. Finally, cells were washed in phosphate-buffered saline, fixed, mounted, and examined with a Deltavision RT microscope.
RESULTS
Virus production efficiency of the 29/31KE MA mutant is markedly impaired in HeLa and 293T cells but not in T cells or MDMs.
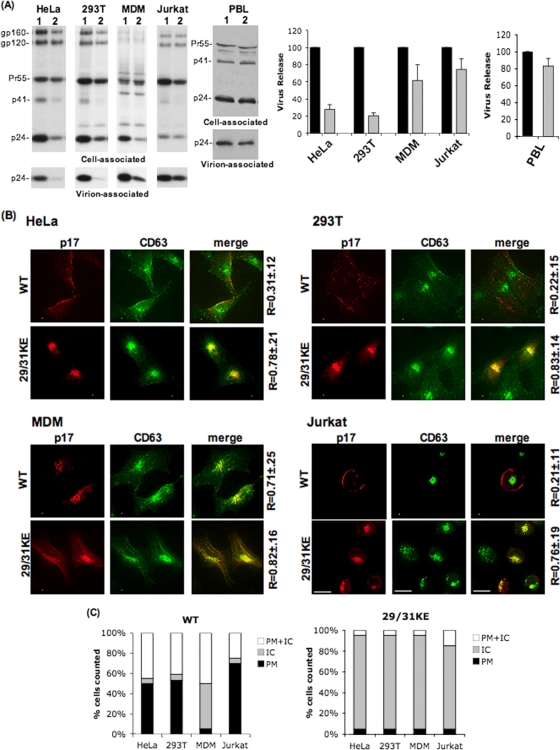
There has been considerable debate as to whether intracellular compartments can support the productive assembly and release of HIV-1 particles. We previously reported that a mutation of two basic residues in the MA domain of Gag to Glu (29/31KE) led to a retargeting of assembly to MVBs and a severe reduction in virus production efficiency in HeLa cells (54, 56). The impact of these mutations on virus production in primary MDMs was markedly less severe than that in HeLa cells (54). To further investigate the consequences of MVB targeting on virus release, we examined the behavior of the 29/31KE mutant in a broader array of cell types. HeLa, 293T, MDM, Jurkat, SupT1, and MT-4 T cells were infected with VSV-G-pseudotyped WT or 29/31KE mutant HIV-1. Infected cells were metabolically labeled with [35S]Met/Cys. Cell and virus lysates were prepared and immunoprecipitated with HIV-Ig followed by SDS-PAGE and PhosphorImager analysis. Consistent with our previous findings (54, 56), the 29/31KE mutant displayed a severe (about four- to fivefold) defect in particle production in HeLa and 293T cells, but its release efficiency was only minimally (∼ 40% reduction) affected in MDMs (Fig. 1A). Surprisingly, in the Jurkat (Fig. 1A), SupT1, and MT-4 (data not shown) T-cell lines, no significant defect in particle production was observed for 29/31KE. We also measured virus release in primary PBLs, and, as in the T-cell lines, we observed no significant effect of the 29/31KE mutations on particle production efficiency relative to that of the WT (Fig. 1A).
FIG. 1.
Release of the MVB-targeted MA mutant 29/31KE is severely impaired in HeLa and 293T cells but is efficient in MDM and Jurkat T cells. The indicated cell types were infected with RT-normalized stocks of VSV-G-pseudotyped WT or 29/31KE mutant virus. (A, left) Twenty-four hours postinfection, cells were metabolically radiolabeled with [35S]Met/Cys, and cell and virus lysates were immunoprecipitated with HIV-Ig and resolved by SDS-PAGE. (Right) Bands were quantified by fluorography, and virus release efficiency was calculated. Lanes 1 and black bars, WT; lanes 2 and gray bars, 29/31KE. Error bars represent means ± standard deviations (n = 3 for HeLa and 293T cells; n = 5 for MDMs). For analysis of virus release in PBLs, cell and virus lysates were subjected to SDS-PAGE and then immunoblotted with HIV-Ig. Bands were quantified by using the AlphaInnotech digital imager, and virus release efficiency was calculated. (B) Twenty-four hours postinfection, cells were fixed and labeled with anti-p17 (MA) and anti-CD63 Abs, followed by image acquisition using a DeltaVision RT deconvolution microscope. Colocalization between Gag and CD63 was determined using the SoftWorx colocalization module. R is the Pearson coefficient of correlation ± standard deviations averaged for 10 to 15 cells. (C) Quantification of Gag localization at the PM or intracellular (IC) sites for WT or 29/31KE Gag in an average of 10 to 15 cells. Gag localization was scored as PM or intracellular only if >90% of the staining was observed on the PM or in internal compartments.
The 29/31KE MA mutant localizes to an internal CD63+ compartment in a wide range of cell types including T cells.
We demonstrated previously that the 29/31KE MA mutant is retargeted to an MVB compartment in HeLa cells and in primary macrophages (29, 54). Because the release efficiency for the 29/31KE mutant was comparable to that of the WT in T-cell lines, we asked whether this mutant is also targeted to MVBs in T cells. Using an anti-p17 (MA) Ab, WT Gag was observed primarily at the PM in HeLa, 293T, and Jurkat cells (Fig. 1B). In contrast, and consistent with data from our previous reports (29, 54), in MDMs, a large amount of internal WT Gag staining was observed, and this staining overlapped significantly with that of the tetraspanin MVB markers CD63 (Fig. 1B) and CD81 (see Fig. S1 in the supplemental material). As we reported previously for HeLa cells and MDMs (29, 54), we observed a marked intracellular staining of 29/31KE Gag in T cells, with a high degree (R ∼ 0.8) of colocalization with CD63 (Fig. 1B) and CD81 (see Fig. S1 in the supplemental material). Similar results were seen with anti-p24 (CA) Ab staining and with a 29/31KE MA mutant derivative bearing the GFP coding region between the MA and CA domains of Gag (HIV-iGFP-29/31KE) (data not shown). In PBLs, as in T-cell lines, we observed predominantly PM staining for WT Gag with limited CD63 colocalization (R = 0.17 ± 0.13), whereas the 29/31KE mutant displayed internal staining with a high degree of overlap with CD63 (R = 0.75 ± 0.21) (data not shown).
To quantify these observations, we scored the Gag distribution pattern in a number of HeLa, 293T, Jurkat, and MDM cells (Fig. 1C). The data demonstrated that >90% of HeLa, 293T, and Jurkat cells infected with WT HIV-1 displayed predominantly PM or mixed PM and intracellular Gag staining. In MDMs, internal staining was markedly more prominent. In contrast, in all cell types examined, >80% of cells expressing the 29/31KE mutant showed a predominantly intracellular localization pattern (Fig. 1C). Together, the data from Fig. 1 indicate that the 29/31KE MA mutant is efficiently released from MDM and T-cell lines despite being localized predominantly to an internal compartment that bears MVB markers. This is in contrast to what is observed for HeLa and 293T cells, from which the 29/31KE mutant is released poorly.
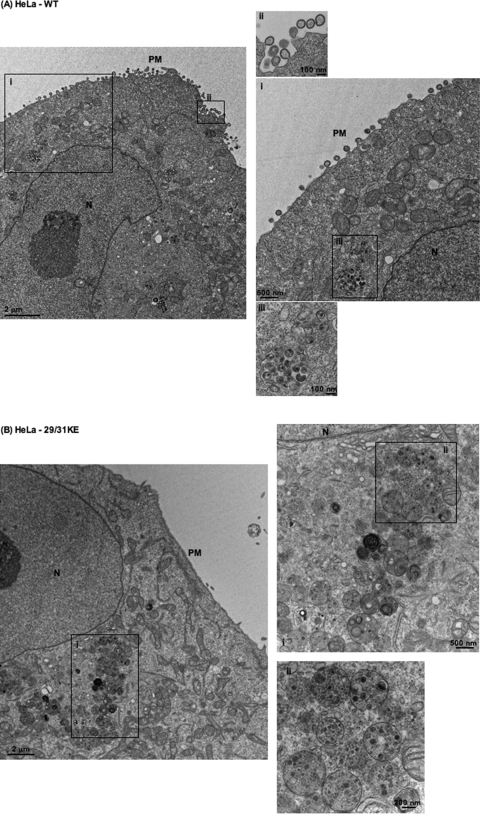
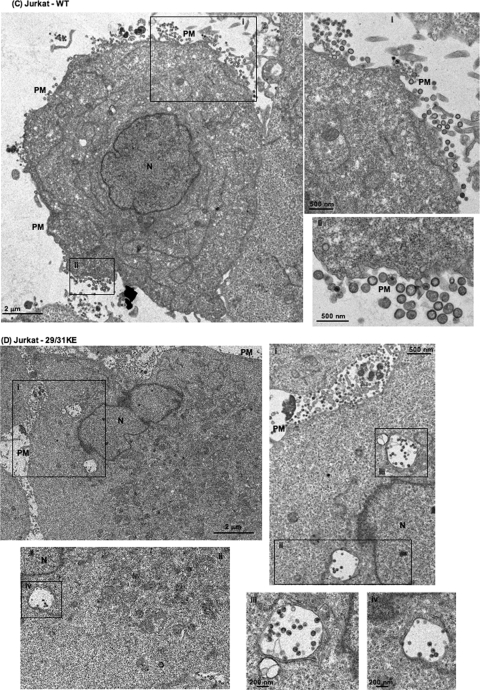
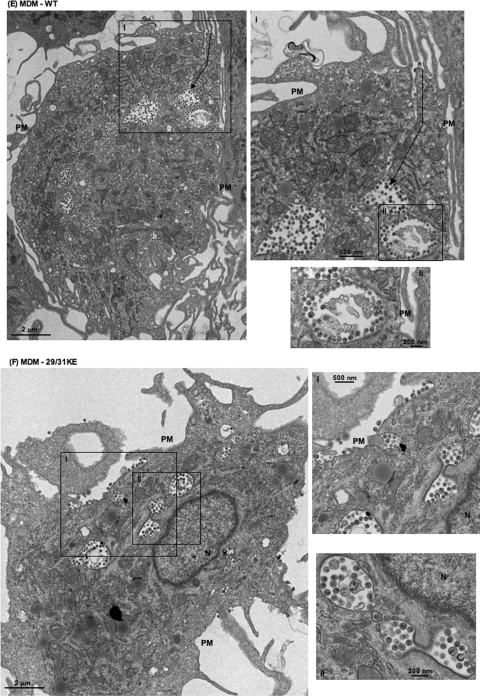
To corroborate the immunofluorescence data presented above, we performed EM analysis comparing WT and 29/31KE localization in HeLa, Jurkat, and MDMs (Fig. 2). In HeLa (Fig. 2A) and Jurkat (Fig. 2C) cells, WT Gag assembly was observed predominantly on the PM, although some intracellular foci of assembly were evident (Fig.2AIII). In contrast, in MDMs, the assembly of WT particles was observed predominantly in an apparently internal compartment (Fig. 2E). Upon careful observation, some of these internal compartments displayed narrow connections to the PM (Fig. 2E, arrow). In contrast to the WT, and consistent with the immunofluorescence data presented above, the 29/31KE mutant showed predominantly intracellular virus assembly irrespective of the cell type (Fig. 2B, D, and F). These EM data confirm the internal location of virus assembly for the 29/31KE MA mutant in all cell types tested.
FIG. 2.
The 29/31KE mutant assembles predominantly in an internal compartment in all cell types tested. Shown is EM analysis of WT HIV-1 and the 29/31KE mutant in HeLa (A and B), Jurkat (C and D), and MDM (E and F) cells. Cells were infected with VSV-G-pseudotyped WT HIV-1 or the 29/31KE mutant. Cells were fixed 24 to 48 h postinfection and analyzed by EM. N, nucleus. Insets represent magnified portions of the image with corresponding numbers (I, II, and III). In E, the arrow indicates the channel connecting the internal virus-containing compartment and the PM.
Evidence that virus released from cells expressing 29/31KE Gag is derived from an internal compartment.
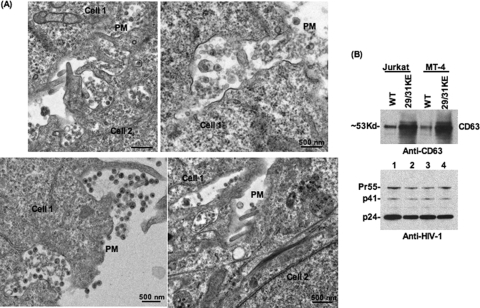
The data presented above demonstrate that the 29/31KE MA mutant is released efficiently from T cells relative to the WT. Both immunofluorescence and EM data indicate that the predominant site for 29/31KE Gag accumulation is in an internal, CD63+ compartment. These results imply that the released 29/31KE mutant Gag assembles in an intracellular, late endosomal compartment. Indeed, in Jurkat cells expressing 29/31KE Gag, involuted virus-containing structures were often seen at the plasma membrane (Fig. 3A); these structures, which were not observed in HeLa cells or in Jurkat cells expressing WT Gag, could potentially represent virus-containing vesicles in the process of releasing their contents. In MDMs, the release of “internal” virions was observed for both WT and 29/31KE mutant Gag (data not shown), consistent with data from previous studies that examined WT Gag in MDMs (59).
FIG. 3.
Evidence that released 29/31KE virions assemble in an internal CD63+ compartment. (A) Jurkat T cells were infected with VSV-G-pseudotyped 29/31KE virus stocks. Cells were fixed 24 to 48 h postinfection and analyzed by EM. Adjacent cells are indicated as cell 1 and cell 2. (B) Jurkat or MT-4 cells were infected with RT-normalized VSV-G-pseudotyped WT or 29/31KE virus stocks. Culture supernatants were harvested 24 h postinfection, ultracentrifuged, and analyzed for CD63 incorporation into virions by Western blotting (WB) (top). The blot was stripped and reprobed with HIV-Ig (bottom).
We also pursued a more biochemical approach to define the subcellular origin of 29/31KE mutant virions in T-cell lines. CD63 is known to be incorporated into virus particles (42, 50, 58, 59); one might therefore predict that virions originating in CD63-enriched MVBs would incorporate more CD63 than particles that assembled on the PM. To test this prediction, Jurkat or MT-4 T cells were transduced with VSV-G-pseudotyped WT and 29/31KE mutant virions. Virus preparations were then analyzed for CD63 incorporation and Gag levels by Western blotting. As shown in Fig. 3B, 29/31KE virions from both Jurkat and MT-4 cells incorporated markedly higher levels of CD63 than did WT virions (Fig. 3B, top). Levels of Gag were similar, consistent with the results shown in Fig. 1A (Fig. 3B, bottom). These data suggest that the released 29/31KE virions are assembled in an intracellular CD63+ compartment.
Inhibition of the cellular endocytic pathway does not prevent accumulation of 29/31KE Gag in a late endosomal compartment.
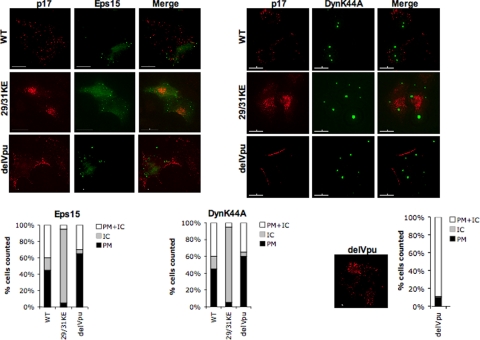
It is clear from the data presented above and from our previous reports (54) that 29/31KE mutant Gag accumulates in a late endosomal/MVB compartment in a diverse array of cell types. In principle, 29/31KE Gag could be targeted directly to MVBs or could arrive there after being internalized from the PM. In the latter case, disruption of the cellular endocytic machinery would be predicted to prevent MVB accumulation of 29/31KE Gag. To address this question, we made use of several cellular protein expression constructs that disrupt the endocytic pathway. It has been shown that the expression of dominant-negative dynamin (K44A) and Eps15 disrupts clathrin-mediated endocytosis. To confirm a defect in clathrin-dependent endocytosis with these inhibitors, we compared the levels of uptake of a fluorescently tagged transferrin receptor in HeLa cells that were untransfected or were transfected with the dynamin-K44A or Eps15 expression vector. We observed a marked defect in the uptake of the transferrin receptor into cells expressing dominant-negative dynamin or Eps15 relative to control (untransfected) cells (see Fig. S2 in the supplemental material), confirming the functionality of these inhibitors. To examine the effects on Gag localization, HeLa cells were cotransfected with WT pNL4-3 or the 29/31KE mutant derivative and yellow fluorescent protein- or GFP-tagged protein expression constructs. Cells were then fixed approximately 24 h posttransfection, stained with anti-p17 (MA) Ab, and analyzed by fluorescence microscopy. Several studies previously reported that Gag expressed in the absence of Vpu in HeLa cells undergoes endocytosis from the PM (27, 32, 33, 40, 49); thus, as a control, we examined the localization of Gag expressed from a Vpu(−) molecular clone (67). As shown in Fig. 4, the disruption of clathrin-mediated endocytosis by the expression of dynamin-K44A or dominant-negative Eps15 did not alter the intracellular localization of 29/31KE Gag. The localization pattern of WT Gag was also not affected by dynamin-K44A or dominant-negative Eps15 (compare the Gag localization profiles in Fig. 4 with those presented in Fig. 1C). In contrast, in the absence of Vpu expression, both dynamin-K44A and dominant-negative Eps15 increased the fraction of PM-associated Gag and reduced intracellular Gag localization (Fig. 4), consistent with data from a previous report (49). These results suggest that MVB localization of 29/31KE Gag is not a result of dynamin- or clathrin-dependent internalization of Gag from the PM.
FIG. 4.
29/31KE Gag is targeted directly to MVBs rather than being internalized from the PM. HeLa cells were transfected with HIV-WT, 29/31KE, and delVpu molecular clones along with vectors expressing dominant-negative Eps15 or GFP-tagged dynamin-K44A (DynK44A). Cells were fixed 24 h posttransfection, stained with anti-p17 Ab, and subjected to immunofluorescence analysis. Graphs under each panel represent PM, intracellular (IC), or mixed PM and IC Gag localization in an average of 10 to 15 cells. The staining pattern for delVpu in the absence of any inhibitors along with the PM or IC localization pattern averaged from 15 cells is depicted as a graph at the bottom right.
The 29/31KE mutant can replicate efficiently in the MT-4 T-cell line following truncation of the gp41 cytoplasmic tail and acquisition of a 16EK MA mutation.
As demonstrated above, the 29/31KE mutant was efficiently released in T cells despite its MVB Gag localization phenotype. However, this mutant failed to initiate a spreading, multiple-round infection in Jurkat or MT-4 T cells (data not shown). These results suggested that the 29/31KE mutations may impose defects that lead to entry or postentry defects in virus replication. We have previously shown that point mutations in MA can block Env incorporation into virions (20, 21, 55) and can disrupt postentry events (39). Env incorporation defects imposed by MA mutations (21, 41) and infectivity defects resulting from a large MA deletion (60) can be reversed by truncating the long cytoplasmic tail of the TM glycoprotein gp41. In an attempt to obtain a 29/31KE derivative capable of initiating a spreading infection, we introduced the gp41 truncation mutation CTdel-144-2, which removes 144 amino acids from the C terminus of gp41 (48), into pNL4-3/29/31KE. MT-4 cells were selected as the target T-cell line because they are permissive for replication of the CTdel-144-2 mutant (48).
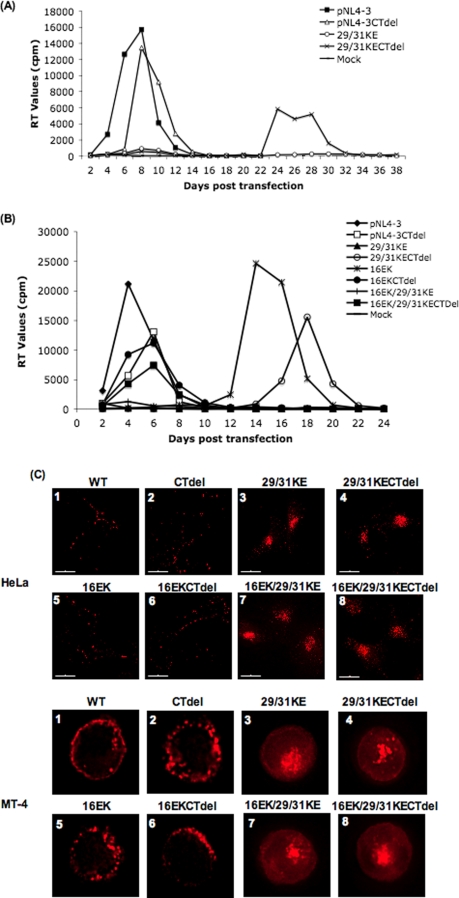
MT-4 T cells were transfected with WT pNL4-3, pNL4-3/29/31KE, and derivatives containing the gp41 CTdel-144-2 mutation. Virus replication was monitored by RT assay. Cultures transfected with WT or CTdel-144-2 molecular clones showed a peak of virus replication at approximately 1 week posttransfection (Fig. 5A). The 29/31KE mutant failed to replicate. Interestingly, the 29/31KE derivative bearing the gp41 truncation showed a peak in virus replication at approximately 4 weeks posttransfection, suggesting the emergence of a viral revertant. Sequencing analysis of DNA derived from cultures infected with the 29/31KE CTdel-144-2 mutant revealed the maintenance of the 29/31KE mutations and the appearance of a Glu-to-Lys change at MA amino acid 16 (16EK). To test the ability of the 16EK mutation to reverse the replication defect imposed by the 29/31KE mutations, the triple mutant 16EK/29/31KE was constructed. The 16EK change was also introduced by itself into pNL4-3. Again, the 29/31KE mutant failed to replicate unless coupled with the gp41 truncation (Fig. 5B). The 16EK single mutant also replicated with a pronounced delay; this defect was largely corrected by the CTdel-144-2 mutation. Most interestingly, the 16EK/29/31KE triple mutant, when coupled with the gp41 truncation mutation, peaked on day 6 posttransfection, the same day as the CTdel-144-2 clone expressing WT Gag.
FIG. 5.
The 29/31KE mutant replicates in the MT-4 T-cell line after truncation of the gp41 cytoplasmic tail and acquisition of the 16EK compensatory mutation despite being MVB localized. (A and B) MT-4 T cells were transfected with WT pNL4-3, pNL4-3 bearing the CTdel-144-2 gp41 truncation (CTdel), and/or the 29/31KE, 16EK, or 16EK/29/31KE MA mutation. Cells were split every 2 days, and supernatants were reserved at each time point for RT assay. (C) HeLa (top) or MT-4 (bottom) cells were infected with the indicated VSV-G-pseudotyped virus stocks. Cells were fixed 24 to 48 h postinfection and stained with anti-p17 Ab.
We next examined the localization of the 16EK/29/31KE mutant in both HeLa and MT-4 cells (Fig. 5C). In both cell types, the WT displayed predominantly PM localization. In contrast, as demonstrated above, 29/31KE Gag showed internal staining. The 16EK mutant was localized primarily to the PM, similar to the WT. Significantly, the 16EK/29/31KE triple mutant showed a localization pattern that was indistinguishable from that of the 29/31KE double mutant. Localization patterns were not affected in any case by the gp41 CTdel-144-2 truncation. These data demonstrate that MVB-targeted Gag mutants not only can be released efficiently in T-cell lines but also can establish a productive spreading infection.
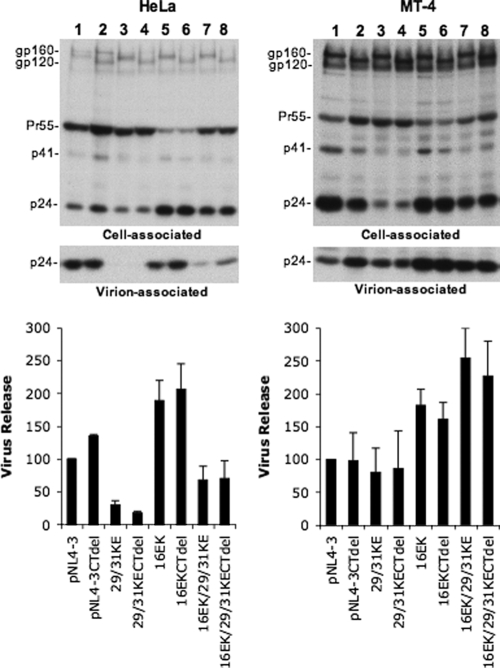
To understand in more detail the mechanism by which the 16EK change in MA promotes the spreading infection of the 29/31KE mutant, we examined the effect of the 16EK substitution on virus particle production in HeLa and MT-4 cells. The 29/31KE mutant was again severely defective for particle production in HeLa cells but not in the MT-4 T-cell line (Fig. 6). The 16EK substitution either by itself or in the context of the 29/31KE mutations markedly enhanced (by a factor of two- to threefold) the yield of cell-free virus. The gp41 truncation had no effect on the efficiency of particle release in any context. These results suggest that the 16EK mutation is acquired at least in part to promote the production of cell-free virus. We again note that although the 16EK mutation enhances particle production (Fig. 6) and virus replication (Fig. 5A), it does not change the internal assembly phenotype conferred by the 29/31KE substitutions (Fig. 5C).
FIG. 6.
The 16EK mutation significantly increases the level of production of cell-free virus. HeLa and MT-4 cells expressing the indicated pNL4-3 molecular clones, encoding WT or truncated (CTdel) Env and WT or mutant MA domains, were metabolically labeled with [35S]Met/Cys. Cell and virion lysates were immunoprecipitated and analyzed as described in the Fig. 1 legend.
DISCUSSION
In this study, we investigated the consequences of intracellular HIV-1 assembly by using a previously described MA mutant, 29/31KE, that is targeted to MVBs (54, 56). We confirmed that in HeLa cells, the release of this MA mutant is severely inhibited, whereas in primary MDMs, only a modest reduction in the level of particle production is measured. Unexpectedly, we observed that in T cells, the 29/31KE mutant is released with efficiencies only slightly lower than those of the WT. Disrupting the endocytic pathway with dominant-negative dynamin or Eps15 did not alter the MVB localization of 29/31KE Gag, suggesting that the mutant Gag is targeted directly to MVBs rather than trafficking there after internalization from the PM. EM and CD63 incorporation data support the hypothesis that the 29/31KE particles that are released from the cell are derived from MVB-assembled Gag. Vpu-deficient virus provided an important control for these experiments; in the case of delVpu, we observed a loss of endosomal localization in cells expressing dominant-negative dynamin and Eps15. Although 29/31KE mutant Gag can be released efficiently in T cells, the establishment of a spreading infection is impaired due to a block early in the replication cycle. This early block could be bypassed by the deletion of the gp41 cytoplasmic tail and by the acquisition of a 16EK change in MA. Importantly, the resulting virus could replicate efficiently in the MT-4 T-cell line despite maintaining MVB-associated assembly. These data suggest that intracellular compartments (e.g., MVBs) are capable of serving as sites for productive HIV-1 assembly in MDMs and T cells.
The results obtained here with the 29/31KE MA mutant differ in some respects from those of a recent study in which Gag was targeted to endosomal compartments by replacing the globular head of the MA domain with foreign endosome-targeting signals (38). In that study, it was observed that endosomal targeting severely reduced particle production not only in 293T cells (consistent with our findings) (54; this study) but also in MDMs. Explanations for these divergent results with MDMs include the following: (i) the compartment in MDMs to which 29/31KE Gag is targeted could differ from the site of assembly of the MA-deleted Gag chimeras, and/or (ii) in MDMs, replacement of the MA globular head with foreign endosome-targeting domains may disrupt some aspect of assembly or release distinct from Gag mislocalization. Jouvenet et al. (38) did not examine the effect of endosomal targeting on particle production in T cells; the behavior of these Gag chimeras in this cell type therefore remains to be determined.
We observed an efficient release of the 29/31KE mutant in T cells and MDMs but not HeLa or 293T cells despite the uniform MVB localization of this mutant Gag in all cell types tested. These findings suggest the existence of cell-type-specific differences in virus release pathways in HeLa and 293T cells versus T cells and MDMs. T cells and MDMs may possess a more active secretory pathway than HeLa and 293T cells (25, 30, 43, 44, 69) and, hence, are more capable of releasing virions assembled in intracellular compartments. This possibility is supported by our EM data showing what might be internal virus-containing compartments fusing with the PM in T cells and MDMs. These structures were not observed in HeLa cells. We also observed an increase in levels of the MVB-localized tetraspanin CD63 in 29/31KE virions produced from Jurkat cells, again suggesting an internal origin for this virus. Interestingly, although 29/31KE mutant Gag is efficiently released from MDMs as cell-free virus, we recently demonstrated that this mutant Gag does not readily move to the virological synapse formed between infected MDMs and uninfected T cells or MDMs (29). These results suggest that the WT and 29/31KE virus-positive compartments in MDMs are not functionally equivalent and that the signals that direct Gag movement to the surface to regulate particle release in T cells and MDMs differ from those that direct Gag/virus-like particle movement to the virological synapse.
Mutations in the MA domain of HIV-1 Gag have been shown to disrupt interactions with the viral Env glycoproteins, leading to defects in Env incorporation into virions (14, 20, 21, 74) or impaired gp120-gp41 associations (9). Interactions between Gag and the gp41 cytoplasmic tail have also been shown to suppress the fusogenic activity of Env (47, 72). These defects can be reversed by deleting the gp41 cytoplasmic tail (9, 20, 21, 41, 46, 47, 60, 72). While gp41 truncation blocks the establishment of a spreading infection in most T-cell lines, gp41 tail truncation mutants can replicate efficiently in the MT-4 T-cell line (48). Here we observed that the 29/31KE mutant is unable to establish a spreading infection in T-cell lines; however, truncation of the gp41 cytoplasmic tail allows replication to occur in MT-4 cells with a pronounced delay relative to the WT. Analysis of the virus replicating in the cultures infected with the gp41-truncated 29/31KE mutant led to the identification of a second-site compensatory mutation (16EK) that facilitated replication such that the gp41-truncated 16EK/29/31KE triple mutant replicated efficiently in MT-4 cells. Interestingly, the 29/31KE mutant did not display a defect in Env incorporation but was impaired in single-cycle infectivity assays (A. Joshi and E. Freed, unpublished data). This infectivity defect could be reversed by a gp41 truncation. These findings indicate that the 29/31KE mutant exhibits a complex entry or postentry defect that involves the gp41 cytoplasmic tail and can be corrected by the 16EK change. We also observed that the 16EK substitution markedly increases the efficiency of cell-free virus particle production, providing an explanation for its selection during propagation of the 29/31KE mutant. Importantly, the gp41-truncated 16EK/29/31KE mutant efficiently establishes a spreading infection in MT-4 cells despite being MVB localized.
In conclusion, we demonstrate the efficient assembly and release of an MVB-targeted mutant in MDMs and T cells but not in HeLa or 293T cells. Although we favor the model that the PM serves as the primary site for HIV-1 assembly in most if not all cell types, these results indicate that internal CD63+ compartments can serve as sites for productive HIV-1 assembly in physiologically relevant cell types. The role that such compartments play in the replication of WT HIV-1 warrants further study.
Supplementary Material
Acknowledgments
We thank members of the Freed laboratory and A. Ono for helpful discussions and critical reviews of the manuscript. We thank M. Caron for kindly providing the GFP-tagged dynamin K44A, P. Bieniasz for the dominant-negative Eps15 construct, K. Strebel for the delVpu (U35) clone, and B. Chen for the HIV iGFP molecular clone. HIV-Ig was obtained from the NIH AIDS Research and Reference Reagent Program.
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH, and by the Intramural AIDS Targeted Antiviral Program. This project was funded in part with federal funds from the National Cancer Institute, NIH, under contract number N01-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 18 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55347-387. [DOI] [PubMed] [Google Scholar]
- 2.Batonick, M., M. Favre, M. Boge, P. Spearman, S. Honing, and M. Thali. 2005. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology 342190-200. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 34455-63. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camus, G., C. Segura-Morales, D. Molle, S. Lopez-Verges, C. Begon-Pescia, C. Cazevieille, P. Schu, E. Bertrand, C. Berlioz-Torrent, and E. Basyuk. 2007. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol. Biol. Cell 183193-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, R., P. D. Uchil, J. Jin, G. Shui, D. E. Ott, W. Mothes, and M. R. Wenk. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 8211228-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 809039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chukkapalli, V., I. B. Hogue, V. Boyko, W.-S. Hu, and A. Ono. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J. Virol. 822405-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, M. R., J. Jiang, J. Zhou, E. O. Freed, and C. Aiken. 2006. A mutation in the human immunodeficiency virus type 1 Gag protein destabilizes the interaction of the envelope protein subunits gp120 and gp41. J. Virol. 802405-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 10687-102. [DOI] [PubMed] [Google Scholar]
- 11.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deneka, M., A. Pelchen-Matthews, R. Byland, E. Ruiz-Mateos, and M. Marsh. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, X., H. Li, A. Derdowski, L. Ding, A. Burnett, X. Chen, T. R. Peters, T. S. Dermody, E. Woodruff, J. J. Wang, and P. Spearman. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120663-674. [DOI] [PubMed] [Google Scholar]
- 14.Dorfman, T., F. Mammano, W. A. Haseltine, and H. G. Gottlinger. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 681689-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facke, M., A. Janetzko, R. L. Shoeman, and H. G. Krausslich. 1993. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 674972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finzi, A., A. Orthwein, J. Mercier, and E. A. Cohen. 2007. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J. Virol. 817476-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 2511-15. [DOI] [PubMed] [Google Scholar]
- 18.Freed, E. O. 2006. HIV-1 Gag: flipped out for PI(4,5)P(2). Proc. Natl. Acad. Sci. USA 10311101-11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 693949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed, E. O., and M. A. Martin. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 691984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 685311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelderblom, H. R. 1991. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS 5617-637. [PubMed] [Google Scholar]
- 24.Gendelman, H. E., J. M. Orenstein, M. A. Martin, C. Ferrua, R. Mitra, T. Phipps, L. A. Wahl, H. C. Lane, A. S. Fauci, D. S. Burke, et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 1671428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goerdt, S., O. Politz, K. Schledzewski, R. Birk, A. Gratchev, P. Guillot, N. Hakiy, C. D. Klemke, E. Dippel, V. Kodelja, and C. E. Orfanos. 1999. Alternative versus classical activation of macrophages. Pathobiology 67222-226. [DOI] [PubMed] [Google Scholar]
- 26.Goff, S. P. 2001. The retroviruses and their replication, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Gottlinger, H. G., T. Dorfman, E. A. Cohen, and W. A. Haseltine. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. USA 907381-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould, S. J., A. M. Booth, and J. E. Hildreth. 2003. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 10010592-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gousset, K., S. D. Ablan, L. V. Coren, A. Ono, F. Soheilian, K. Nagashima, D. E. Ott, and E. O. Freed. 2008. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 4e1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths, G. M. 1995. The cell biology of CTL killing. Curr. Opin. Immunol. 7343-348. [DOI] [PubMed] [Google Scholar]
- 31.Grigorov, B., F. Arcanger, P. Roingeard, J. L. Darlix, and D. Muriaux. 2006. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 359848-862. [DOI] [PubMed] [Google Scholar]
- 32.Handley, M. A., S. Paddock, A. Dall, and A. T. Panganiban. 2001. Association of Vpu-binding protein with microtubules and Vpu-dependent redistribution of HIV-1 Gag protein. Virology 291198-207. [DOI] [PubMed] [Google Scholar]
- 33.Harila, K., I. Prior, M. Sjoberg, A. Salminen, J. Hinkula, and M. Suomalainen. 2006. Vpu and Tsg101 regulate intracellular targeting of the human immunodeficiency virus type 1 core protein precursor Pr55gag. J. Virol. 803765-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 748670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubner, W., P. Chen, A. Del Portillo, Y. Liu, R. E. Gordon, and B. K. Chen. 2007. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 8112596-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi, A., and E. Freed. 2007. HIV-1 Gag trafficking. Future Med. 1427-438. [Google Scholar]
- 37.Joshi, A., H. Garg, K. Nagashima, J. S. Bonifacino, and E. O. Freed. 2008. GGA and Arf proteins modulate retrovirus assembly and release. Mol. Cell 30227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Jouve, M., N. Sol-Foulon, S. Watson, O. Schwartz, and P. Benaroch. 2007. HIV-1 buds and accumulates in “nonacidic” endosomes in macrophages. Cell Host Microbe 285-95. [DOI] [PubMed] [Google Scholar]
- 38.Jouvenet, N., S. J. Neil, C. Bess, M. C. Johnson, C. A. Virgen, S. M. Simon, and P. D. Bieniasz. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 724116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klimkait, T., K. Strebel, M. D. Hoggan, M. A. Martin, and J. M. Orenstein. 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol. 64621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mammano, F., E. Kondo, J. Sodroski, A. Bukovsky, and H. G. Gottlinger. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 693824-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsh, M., and M. Thali. 2003. HIV's great escape. Nat. Med. 91262-1263. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell, D. A., S. K. Nair, and E. Gilboa. 1998. Dendritic cell/macrophage precursors capture exogenous antigen for MHC class I presentation by dendritic cells. Eur. J. Immunol. 281923-1933. [DOI] [PubMed] [Google Scholar]
- 44.Monks, C. R., B. A. Freiberg, H. Kupfer, N. Sciaky, and A. Kupfer. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 39582-86. [DOI] [PubMed] [Google Scholar]
- 45.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20395-425. [DOI] [PubMed] [Google Scholar]
- 46.Murakami, T., S. Ablan, E. O. Freed, and Y. Tanaka. 2004. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J. Virol. 781026-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J. Virol. 743548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neil, S. J., S. W. Eastman, N. Jouvenet, and P. D. Bieniasz. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 27852347-52354. [DOI] [PubMed] [Google Scholar]
- 51.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4902-910. [DOI] [PubMed] [Google Scholar]
- 52.Ono, A., S. D. Ablan, S. J. Lockett, K. Nagashima, and E. O. Freed. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA 10114889-14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 745142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 781552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ono, A., M. Huang, and E. O. Freed. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 714409-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 742855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orenstein, J. M., and F. Jannotta. 1988. Human immunodeficiency virus and papovavirus infections in acquired immunodeficiency syndrome: an ultrastructural study of three cases. Hum. Pathol. 19350-361. [DOI] [PubMed] [Google Scholar]
- 58.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3718-729. [DOI] [PubMed] [Google Scholar]
- 60.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 172699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rudner, L., S. Nydegger, L. V. Coren, K. Nagashima, M. Thali, and D. E. Ott. 2005. Dynamic fluorescent imaging of human immunodeficiency virus type 1 Gag in live cells by biarsenical labeling. J. Virol. 794055-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saad, J. S., S. D. Ablan, R. H. Ghanam, A. Kim, K. Andrews, K. Nagashima, F. Soheilian, E. O. Freed, and M. F. Summers. 2008. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J. Mol. Biol. 382434-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saad, J. S., J. Miller, J. Tai, A. Kim, R. H. Ghanam, and M. F. Summers. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 10311364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4785-801. [DOI] [PubMed] [Google Scholar]
- 65.Shkriabai, N., S. A. Datta, Z. Zhao, S. Hess, A. Rein, and M. Kvaratskhelia. 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 454077-4083. [DOI] [PubMed] [Google Scholar]
- 66.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 683232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strebel, K., T. Klimkait, F. Maldarelli, and M. A. Martin. 1989. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J. Virol. 633784-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 69.Underhill, D. M., M. Bassetti, A. Rudensky, and A. Aderem. 1999. Dynamic interactions of macrophages with T cells during antigen presentation. J. Exp. Med. 1901909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogt, V. M. 1997. Retroviral virions and genomes in “retroviruses.” Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 71.Welsch, S., O. T. Keppler, A. Habermann, I. Allespach, J. Krijnse-Locker, and H. G. Krausslich. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wyma, D. J., J. Jiang, J. Shi, J. Zhou, J. E. Lineberger, M. D. Miller, and C. Aiken. 2004. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 783429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 4399-112. [DOI] [PubMed] [Google Scholar]
- 74.Yu, X., X. Yuan, Z. Matsuda, T. H. Lee, and M. Essex. 1992. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J. Virol. 664966-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan, X., X. Yu, T. H. Lee, and M. Essex. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J. Virol. 676387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 682556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.