Abstract
Poleroviruses are restricted to vascular phloem tissues from which they are transmitted by their aphid vectors and are not transmissible mechanically. Phloem limitation has been attributed to the absence of virus proteins either facilitating movement or counteracting plant defense. The polerovirus capsid is composed of two forms of coat protein, the major P3 protein and the minor P3/P5 protein, a translational readthrough of P3. P3/P5 is required for insect transmission and acts in trans to facilitate long-distance virus movement in phloem tissue. Specific potato leafroll virus mutants lacking part or all of the P5 domain moved into and infected nonvascular mesophyll tissue when the source-sink relationship of the plant (Solanum sarrachoides) was altered by pruning, with the progeny virus now being transmissible mechanically. However, in a period of months, a phloem-specific distribution of the virus was reestablished in the absence of aphid transmission. Virus from the new phloem-limited infection showed compensatory mutations that would be expected to restore the production of full-length P3/P5 as well as the loss of mechanical transmissibility. The data support our hypothesis that phloem limitation in poleroviruses presumably does not result from a deficiency in the repertoire of virus genes but rather results from P3/P5 accumulation under selection in the infected plant, with the colateral effect of facilitating transmission by phloem-feeding aphid vectors.
In nature, a majority of plant viruses are inoculated into epidermal or mesophyll host cells by vectors or, less frequently, by mechanical wounding. Subsequently, virus-encoded movement proteins facilitate cell-to-cell spread of the virus locally among epidermal and mesophyll cells through plasmodesmata, into vascular tissue (phloem) for long-distance systemic transport, and back into nonvascular tissue for further localized spread (22). Virus unloading from phloem into nonvascular cells involves different mechanisms than those used for virus movement into phloem tissue (41). Members of the Luteoviridae (luteovirids), which include poleroviruses, are introduced directly into vascular tissues by aphids, their phloem-feeding insect vectors. The introduction of these viruses into epidermal or mesophyll cells by aphids or mechanical means can result in virus replication, but it does not result in systemic infection (25). Although luteovirids encode at least two movement proteins and move locally and long distance in vascular tissues, they are unable to move between nonvascular cells or between nonvascular and vascular tissues (36). This phloem limitation is generally attributed to a lack of or an inability of luteovirid movement proteins to function in nonvascular tissues or because the virus cannot suppress a silencing-based plant defense mechanism outside of phloem tissues (5, 36).
Luteovirids move as intact virions throughout their plant hosts and their aphid vectors. The two recognized movement proteins of the luteovirids are P4 and P3/P5. P4, which has functional similarity to the tobacco mosaic virus movement protein (41), localizes to plasmodesmata at the companion cell-sieve element boundary and likely facilitates the cell-to-cell movement of assembled virus particles in a host-dependent manner (20, 35). P3/P5, which is a multifunctional protein, is a C-terminal readthrough protein extension of the coat protein (P3) resulting from an inefficient suppression of the P3 amber stop codon (1) (Fig. 1A). In an assembled virion, only a few of the 180 P3 molecules are the P3/P5 readthrough rather than P3. Within the P5 domain, there is a highly conserved N-terminal region and a variable C-terminal region. P3/P5 plays a functional role in movement within plants and within the insect vector, and it is required for aphid transmission (9). When potato leafroll virus (PLRV) circulates through the aphid, virions must be transmitted across the midgut epithelial cell, survive in the hemocoel, and be transported through the accessory salivary gland cells. The P5 N terminus has been shown to be responsible for aphid transmission and aphid endosymbiont interactions, whereas the C terminus was dispensable for aphid transmission (10, 39, 42). However, P3/P5 can affect the efficiency of virus transport across the gut (6, 7, 29), and P3/P5 can determine intestinal tropism when mediating acquisition (8).
FIG. 1.
Genome organization of PLRV and illustration of P5 mutant viruses. (A) The 6-kb monopartite, positive-sense, single-stranded RNA genome consisting of eight ORFs. ORFs P3 to P5, which are involved in structure and movement, are conserved among luteoviruses and are expressed from subgenomic RNA-1. Virions are composed mainly of P3 coat proteins and a small number of P3/P5 readthrough proteins, which are anchored into the virion by the P3 moiety. The P5 domain is exposed on the surface and has a conserved N-terminal region and a variable C-terminal region. (B) Illustration of the P3/P5 translation of the mutant viruses described in this study. Deletion of amino acids is specified as a line, and a lack of protein expression is shown as a dotted line. WT PLRV with expression of the entire P3/P5 is depicted. For the SST and QSS mutant viruses, the 9 nt comprising the SST sequence (nt 4531 to 4539) and the 9-nt QSS sequence (nt 4438 to 4446) were deleted, respectively, while full-length P3/P5 translation was maintained. The resulting P3/P5 protein was incorporated into the virion for the SST mutant virus but not for the QSS mutant virus. The ΔP5 mutant virus contains two stop codons at the end of the P3 sequence followed by a 100-nt deletion in P5 (double Xs); no P3/P5 translation occurs. The SYG mutation is located 18 amino acids from the C terminal region. In addition to the 9-nt deletion to create the SYG deletion (nt 4828 to 4836), a single nucleotide was deleted 14 nt downstream (nt 4850) (triangle). This created a frameshift, which altered the subsequent amino acids (diagonal lines) and eventually produced a stop codon (X) 17 nt downstream from the single-nucleotide deletion. As a result, a truncated P3/P5 was translated. ΔP5 and SYG mutant viruses do not have P3/P5 incorporated into their virions.
P3/P5 is not required for virion assembly, nor is it required for plant infection. Studies to date have examined the role of P3/P5 when incorporated into the virion (in cis). The P5 N and C regions were shown to be involved in viral movement and accumulation in plant hosts, suggesting that both are critical for whole-plant infection (7, 10, 11, 24). Beet western yellows virus (BWYV) and barley yellow dwarf virus-PAV P3/P5 mutants were shown to accumulate to a lower titer in infected plants than in wild-type (WT) virus (9, 11). In subsequent plant tissue immunolocalization experiments, the BWYV P3/P5 mutants had reduced virus movement to new infection sites in Nicotiana clevelandii plants, indicating that P3/P5, when incorporated into the virion, affected the efficiency with which the virus could move systemically (24). Recently, we showed that the P3/P5 readthrough protein of PLRV functions to promote systemic movement of the virus in trans even when it is not incorporated into the virions and that mutations that eliminate the translation of P5 caused a delay in symptom development (27). It is unknown if the P3/P5 proteins of PLRV and BWYV have additional functional differences or if these proteins function similarly in natural and experimental hosts, especially since the PLRV P4 protein is not required for movement in some experimental hosts (20).
MATERIALS AND METHODS
Plant infection and localization of virus.
To introduce virus into Solanum sarrachoides, the midrib and petioles of four fully expanded leaves on plants that were at the 8- to 10-leaf stage (Fig. 2, inset) were inoculated with a suspension of Agrobacterium containing full-length infectious clones of PLRV to initiate a systemic infection as described previously by Peter et al. (27). Those studies focused on WT virus and four previously described PLRV P5 mutants (27), three of which contain single three-contiguous-amino-acid deletions in the P5 N terminus, SST, QSS, and SYG, and a mutant lacking P5 (ΔP5, previously called ΔRTD). In addition to the 9 nucleotides (nt) that result in the SYG deletion, a single additional nucleotide inadvertently deleted 14 nt downstream created a frameshift, which altered the subsequent amino acids and eventually produced a stop codon 17 nt downstream from the single-nucleotide deletion. As a result, the SYG mutant translates a truncated P3/P5 (Fig. 1).
FIG. 2.
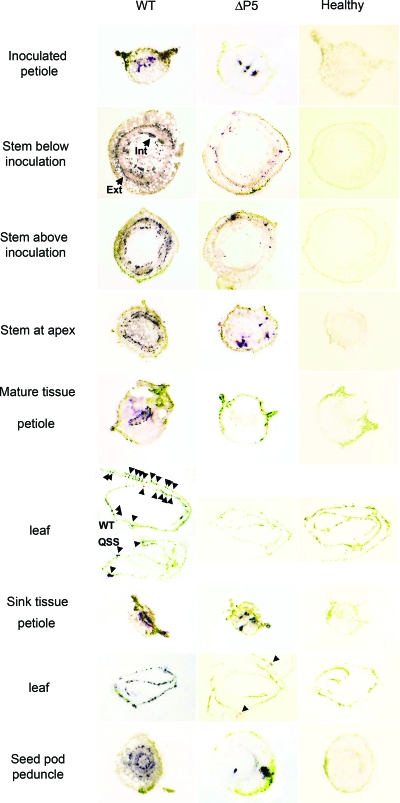
Representative S. sarrachoides plant 3 weeks p.i. Samples were taken for tissue immunoblots. (A) Stem tissue sample below inoculation sites; (B to I) stem samples above the inoculation sites, with stem samples taken at nodes to apex of branches; (J) mature leaf and petiole samples; (K) developing leaf and petiole samples; (L) seed pod peduncles. The inset picture shows a plant (8- to10-leaf stage) at the time of inoculation. Stars indicate the general area where 3- to 4-week-old plants were inoculated. Plants were pruned at 3 to 4 weeks p.i. by removing 7.5 to 10 cm of stem from each branch.
To localize the virus in various tissues over time, a tissue immunoblot analysis was performed on plant tissues infected with the various mutants at 3 and 9 weeks postinoculation (p.i.) as described previously by Lee et al. (20). Briefly, inoculated leaves were marked to determine where the zones above and below the inoculation region were located. Two plants were tested for each sample at each time point. Using a double-edged razor blade, petioles and stems were cut along their axes, and the cut surfaces were pressed onto nitrocellulose membranes for 20 s. Stem tissue prints were taken below and above the inoculation zone at each node for each stem branch including the apex (Fig. 2). Fully expanded and developing leaves were also tissue immunoblotted by rolling the leaf, cutting along the axis, and pressing the cut surfaces to the membranes. In addition, the petioles of these leaves were tissue immunoblotted along with the peduncles of seed pod clusters that were attached along the stem. For whole-leaf tissue immunoblots, leaves were collected and placed into a petri plate with a moist paper towel in order to prevent drying. The epidermis of the abaxial surface of the leaves tested was peeled away to expose the mesophyll tissue. The leaves were sandwiched between two pieces of nitrocellulose and placed between two 10- by 10-cm sheets of 6.35-mm steel. This steel membrane sandwich was placed into a vice and hand tightened to press the sandwich together and print the leaves onto the membrane. Membranes were allowed to air dry, and blots were placed into a plastic bag and stored at 4°C until they were processed. Blots were developed as described previously for Western blots by Lee et al. (20) and Kaplan et al. (18) using monoclonal antibody SCR3, which recognizes an N-terminal domain on the P3 protein (38). After the tissue immunoblots were developed, the cells in each of the stem, petiole, leaf, and seed pod peduncle sections with detectable virus present were observed and counted under a dissecting microscope.
Analysis of virus in infected tissue.
Three leaves from each WT-, ΔP5-, and SYG-infected plant were removed and divided into two halves. For each infecting virus, the P3 protein was detected in total protein extracts in half of the leaf using Western blot analysis as previously described (18, 19, 27). The remaining half of the leaf was used to measure virus accumulation using double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) as described previously by Lee et al. (19). The progeny viruses from systemically infected plants were analyzed by reverse transcription-PCR (RT-PCR) and direct sequencing of the RT-PCR products as described previously by Peter et al. (27).
Electron microscopy.
WT PLRV- and SYG-infected plant tissues at 4 and 10 weeks p.i., respectively, were fixed for electron microscopy in 1% formaldehyde-2% glutaraldehyde in 0.02 M sodium cacodylate buffer (pH 7.4) containing 10 mM CaCl2 and 0.05% sodium azide. Plant leaf tissues consisting of lamina leaf epidermal, parenchyma, and mesophyll tissue and minor veins adjacent to areas showing chlorotic symptoms were immersed in fixative, cut into 2- to 3-mm squares with a razor blade, and then transferred to fresh fixative and incubated overnight at 4°C. Tissues were then incubated for 30 min in three changes of 0.1 M sodium cacodylate buffer and postfixed for 60 min in 1% osmium tetroxide in 0.1 M buffer at 4°C. Following two 5-min rinses in buffer and two rinses in water, tissues were incubated for 60 min in 2% aqueous uranyl acetate at 4°C for en bloc staining and fixation. The leaf tissue was then dehydrated in a 12-to-100% ethanol series, followed by 1:1 ethanol-propylene oxide to 100% propylene oxide. The tissue was embedded in Spurr's plastic (Electron Microscopy Sciences, Ft. Washington, PA) and hardened at 60°C for 24 h. Semithick 0.5-μm sections were stained with azure B for tissue identification and orientation by Nomarski interference microscopy. Ultrathin sections, approximately 60 nm thick, on Formvar-carbon-coated grids were contrasted for 45 min in 2% uranyl acetate with 10% isobutanol (30) and for 15 min in aqueous 0.4% lead citrate. Grids were viewed on a Jeol 1200 transmission electron microscope at the Electron Microscope Facility for the Life Sciences at the Pennsylvania State University.
Mechanical inoculation of healthy plants using infected S. sarrachoides tissue.
Healthy S. sarrachoides plants were inoculated mechanically using sap from S. sarrachoides tissue infected with WT PLRV, ΔP5, and SYG and also SYG-infected leaf tissue exhibiting WT-like symptoms. Infected leaf tissue (0.1 g) was homogenized in 2 to 3 ml of 1× phosphate-buffered saline using a mortar and pestle, and the suspension was used to rub inoculate two leaves on each of three plants. Leaves were inoculated by gently swabbing the area where Carborundum was lightly dusted. Plants were monitored for symptoms and tested 5 to 10 days after inoculation for infection by DAS-ELISA as described previously by Kaplan et al. (18).
Virus detection in aphids.
Healthy Myzus persicae aphids were allowed 24- and 72-h acquisition access periods on S. sarrachoides tissue systemically infected with WT PLRV, QSS, ΔP5, and SYG. Six aphids were removed, divided randomly into two groups of three aphids, and stored at −80°C in 50 μl nuclease-free water in RNase-free microcentrifuge tubes. Total RNA was isolated and analyzed as described previously by Peter et al. (27) except that primers PLRV 5′ P3 3592-3614 (5′-ATGAGTACGGTCGTGGTTAAAGGAAAT-3′) and PLRV 3′ P3 4215-4191 (5′-ATTTGGGGTTTTGCAAAGCCACC-3′) were used to amplify a 624-bp fragment, which included the sequences encoding the P3 protein that was used to indicate the presence of virus. This experiment was repeated twice.
RESULTS
P3/P5 works in trans to affect the dynamics of systemic movement of virus and symptom expression.
In nature, aphids are required to inoculate plants with PLRV, and only virions that have the P5 domain incorporated into the particle are transmitted. The use of agroinfection allowed us to bypass the aphid requirement and efficiently inoculate plants with different PLRV mutants that do not translate normal P3/P5 proteins or that do not incorporate P5 into the virion. The SST mutant translates a full-length P3/P5 (minus the SST amino acid triplet) that is incorporated into virions, QSS translates a full-length P3/P5 (minus the QSS triplet) that does not incorporate into virions, SYG translates a truncated P3/P5 (minus the SYG triplet in the N-terminal half) that is missing the C-terminal half of the P5 domain and is not incorporated into virions, and ΔP5 does not translate P3/P5; it translates only P3.
Tissue immunoblot analysis (20) was then used to observe virus distribution in S. sarrachoides plants infected with each mutant. Virus was localized in sections of the stems at each node, leaves and their respective petioles, and seed pod peduncles (Fig. 2). At 3 weeks p.i., all of the mutants were detected in stem sections above and below the inoculated leaves and in developing tissues; however, the presence of P3/P5, its size, and whether it was incorporated into virions all affected the number of infection foci in all tissues and the ability of virus to move into mature tissues. The WT and SST and QSS mutants were detected at similar levels in internal and external phloem bundles of the stem sections and sink tissues (developing leaves, petioles, and seed pod peduncles) (Table 1 and Fig. 3). However, QSS, the mutant that retained the ability to translate a near-complete P3/P5 but did not incorporate the P3/P5 into virions, was observed consistently in fewer phloem bundles in mature leaves that had transitioned from sink to source tissue (Table 1 and Fig. 3). Furthermore, at 3 weeks p.i., interveinal chlorosis was observed on WT- and SST-infected plants, whereas these symptoms were not observed on QSS-infected plants for an additional 7 to 10 days. In previous studies (27), several other mutants that contained 3-amino-acid deletions distinct from QSS but that also did not incorporate P3/P5 into virions were similarly delayed in symptom expression. Therefore, it is likely that the lack of incorporation and not the specific 3-amino-acid deletion was responsible for this phenotype. Deleting or truncating P5 in the ΔP5 and SYG mutants, respectively, significantly reduced the movement and accumulation of virus in all tissues. The SYG and ΔP5 mutants were detected in stem and developing leaf tissues at 3 weeks p.i.; however, compared to the WT, few infected cells were observed in the internal and external phloem bundles (Fig. 3). These mutant viruses were not detected in mature leaves or their respective petioles (Fig. 3), and WT-like symptoms were absent. Although the number of infection foci was reduced, the intensity of the staining was similar to that of the infection foci observed in WT-infected tissue, suggesting that virus replication was not affected. This is consistent with previous data indicating that virus replication and P3/P5 translation efficiency were not affected by the P5 mutations (10, 27).
TABLE 1.
Numbers of S. sarrachoides tissue samples with virus and average number of infected cells detected for each virus sample at 3 weeks p.i.
| Tissue sample | Infecting virus
|
|||||||
|---|---|---|---|---|---|---|---|---|
| WTa
|
QSS
|
SYG
|
ΔP5
|
|||||
| No. of samples with detectable infected cells/total no. of tissue samples taken | Avg no. of cells with virus for each tissue sample | No. of samples with detectable infected cells/total no. of tissue samples taken | Avg no. of cells with virus for each tissue sample | No. of samples with detectable infected cells/total no. of tissue samples taken | Avg no. of cells with virus for each tissue sample | No. of samples with detectable infected cells/total no. of tissue samples taken | Avg no. of cells with virus for each tissue sample | |
| Inoculated petiole | 8/8 | 35 | 8/8 | 30 | 7/7 | 3 | 8/8 | 2 |
| Stem, below area of inoculation | 2/2 | 237 | 2/2 | 216 | 2/2 | 11 | 2/2 | 3 |
| Stem, above area of inoculation | 53/53 | 97-175b | 48/48 | 114-181b | 16/49 | 4 | 42/44 | 4 |
| Stem, apex | 11/11 | 57 | 9/9 | 40 | 1/9 | 4 | 10/10 | 3 |
| Mature leaves | 8/8 | 59 | 4/9 | 13 | 0/11 | 0 | 0/9 | 0 |
| Petioles from mature leaves | 8/8 | 32 | 9/9 | 2 | 0/11 | 0 | 0/9 | 0 |
| Young leaves | 10/10 | 48 | 10/12 | 47 | 3/10 | 3 | 1/8 | 2 |
| Petioles from young leaves | 10/10 | 21 | 11/12 | 21 | 3/10 | 3 | 2/8 | 2 |
| Seed pods peduncles | 6/6 | 56 | 7/7 | 47 | 2/7 | 2 | 1/5 | 1 |
The data for the WT and SST samples were nearly identical.
The range includes the average numbers of cells in the smaller-diameter sections and the larger-diameter stem sections.
FIG. 3.
S. sarrachoides stem and leaf tissue immunoblots. Plants were agroinoculated with WT virus or the ΔP5 mutant 3 weeks prior to destructive sampling from many sites on the plant. Samples from similar regions on healthy plants were also collected. Virus antigen is detected as a purplish-blue immunostained region, some of which are indicated by arrowheads. The internal (Int) and external (Ext) phloem bundles are labeled. Immunoblots on tissue collected from plants infected with the SST, QSS, and SYG mutants were also completed (data not shown). The level and distribution of virus for SST and QSS were similar to those of the WT, and SYG was similar to ΔP5.
The P5 domain affects the tissue specificity of PLRV infection.
A loss of the P5 domain or the C-terminal portion of the P5 domain had a much more profound impact on virus movement than a reduction in the number of phloem bundles invaded, but this became evident only many weeks after plants were inoculated. SYG- and ΔP5-infected plants began to develop symptoms 8 to 10 weeks p.i.; however, instead of the interveinal chlorosis typical of WT infection (Fig. 4A), the symptoms were characterized by localized, slowly expanding, chlorotic lesions (Fig. 4E), which developed only on the youngest leaves. Furthermore, these symptoms were observed only when 7.5 to 10 cm of each branch was pruned from the plants at 3 to 4 weeks p.i. Pruning facilitated symptom expression in the new growth 4 to 6 weeks later in SYG- and ΔP5-infected plants, whereas plants that were not pruned remained asymptomatic. Pruning did not affect either symptom development or the type of symptoms in WT-, QSS-, and SST-infected plants (data not shown). Tissue immunoblots comparing tissues from pruned and nonpruned plants infected with the WT or the QSS mutant also indicated no differences in numbers of infected cells or distribution of virus in tissue, whereas major differences were apparent between pruned and nonpruned plants infected with the SYG or ΔP5 mutant (Table 2). Tissues from pruned SYG- or ΔP5-infected plants contained numbers of infection foci and a distribution of virus that were similar to those of WT-infected tissue (Table 2 and Fig. 5) with the exception of mature leaf and associated petiole tissues. No virus was detected in these tissues from pruned SYG- or ΔP5-infected plants. Similar to the above-described results (Table 1 and Fig. 3), few infection foci were observed in stem tissues of SYG- or ΔP5-infected plants that were not pruned (Table 2 and Fig. 5). In contrast to results from samples taken at 3 weeks p.i. (Table 1 and Fig. 3), no virus was detected in sink leaf or petiole tissues from nonpruned SYG- or ΔP5-infected plants tested at 9 weeks p.i. (Table 2 and Fig. 5).
FIG. 4.
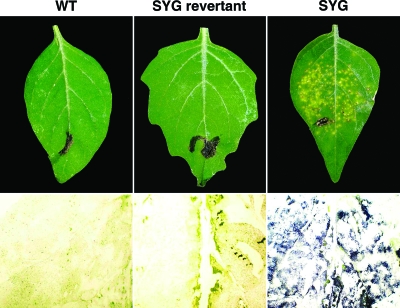
Symptoms observed on S. sarrachoides leaves infected systemically with WT (A) and SYG (E) viruses at 4 and 10 weeks p.i., respectively. These leaves were used to generate whole-leaf tissue immunoblots of the abaxial surfaces. (B) Enlarged view of the immunoblot of WT-infected leaf tissue showing virus associated with only the veins (arrows pointing to small purple regions). The area shown is approximately a ×2.5 magnification of what is viewed in F. (C) Transmission electron microscopy analysis of similar WT-infected tissues (magnification, ×3,000) illustrating virions present in phloem-related cells, companion cells (CC), and sieve elements (SE). (D) Higher magnification (magnification, × 30,000) of the boxed region in C displaying virions (arrows) within the cell cytoplasm. (F) Enlarged view of the whole-leaf immunoblot of SYG-infected tissue revealing extensive spread of virus outside the phloem tissues and into mesophyll tissues (purple regions). Similar results were observed for ΔP5-infected leaves (not shown). (G) Transmission electron microscopy analysis of SYG-infected tissue (magnification, ×2,000) confirming immunoblot data for the presence of virions in mesophyll cells (Me). (H) Higher magnification (magnification, ×30,000) of the boxed region in G where crystalline arrays of virions (arrows) were commonly observed to be associated with the chloroplast outer membranes.
TABLE 2.
Average numbers of infected cells in pruned versus nonpruned plants at 9 weeks p.i.
| Tissue sample | Avg no. of cells with virus in pruned plants/avg no. of cells with virus in nonpruned plantsa
|
|||
|---|---|---|---|---|
| WT | QSS | SYG | ΔP5 | |
| Stem, below inoculation | 488/450 | 442/415 | 333/148 | 375/180 |
| Stem, above inoculation | 200/188 | 190/175 | 170/6 | 158/3 |
| Stem, apex | 81/73 | 79/85 | 69/4 | 67/4 |
| Mature leaves | 77/65 | 68/59 | 0/0 | 0/0 |
| Petioles from mature leaves | 38/35 | 34/37 | 0/0 | 0/0 |
| Young leaves | 65/59 | 69/62 | ++/0 | ++/0 |
| Petioles from young leaves | 35/37 | 30/34 | 28/0 | 25/0 |
| Seed pod peduncles | 70/77 | 83/71 | 85/0 | 72/0 |
++, individual cells were not discernible since a large amount of virus was present in the youngest leaves of pruned plants.
FIG. 5.
Immunoblots of various tissue sections from pruned or nonpruned S. sarrachoides plants infected previously with WT or the ΔP5 mutant at 9 weeks p.i. Areas positive for virus are indicated by purplish-blue staining. Virus levels and distribution were similar for pruned and nonpruned (not shown) plants infected with WT and QSS virus (pruned WT samples are shown). Virus levels and distribution for the SYG mutant (not shown) were similar to those of images shown for ΔP5. Similar to the images of healthy tissue shown in Fig. 3, no labeling was observed in healthy control tissue used in this experiment (not shown).
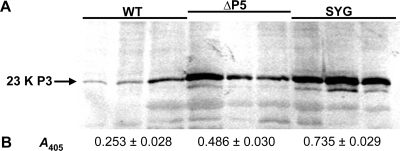
The expanding chlorotic lesion symptoms and the increased number of infected cells in leaf tissue from SYG- or ΔP5-infected plants were suggestive of virus invasion beyond the phloem. DAS-ELISA and Western blot analysis comparing virus titers among leaf tissues from WT-, SYG-, and ΔP5-infected plants found significantly higher virus antigen levels in SYG- and ΔP5-infected tissue than in WT-infected tissue (Fig. 6). Whole-leaf tissue immunoblots comparing WT-, SYG-, and ΔP5-infected plants revealed that SYG (Fig. 4F) and ΔP5 (data not shown) viruses were unloading from the vascular tissue and translocating into and replicating in the mesophyll tissue. This was in stark contrast to the punctate distribution of WT virus that was limited to phloem cells (Fig. 4B). Transmission electron microscopy of leaf tissue infected with WT virus revealed virions only in the cytoplasm of phloem-associated cells (Fig. 4C and D), whereas a substantial accumulation of SYG virions was observed in mesophyll tissue associating with chloroplast membranes (Fig. 4G and H). This chloroplast association, particularly in phloem tissues, has been described previously for luteovirids and was suggested to be a potential site for replication (16, 24, 33).
FIG. 6.
Measurement of virus and P3 protein accumulation in WT-, ΔP5-, and SYG-infected leaves using Western blot analysis and DAS-ELISA. Three leaves each from pruned WT-, ΔP5-, and SYG-infected plants were divided into two halves. (A) The P3 protein was detected in total protein extracts in one half of the leaf by Western blot analysis. (B) Virus accumulation was detected in the other half of the leaf using DAS-ELISA. The mean absorbance values (A405) ± standard errors are shown. One-way analysis of variance indicated a significant effect of virus type on antigen accumulation (F = 69.53; df = 2; P < 0.001), and treatment means were all significantly different from each other according to the Newman-Keuls test (P = 0.05).
Lack of the P5 C-terminal domain permits mechanical transmission of PLRV.
Since SYG and ΔP5 mutant viruses were no longer phloem limited in the youngest leaves and were able to move into and successfully replicate in mesophyll tissue, we hypothesized that it may be possible to initiate plant infection by rub inoculation using plant sap containing these mutants. Leaf sap of SYG- and ΔP5-infected S. sarrachoides was used to inoculate mechanically healthy S. sarrachoides plants. After 5 to 10 days, localized chlorotic lesions that were similar to those described above and shown in Fig. 4 were observed (Fig. 7), and virus was distributed in mesophyll tissue (Fig. 7). The nucleotide sequence of RNA from the progeny virus extracted from these leaves was identical to that of the inoculating virus. In contrast, mechanical inoculation of WT virus failed to induce symptoms on inoculated leaves, and virus was not detected at 5 to 10 days after inoculation (Fig. 7).
FIG. 7.
Mechanically inoculated S. sarrachoides leaves and their respective immunoblots approximately 5 days p.i. using WT-, SYG revertant (exhibiting WT-like symptoms)-, or SYG-infected S. sarrachoides leaves as the source of plant sap for virus inoculation. The immunoblots for the WT and the SYG revertant are approximately a ×2.5 magnification of the SYG immunoblot.
Phloem restriction of PLRV appears to be a self-imposed trait.
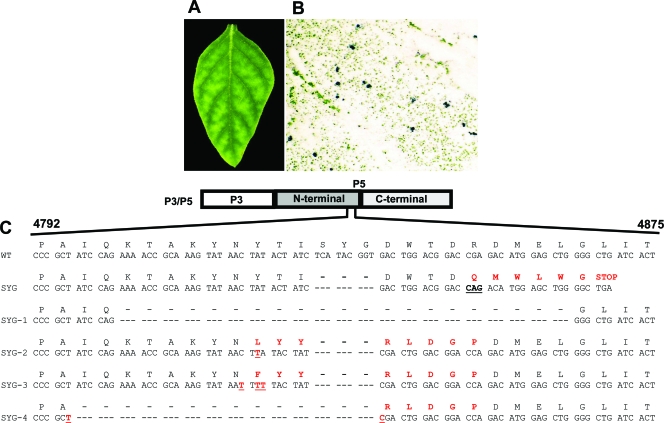
Approximately 4 months p.i., multiple young leaves on several SYG-infected plants manifested WT-like symptoms. Whole-leaf tissue immunoblots of four of these leaves indicated that the virus distribution in these leaves was confined to phloem tissue and no longer moving out into mesophyll cells (Fig. 8A and B). The P5 open reading frame (ORF) was sequenced from progeny viral RNA isolated from these leaves. The consensus sequence from RNA isolated from each of the four leaves contained a different intragenic mutation or pseudorevertant mutation, either nucleotide insertions or larger deletions, all of which would result in a frameshift and would be expected to restore the translation of the P5 C terminus (Fig. 8C). The low virus titer and the antibodies available do not allow Western blot detection of P3/P5 in systemically infected leaves, data which would have allowed us to verify that the P5 C terminus was translated. The revertant SYG virus from these leaves was not sap transmissible (Fig. 7), further supporting the observation that the virus was again phloem specific and likely translating the P5 C terminus. Continuing experiments will address this issue directly using a reverse-genetics approach and complementation of P5 domains in transgenic plants.
FIG. 8.
Analysis of SYG revertant-infected leaf tissue. (A) SYG-infected S. sarrachoides leaf exhibiting WT-like symptoms. (B) Enlarged view of a SYG-infected whole-leaf tissue immunoblot from the same leaf showing virus confined to phloem tissue. (C) SYG revertant sequences (sequences SYG-1 to -4) obtained from progeny virus in four different infected leaves. WT and SYG sequences are shown for comparison. In the SYG sequence, the codon containing a deleted nucleotide (14 nt downstream from the SYG deletion) that resulted in a frameshift mutation is underlined. Amino acids and nucleotides that were altered in the pseudorevertants are shown in red type. Sequences SYG-1 to -4 originate from leaves manifesting WT-like symptoms.
Distribution of virus in the plant affects the ability of aphids to ingest virus.
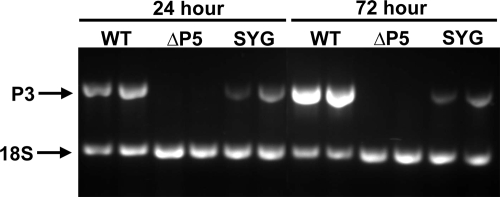
PLRV and other luteovirids are normally ingested by aphids during prolonged phloem feeding. Aphids also probe briefly on mesophyll and epidermal cells and can acquire sufficient virus for stylet-borne transmission, but it is unknown if they can acquire sufficient virus for circulative transmission to occur. To determine if virus distribution in nonvascular tissues, rather than being confined to the phloem, was disadvantageous to the transmission of PLRV, we compared the ingestion efficiencies of the ΔP5 and SYG mutants to that of the WT by allowing aphids a 24- or 72-h feeding access period on infected leaves. Although the ΔP5 and SYG virions lack an incorporated P5, which is required for virus transmission, the lack of P5 is not expected to affect the ingestion of virus during probing or feeding. WT virus was detected by RT-PCR in all individual aphids following either a 24- or 72-h feeding period. SYG was also detected in all aphids, but the concentration was much lower than that for the WT. ΔP5 virus was not detected in any aphids tested (Fig. 9).
FIG. 9.
RT-PCR to detect the presence of virus in aphids after 24- and 72-h periods of feeding on virus-infected tissue to determine if aphids ingested virus from infected leaves. Primers amplify a 624-bp fragment of the P3 ORF. Primers that amplify a 301-bp fragment of the 18S rRNA were used as an RNA control. Aphids were fed on tissue systemically infected with WT, QSS, ΔP5, or SYG virus. WT and QSS viruses were identical, with the WT being the representative sample shown. Two groups of three aphids were collected at each time point for each sample.
DISCUSSION
The prevailing hypotheses to explain the phloem limitation of poleroviruses such as PLRV are that the virus is unable to suppress plant defense mechanisms outside of the phloem or that the viruses lack a movement protein that can function in nonvascular tissues (36). Our current study has demonstrated an alternative explanation: the virus encodes a protein, P5, which confines the virus to phloem tissues. Previous work has shown that P3/P5, when incorporated into the virion, has a functional role during movement. Reduced systemic movement and reduced virus accumulation in systemically infected tissues were reported for mutations in other luteovirids that affected P3/P5 incorporation into the virion or translation of a full-length P3/P5 (9-11, 24). Our data indicate that P3/P5 acts primarily as a nonstructural movement protein in trans and does not need to be incorporated into virus particles to move throughout the plant. In the host plant S. sarrachoides, the P3/P5 present in trans affects virus movement among the internal and external phloem bundles, which affects the overall accumulation of virus within the plant. However, P3/P5 functions differently in tissues that differ in sink-source status. P3/P5 incorporation into the virion appears to be required for virus to move into mature tissues. The QSS mutant translated a nearly complete P3/P5 that was not incorporated into virions. QSS was able to move into younger sink tissues similarly to WT virus, but the QSS virus was not detected in mature source tissues. The combined effect of P3/P5 incorporation and P3/P5 functioning in trans contributes to successful virus spread within phloem tissues throughout the plant.
The role of P3/P5, particularly the P5 C terminus, in determining the tissue specificity of PLRV in S. sarrachoides plants was not observed for several months p.i. and only when plants had been pruned. Pruning facilitated the movement of ΔP5 and SYG viruses to infect extensively the internal and external phloem bundles within the stem and sink petiole tissues, comparable to viruses translating full-length P3/P5 (the WT and the SST and QSS mutants). As a result of the increased accumulation of virus in the ΔP5- and SYG-infected plants, symptoms manifested albeit only in the youngest tissue. When sink tissue is removed, the turgor pressure increases in the phloem to push photoassimilates, which include virions, to aid in producing new sink areas (37). This increase in turgor pressure may help overcome the movement barrier imposed by the P5 mutants by allowing even the SYG and ΔP5 mutants to spread among internal and external phloem bundles, which provides more opportunity for movement into and accumulation in younger leaves. Pruning plants also allowed a coat-protein-less mutant of TMV to move systemically more rapidly (21). This was attributed to rapid cell-to-cell movement of virus that was accumulating in side-shoot meristems of the pruned plant. Mutterer et al. (24) previously proposed that poleroviruses may move systemically by two distinct mechanisms: one that is P5 dependent and moves the virus in sieve elements and a second that is P5 independent and moves virus cell to cell in nucleate phloem companion or phloem parenchyma cells. In pruned plants, the SYG and ΔP5 mutants may be moving rapidly between the living phloem-associated cells of the expanding side shoots rather than the typical pathway within sieve elements and then into adjacent living cells. In the nonpruned plants, little to no virus was detected in internal phloem, particularly above the area of inoculation, which could be attributed to the plant growing faster than the virus can move; however, virus was detected throughout the external phloem below the inoculation area. A similar virus distribution was observed in resistant potato varieties where virus was restricted to internal phloem as opposed to external phloem (3, 4, 13).
Our results showing that P3/P5 regulates the tissue specificity of virus infection are in contrast to the findings described previously by Mutterer et al. (24). The deletion of P5 of BWYV did not allow the virus to move into mesophyll tissue. However, those authors were studying infection at earlier time points in a Nicotiana sp., and it is unknown if plants were pruned in that study. Infection of Nicotiana benthamiana or N. clevelandii with our PLRV mutants did not result in a loss of tissue specificity, but other natural hosts of PLRV have not been tested. Similar to our findings showing that P4 is a host-specific movement protein (18, 20, 27), P3/P5 may also act in a host-specific manner for restricting the virus to the phloem during infection.
The local lesion symptoms which we observed in the youngest leaves indicated that the virus was no longer confined to the phloem. Transmission electron microscopy and tissue immunoblot analysis verified that the P5 C terminus mutant viruses had translocated into the mesophyll tissue where the virus was replicating and that infection spread among many mesophyll cells beyond phloem tissue. The current study is the first report of an unaided luteovirus moving cell to cell outside phloem cells.
Infection by the phloem-limited luteovirids cannot be propagated mechanically, such that the infection expands by translocating cell to cell beyond initially infected epidermal or mesophyll cells. Although luteovirids are capable of replicating in epidermal and mesophyll cells (25, 28), and PLRV can infect mesophyll cells neighboring minor phloem vessels, the spread of the virus from cell to cell in the mesophyll has not been observed (40). The one exception in the family Luteoviridae is pea enation mosaic virus (PEMV), which is transmitted mechanically and by aphids and can be found in most tissues of infected plants. PEMV is a bipartite virus composed of two autonomously replicating, unrelated, positive-sense RNAs, the polerovirus-like RNA-1 and the umbravirus-like RNA-2 (12, 14). Both RNAs are required for successful infection. RNA-1 is similar to the PLRV genome except that it lacks the homologous P4, P6, and P7 ORFs (Fig. 1A). PEMV RNA-1 encodes proteins for replication, encapsidation, and aphid transmission. Both PEMV proteins that are homologous to P3 and P5 have sequence similarity to PLRV; however, PEMV P5 effectively lacks the C-terminal domain found in other luteovirids. RNA-1 is unable to infect plants systemically since it lacks a P4 movement protein; therefore, RNA-2 provides the functions for systemic movement and mechanical transmission. Interestingly, if an umbravirus RNA is combined with PLRV RNA, mechanical transmission and subsequent systemic infection of PLRV are possible (23, 31). Similarly, mixed infections of PLRV and either potato virus Y or potato virus A, members of the Potyviridae, allowed PLRV to overcome its phloem limitation and invade mesophyll tissues to a limited extent (2, 32), presumably because the helper virus provided a movement protein or disabled plant defenses. Interestingly, PLRV infection of transgenic plants expressing the potyvirus suppressor of gene silencing (HC-Pro) resulted in an increase in the numbers of PLRV-infected phloem cells but did not allow detectable egress of PLRV from phloem (32), suggesting that the suppression of general plant defenses against RNA viruses is not solely responsible for phloem limitation.
The observation that PLRV containing a truncated P3/P5 can be inoculated mechanically suggests that the phloem limitation of luteovirids is not due to a lack of a movement protein that allows the virus to move in nonvascular tissue but rather that the virus has evolved a protein-mediated mechanism conferred by the C-terminal portion of P5 that prevents the virus from escaping the phloem tissue. We hypothesize that the P4 movement protein may contribute to virus movement between mesophyll cells, which is supported by results showing that the PLRV P4 protein expressed in transgenic tobacco is localized in plasmodesmata of mesophyll cells and is able to promote the movement of fluorescent dextrans between mesophyll cells (17). PEMV P3/P5 shares amino acid sequence similarity with the truncated SYG P3/P5. PEMV was suggested to represent an evolutionary permutation of the luteovirus group that has circumvented phloem limitation (12), presumably by the acquisition of umbravirus RNA-2; however, the loss of the P5 C terminus may also be a key feature of this adaptation.
When SYG-infected plants were maintained for several months, mutations that restored the translation of the P5 C terminus were selected. A similar situation was noted in our previous study (27) when the consensus sequence of progeny virus in systemically infected leaves of SYG-infected Physalis floridana plants contained pseudoreversions that would allow the translation of the P5 C terminus. Brault et al. (9) also described similar pseudorevertants of a P5 C-terminal translation mutant of BWYV that restored the ability of the virus to move efficiently in phloem and accumulate to WT levels in plants. They did not report any alteration in the tissue specificity of their mutants. Taken together, these data indicate that mutations that restore the translation of the P3/P5 protein and, by extension, phloem limitation and efficient movement in phloem are favored.
This positive selection occurs in the absence of selective pressure for aphid transmission, but this selection is advantageous for aphids. The colateral effect of virus being concentrated in phloem tissues is that it is more readily available for uptake and dispersal by phloem-feeding aphid vectors. The circulative mechanism of polerovirus transmission requires lengthy phloem feeding by aphids to acquire sufficient quantities of virus for efficient transmission (15). Aphids do not initiate long-term feeding in mesophyll cells, and virus acquired during brief probes of mesophyll cells prior to initiating phloem feeding would be limited in quantity and not likely be transmitted efficiently. Furthermore, phloem infection results in a restriction of photoassimilates, which often leads to a general chlorosis or yellowing of the plant (luteolus is Latin for yellow). Many species of aphids are attracted to yellow, resulting in increased rates of landing of migrant aphids on diseased plants. In addition, virus-infected plants are often much better nutritional sources for aphids due to altered phloem amino acid and sugar compositions, resulting in aphid reproduction and distribution. Thus, there would seem to be an evolutionary advantage for the virus to remain in the phloem to facilitate its dispersal to other potential hosts (34). Indeed, we saw this advantage in aphid ingestion experiments. When the virus was not confined exclusively to the phloem, as in the SYG- or ΔP5-infected leaves, there was a significant reduction in the amount of virus ingested (Fig. 9). These results support our conclusion that phloem limitation in poleroviruses should not be considered a deficiency in movement or tissue invasion but rather an adaptive strategy to benefit some undefined aspect of virus survival in the plant since the selection occurs in the absence of aphid transmission selection pressure. However, this phloem preference has the colateral effect of enhancing transmission by aphids and ensuring continued dispersal to new host plants.
Based on these results, we propose a new model of P5 function during luteovirid movement. P3/P5 incorporated into the virion is not required for systemic movement or movement into developing leaf tissues; however, an incorporated P3/P5 may help the virus efficiently invade mature leaf phloem tissue that has made the transition from sink to source. Perhaps this is related to the morphology of plasmodesmata. In sink tissues, plasmodesmata are simple (not branched) and possess a high-molecular-mass exclusion limit (<50 kDa), which may permit the nonspecific trafficking of macromolecules (26). During the sink-source transition, the plasmodesmata switch from simple to branched forms, and having the P3/P5 associated with the virion may facilitate transport across the plasmodesmata or interaction with other viral or host factors that mediate the transport. Soluble P3/P5, and, more specifically, the P5 C terminus, facilitates virus movement among phloem bundles within all tissues, but importantly, it also actively restricts the virus to phloem-associated cells through an as-yet-unknown mechanism even when it is not incorporated into virions. It is unclear if the P5 C terminus interacts with other host or viral proteins, but when this domain is absent, the virus is able to invade nonvascular tissues in leaves and perhaps in the stem and petioles. More often than not, SYG or ΔP5 virus infection foci were multicellular in stem and petiole sections (Fig. 3), whereas in most cases, WT infection foci were localized to phloem bundles. P5-mediated phloem restriction does not preclude the idea that luteovirids are unable to suppress plant defense mechanisms outside of the phloem tissue (5). In fact, the local lesion type of symptoms and the lack of an even distribution of SYG or ΔP5 in mesophyll tissue argue that the virus is not completely free to invade mesophyll tissue once its phloem restriction protein is eliminated. However, as a virus that has evolved to be phloem restricted, it would not be expected to have evolved (or maintained) a mechanism to evade nonvascular plant defenses.
Acknowledgments
We thank T. Hammond and D. Smith for their assistance with greenhouse work and plant maintenance, K. Loeffler for photographing infected leaves and assistance with formatting figures, and G. Bruening, J. Murphy, and R. Turgeon for helpful and thoughtful discussions.
This work was partially supported by USDA CSREES NRI grants 96-01120 and 2005-35604-15446.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Bahner, I., J. Lamb, M. Mayo, and R. Hay. 1990. Expression of the genome of potato leafroll virus: readthrough of the coat protein termination codon in vivo. J. Gen. Virol. 712251-2256. [DOI] [PubMed] [Google Scholar]
- 2.Barker, H. 1987. Invasion of non-phloem tissue in Nicotiana clevelandii by potato leafroll virus is enhanced in plants also infected with potato Y potyvirus. J. Gen. Virol. 681223-1227. [Google Scholar]
- 3.Barker, H., and B. D. Harrison. 1985. Restricted multiplication of potato leafroll virus in resistant potato genotypes. Ann. Appl. Biol. 107205-212. [Google Scholar]
- 4.Barker, H., and J. A. T. Woodford. 1992. Spread of potato leafroll virus is decreased from plants of potato clones in which virus accumulation is restricted. Ann. Appl. Biol. 121345-354. [Google Scholar]
- 5.Baumberger, N., C. H. Tsai, M. Lie, E. Havecker, and D. C. Baulcombe. 2007. The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 171609-1614. [DOI] [PubMed] [Google Scholar]
- 6.Brault, V., E. Herrbach, and C. Reinbold. 2007. Electron microscopy studies on luteovirid transmission by aphids. Micron 38302-312. [DOI] [PubMed] [Google Scholar]
- 7.Brault, V., J. Mutterer, D. Scheidecker, M. T. Simonis, E. Herrbach, K. Richards, and V. Ziegler-Graff. 2000. Effects of point mutations in the readthrough domain of the beet western yellows virus minor capsid protein on virus accumulation in planta and on transmission by aphids. J. Virol. 741140-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brault, V., S. Perigon, C. Reinbold, M. Erdinger, D. Scheidecker, E. Herrbach, K. Richards, and V. Ziegler-Graff. 2005. The polerovirus minor capsid protein determines vector specificity and intestinal tropism in the aphid. J. Virol. 799685-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brault, V., J. F. J. M. van den Heuvel, M. Verbeek, V. Ziegler-Graff, A. Reutenauer, E. Herrbach, J. C. Garaud, H. Guilley, K. Richards, and G. Jonard. 1995. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 14650-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruyere, A., V. Brault, V. Ziegler-Graff, M. T. Simonis, J. F. J. M. van den Heuvel, K. Richards, H. Guilley, G. Jonard, and E. Herrbach. 1997. Effects of mutations in the beet western yellows virus readthrough protein on its expression and packaging and on virus accumulation, symptoms, and aphid transmission. Virology 230323-334. [DOI] [PubMed] [Google Scholar]
- 11.Chay, C. A., U. B. Gunasinge, S. P. Dinesh-Kumar, W. A. Miller, and S. M. Gray. 1996. Aphid transmission and systemic plant infection determinants of barley yellow dwarf luteovirus-PAV are contained in the coat protein readthrough domain and 17-kDa protein, respectively. Virology 21957-65. [DOI] [PubMed] [Google Scholar]
- 12.Demler, S. A., and G. A. de Zoeten. 1991. The nucleotide sequence and luteovirus-like nature of RNA 1 of an aphid non-transmissible strain of pea enation mosaic virus. J. Gen. Virol. 721819-1834. [DOI] [PubMed] [Google Scholar]
- 13.Derrick, P., and H. Barker. 1997. Short and long distance spread of potato leafroll luteovirus: effects of host genes and transgenes conferring resistance to virus accumulation in potato. J. Gen. Virol. 78243-251. [DOI] [PubMed] [Google Scholar]
- 14.de Zoeten, G. A., and J. S. Skaf. 2001. Pea enation mosaic and the vagaries of a plant virus. Adv. Virus Res. 57323-350. [DOI] [PubMed] [Google Scholar]
- 15.Gray, S., and F. E. Gildow. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41539-566. [DOI] [PubMed] [Google Scholar]
- 16.Herbers, K., E. Tacke, M. Hazirezaei, K. P. Krause, M. Melzer, W. Rohde, and U. Sonnewald. 1997. Expression of a luteoviral movement protein in transgenic plants leads to carbohydrate accumulation and reduced photosynthetic capacity in source leaves. Plant J. 121045-1056. [DOI] [PubMed] [Google Scholar]
- 17.Hofius, D., K. Herbers, M. Melzer, A. Omid, E. Tacke, S. Wolf, and U. Sonnewald. 2001. Evidence for expression level-dependent modulation of carbohydrate status and viral resistance by the potato leafroll virus movement protein in transgenic tobacco plants. Plant J. 28529-543. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, I. B., L. Lee, D. R. Ripoll, P. Palukaitis, F. Gildow, and S. M. Gray. 2007. Point mutations in the potato leafroll virus major capsid protein alter virion stability and aphid transmission. J. Gen. Virol. 881821-1830. [DOI] [PubMed] [Google Scholar]
- 19.Lee, L., I. B. Kaplan, D. R. Ripoll, D. Liang, P. Palukaitis, and S. M. Gray. 2005. A surface loop of the potato leafroll virus coat protein is involved in virion assembly, systemic movement, and aphid transmission. J. Virol. 791207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, L., P. Palukaitis, and S. M. Gray. 2002. Host-dependent requirement for the potato leafroll virus 17-kDa protein in virus movement. Mol. Plant-Microbe Interact. 151086-1094. [DOI] [PubMed] [Google Scholar]
- 21.Lindbeck, A. G. C., D. J. Lewandowski, J. N. Culver, W. W. Thomson, and W. O. Dawson. 1992. Mutant coat protein of tobacco mosaic virus induces acute chlorosis in expanded and developing tobacco leaves. Mol. Plant-Microbe Interact. 5235-241. [Google Scholar]
- 22.Lucas, W. J. 2006. Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344169-184. [DOI] [PubMed] [Google Scholar]
- 23.Mayo, M., E. Ryabov, G. Fraser, and M. Taliansky. 2000. Mechanical transmission of potato leafroll virus. J. Gen. Virol. 812791-2795. [DOI] [PubMed] [Google Scholar]
- 24.Mutterer, J. D., C. Stussi-Garaud, P. Michler, K. E. Richards, G. Jonard, and V. Ziegler-Graff. 1999. Role of the beet western yellows virus readthrough protein in virus movement in Nicotiana clevelandii. J. Gen. Virol. 802771-2778. [DOI] [PubMed] [Google Scholar]
- 25.Nurkiyanova, K. M., E. V. Ryabov, U. Commandeur, G. H. Duncan, T. Canto, S. M. Gray, M. A. Mayo, and M. E. Taliansky. 2000. Tagging potato leafroll virus with the jellyfish green fluorescent protein gene. J. Gen. Virol. 81617-626. [DOI] [PubMed] [Google Scholar]
- 26.Oparka, K. J., A. G. Roberts, P. Boevink, S. Santa Cruz, I. M. Roberts, K. S. Pradel, A. Imlau, G. Kotlizky, N. Sauer, and B. L. Epel. 1999. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97743-754. [DOI] [PubMed] [Google Scholar]
- 27.Peter, K. A., D. Liang, P. Palukaitis, and S. M. Gray. 2008. Small deletions in the potato leafroll virus readthrough protein affect particle morphology, aphid transmission, virus movement and accumulation. J. Gen. Virol. 892046-2054. [DOI] [PubMed] [Google Scholar]
- 28.Prüfer, D., J. Schmitz, E. Tacke, B. Kull, and W. Rohde. 1997. In vivo expression of a full-length cDNA copy of potato leafroll virus (PLRV) in protoplasts and transgenic plants. Mol. Gen. Genet. 253609-614. [DOI] [PubMed] [Google Scholar]
- 29.Reinbold, C., F. E. Gildow, E. Herrbach, V. Ziegler-Graff, M. C. Gonçalves, J. F. J. M. van den Heuvel, and V. Brault. 2001. Studies on the role of the minor capsid protein in transport of beet western yellows virus through Myzus persicae. J. Gen. Virol. 821995-2007. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, I. M. 2002. Iso-butanol saturated water: a simple procedure for increasing staining intensity of resin sections for light and electron microscopy. J. Microsc. 20797-107. [DOI] [PubMed] [Google Scholar]
- 31.Ryabov, E. V., G. Fraser, M. A. Mayo, H. Barker, and M. Taliansky. 2001. Umbravirus gene expression helps potato leafroll virus to invade mesophyll tissues and to be transmitted mechanically between plants. Virology 286363-372. [DOI] [PubMed] [Google Scholar]
- 32.Savenkov, E. I., and J. P. T. Valkonen. 2001. Potyviral helper-component proteinase expressed in transgenic plants enhances titers of potato leaf roll virus but does not alleviate its phloem limitation. Virology 283285-293. [DOI] [PubMed] [Google Scholar]
- 33.Schmitz, J., C. Stussi-Garaud, E. Tacke, D. Prüfer, W. Rohde, and O. Rohfritsch. 1997. In situ localization of the putative movement protein (pr17) from potato leafroll luteovirus (PLRV) in infected and transgenic potato plants. Virology 235311-322. [DOI] [PubMed] [Google Scholar]
- 34.Stout, M. J., J. S. Thaler, and B. P. H. J. Thomma. 2006. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu. Rev. Entomol. 51663-689. [DOI] [PubMed] [Google Scholar]
- 35.Tacke, E., D. Prüfer, J. Schmitz, and W. Rohde. 1991. The potato leafroll luteovirus 17k protein is a single-stranded nucleic acid-binding protein. J. Gen. Virol. 722035-2038. [DOI] [PubMed] [Google Scholar]
- 36.Taliansky, M., M. A. Mayo, and H. Barker. 2003. Potato leafroll virus: a classic pathogen shows some new tricks. Mol. Plant Pathol. 481-89. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, M. V. 2006. Phloem: the long and the short of it. Trends Plant Sci. 1126-32. [DOI] [PubMed] [Google Scholar]
- 38.Torrance, L. 1992. Analysis of epitopes on potato leafroll virus capsid protein. Virology 191485-489. [DOI] [PubMed] [Google Scholar]
- 39.van den Heuvel, J. F., A. Bruyere, A. Hogenhout, V. Ziegler-Graff, V. Brault, M. Verbeek, F. van der Wilk, and K. Richards. 1997. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J. Virol. 717258-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Heuvel, J. F., C. M. de Blank, D. Peters, and J. W. M. van Lent. 1995. Localization of potato leafroll virus in leaves of secondarily-infected potato plants. Eur. J. Plant Pathol. 101567-571. [Google Scholar]
- 41.Waigmann, E., S. Ueki, K. Trutnyeva, and V. Citovsky. 2004. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit. Rev. Plant Sci. 23195-250. [Google Scholar]
- 42.Wang, J. Y., C. Chay, F. E. Gildow, and S. M. Gray. 1995. Readthrough protein associated with virions of barley yellow dwarf luteovirus and its potential role in regulating the efficiency of aphid transmission. Virology 206954-962. [DOI] [PubMed] [Google Scholar]