Abstract
Avian bornaviruses (ABV), representing a new genus within the family Bornaviridae, were recently discovered in parrots from North America and Israel with proventricular dilatation disease (PDD). We show here that closely related viruses are also present in captive European parrots of various species with PDD. The six ABV strains that we identified in clinically diseased birds are new members of the previously defined ABV genotypes 2 and 4. Viruses of both genotypes readily established persistent, noncytolytic infections in quail and chicken cell lines but did not grow in cultured mammalian cells in which classical Borna disease virus strains replicate very efficiently. ABV antigens were present in both the cytoplasm and nucleus of infected cells, suggesting nuclear replication of ABV. The genome organization of avian and mammalian bornaviruses is highly conserved except that ABV lacks a distinct control element in the 5′ noncoding region of the bicistronic mRNA encoding the viral proteins X and P. Reverse transcription-PCR analysis demonstrated the presence of virus in many, if not all, organs of birds with PDD. Viral nucleic acid was also found in feces of diseased birds, suggesting virus transmission by the fecal-oronasal route. Immunohistochemical analysis of organs from birds with PDD revealed that infection with ABV is not restricted to cells of the nervous system. Thus, ABV exhibits a broad tissue and cell tropism that is strikingly different from classical Borna disease virus.
Proventricular dilatation disease (PDD) is an inflammatory disease of birds first described in the 1970s as macaw wasting disease during an outbreak among macaws (4, 6, 12, 22). PDD primarily affects the autonomic nerves of the upper and middle digestive tract, including the esophagus, crop, proventriculus, ventriculus, and duodenum. Clinically, PDD cases usually present with gastrointestinal tract dysfunction such as dysphagia, regurgitation, and presence of undigested food in feces. A dilated proventriculus and ventriculus are typical radiographic findings, and paper-thin walls and spontaneous ruptures of these parts of the digestive tract are frequently seen during necropsy (2). Neurological symptoms such as ataxia and abnormal gait may accompany the gastrointestinal tract symptoms. PDD is generally fatal although the disease may not always progress fast. Microscopically, PDD is recognized by the presence of immune cell infiltrates at enteric ganglia and nerves. Similar infiltrates may also be present in the brain, spinal cord, peripheral nerves, conductive tissue of the heart, smooth and cardiac muscle, and adrenal glands (1). At present, clinical diagnosis is based on biopsy of crop wall and demonstration of inflammatory cell infiltrates at ganglion cells (11).
Two independent searches for a viral etiology of PDD led to the identification of a group of novel viruses that form a new genus in the family Bornaviridae (17, 19). RNA products derived from these viruses, now collectively designated avian bornaviruses (ABV), were found in tissue samples from parrots with PDD but not in healthy birds from North America and Israel.
Until the discovery of ABV, the Bornaviridae family consisted of a single genus, classical Borna disease virus (BDV). BDV is the causative agent of a progressive neurological disorder that primarily affects horses, sheep, and some other farm animals in central Europe (7, 8, 20, 21, 27, 29). BDV exhibits a high tropism for the central nervous system in natural and experimental hosts where it can establish persistent, noncytolytic infections of neurons and astrocytes (3, 5, 10, 15, 30). The clinical symptoms of Borna disease are accompanied by strong immune cell infiltrations into infected brains (13, 18, 26, 28). Isolates of BDV readily infect mammalian cell cultures of neuronal and nonneuronal origin. Infection of these cells usually leads to noncytolytic viral persistence, with high levels of viral products accumulating in infected cells but few infectious particles being formed (15, 23). The epidemiology of BDV is not well understood, but recent findings indicate that farm animals may acquire the infection from infected insectivores which presumably contaminate the feed (16).
Sequence information from three different parts of the viral genome revealed the existence of at least five distinct ABV genotypes (19). The complete sequences of one ABV genotype 2 strain and large parts of genotype 3 and genotype 4 strains are available (17, 19). Although the ABV genome exhibits only about 65% sequence identity to reference strains of classical BDV, a striking conservation of the genome organization was noted. Conservation includes the number and order of genes on the linear viral genome, the structure of transcription initiation and termination signals, and the location of splice donor and acceptor sites.
Many important questions regarding the biology and epidemiology of ABV remain to be answered. Here, we report that ABV is also present in captive European parrots with PDD. We further report that ABV infects a surprisingly broad spectrum of cell types and organs of diseased birds and that it can readily be isolated using established quail or chicken cell lines.
MATERIALS AND METHODS
Birds with clinical signs of PDD.
Thirteen psittacine birds belonging to 10 species (two Amazona festiva, one Amazona ventralis, one Ara ararauna, one Ara macao, two Psittacus erithacus, two Cacatua galerita, one Cacatua alba, one Pionis menstruus, one Pionites leucogaster, and one Psittacula eupatria) were included in the study. Nine animals were males, and three were females while the sex of one bird was not known. Age varied between less than 1 year and 8 years in nine birds and was unknown for four animals. Three birds included in this study were sent to the Clinic for Birds for necropsy, while 10 other birds were brought live for diagnosis and treatment between July and September 2008. All birds showed clinical and/or pathological signs of PDD related to the digestive tract (reduced body weight, diarrhea, undigested food in feces, or dilated proventriculus) or to the nervous system (tremor, ataxia, or convulsions).
PCR-based screening for ABV.
Crop biopsies, blood samples, or brain tissues were initially screened for the presence of ABV by reverse transcription-PCR (RT-PCR). RNA extraction using an RNeasy kit (Qiagen, Hilden, Germany) was followed by reverse transcription with random hexamer primers and PCR targeting conserved regions of the L and N genes with published primers (19). Positive samples were further examined by PCR with primers targeting the M gene of ABV as described previously (19). PCR products were purified from agarose gels using a QIAquick gel extraction kit (Qiagen) and were sequenced with the same primers. All polymorphic sites were confirmed by at least one additional DNA sequence originating from an independent PCR amplification to rule out PCR polymerase artifacts.
Available organs of ABV-positive parrots were investigated for the presence of bornavirus RNA by real-time RT-PCR with primers and probes as described previously (17) or by conventional RT-PCR with primers targeting the L gene (19).
Construction of phylogenetic trees.
Sequences included in the phylogenetic analysis are summarized in Table S1 in the supplemental material. Alignments were based on nucleotide positions 657 to 1003, 1932 to 2241, and 3793 to 4203 for the N, M, and L genes, respectively, with base numbering according to the full genome sequence of strain bil (NCBI GenBank accession number EU781967). Sequences were aligned using the Clustal W algorithm (http://www.ebi.ac.uk/clustalw). Phylogenetic analysis was done employing distance and maximum-likelihood and-parsimony methods using the DNADIST and NEIGHBOR as well as the DNAML and the DNAPARS algorithms included in the PHYLIP software, version 3.68 (http://evolution.gs.washington.edu/phylip.html). Support for phylogenies was measured by bootstrapping 1,000 replicates with the programs SEQBOOT and CONSENSE. Phylogenetic trees were drawn with the programs RETREE and DRAWGRAM of the same PHYLIP package.
Virus isolation.
Frozen organs of diseased birds were homogenized with a mortar and sterile quartz sand before phosphate-buffered saline was added to make 10% (wt/vol) tissue suspensions. Sand and tissue debris were removed by low-speed centrifugation, and the virus-containing suspensions were stored at −80°C until use. Samples of the suspensions were added to the medium of semiconfluent cell cultures. After 3 h at 37°C, the medium was exchanged, and the infected cultures were passaged by splitting twice or trice weekly. Cell lines used for the isolation attempts included the quail fibroblast cell line CEC32 (31), the quail skeletal muscle cell line QM7 (ATCC CRL-1632), the chicken LMH hepatoma cell line (ATCC CCL-2117), and monkey Vero (ATCC CCL-81), rat C6 (ATCC CCL-107), and canine MDCK (ATCC CCL-34) cells. Productive virus infection of the cultures was monitored by indirect immunofluorescence as described for BDV (14). Antibodies used included cross-reactive rabbit antiserum raised against the N and P proteins of BDV (9).
Full genome sequencing.
RNA from infected CEC32 cells was reverse transcribed using random hexamer primers and standard protocols. Fragments of the viral genome were amplified by PCR using primers derived from published ABV sequences. A company (GATC, Konstanz, Germany) sequenced the gel-purified PCR products.
Immunohistochemistry.
Thin sections were prepared from formalin-fixed and paraffin-embedded tissue samples. After the paraffin was removed, the sections were rehydrated, permeabilized with Triton X-100, and incubated with 1:300 dilutions of rabbit antiserum in blocking solution (phosphate-buffered saline containing 5% goat serum) for 3 h at room temperature. Specifically bound antibodies were visualized using biotinylated goat anti-rabbit antibodies and a Vectastain ABC Elite detection kit (Vector Laboratories).
Nucleotide sequence accession numbers.
Partial ABV genome sequences reported here have been deposited in the NCBI GenBank database (http://www.ncbi.nlm.nih.gov) under the accession numbers FJ603671 to FJ603688. The accession number of the full-length sequence of strain 6609 is FJ620690.
RESULTS
Detection of ABV in captive parrots with PDD.
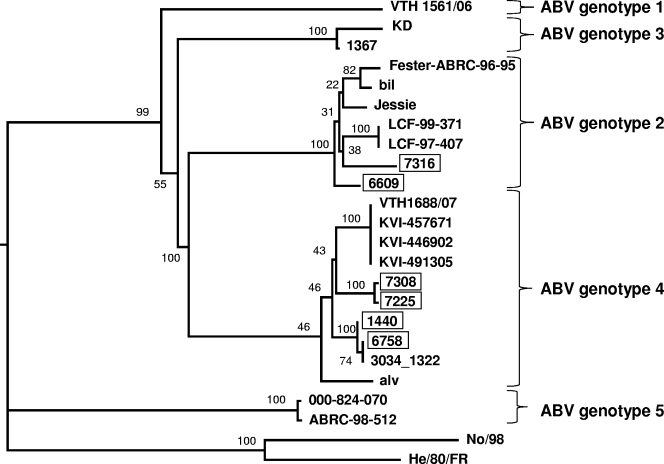
Thirteen parrots with clinical signs of PDD that were brought to the Clinic for Birds of the University of Munich for necropsy or diagnosis and treatment between July and September 2008 were included in this study. Screening by RT-PCR for the presence of nucleic acids derived from highly conserved regions of the L and the N gene of ABV identified six ABV-infected animals that scored positive in both assays, namely, two Cacatua galerita birds and one Cacatua alba, one Amazona ventralis, one Amazona festiva, and one Ara ararauna bird (Table 1). The other seven birds were negative in these assays. All six ABV-infected birds further scored positive if PCR primers were employed that target the M gene of ABV. Phylogenetic analyses based on partial sequences of the L, M, or N gene resulted in essentially identical trees and revealed that four infected birds carried ABV strains of genotype 4, while two other birds carried strains of genotype 2 (Fig. 1; see also Fig. S1 in the supplemental material). A very close genetic relationship was detected between viruses from birds 7225 and 7308, which had lived in the same household as partner birds.
TABLE 1.
Detection of ABV nucleic acids by RT-PCR in organs from birds with PDD
| ABV infection group and bird no. | Species | Age (yr)/sexa | Detection of ABV nucleic acids inb:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | Crop | Esophagus | Proventriculus | Gizzard | Duodenum | Heart | Liver | Kidney | Lung | Spleen | Feces | |||
| Genotype 4 | ||||||||||||||
| 1440 | Amazona festiva | 7/M | ++++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++++ | +++ | ND | + |
| 6758 | Ara ararauna | 1/F | +++ | ++ | ND | ++ | ++ | ++ | + | + | + | + | ND | ND |
| 7225 | Cacatua galerita | Unknown/F | +++ | +++ | ++ | +++ | +++ | +++ | +++ | ++ | ++++ | +++ | ++ | ND |
| 7308 | Cacatua galerita | 6/M | ND | +++ | ND | ND | ND | ND | ND | ND | ND | ND | ND | + |
| Genotype 2 | ||||||||||||||
| 6609 | Amazona ventralis | 7/M | +++ | +++ | ND | ND | ++ | ND | ND | ND | ND | ND | ND | ND |
| 7316 | Cacatua alba | Unknown/F | +++ | +++ | +++ | + | +++ | +++ | ++ | +++ | +++ | +++ | +++ | ND |
M, male; F, female.
Birds infected with ABV genotype 4 were screened by real-time RT-PCR, and scoring in terms of threshold cycle values was as follows: +, 35 to 40; ++, 30 to 35; +++, 25 to 20; ++++, <20. Birds infected with ABV genotype 2 were analyzed by semiquantitative RT-PCR, and scoring was as follows: +, faint band visible on gel after 35 cycles; ++, moderately strong band visible on gel after 35 cycles; +++, very strong band visible on gels after 35 cycles. ND, not done due to lack of material.
FIG. 1.
Phylogenetic tree based on partial N gene sequences of avian bornaviruses. The tree was constructed using a distance method (neighbor joining) and with two BDV sequences as outgroups. Sequences of this study are boxed, and bootstrap values are given as percentages for the main nodes.
Abundant presence of ABV-specific nucleic acids in diverse organs of infected parrots.
One of the ABV-positive parrots described above was already dead when it arrived at the Clinic for Birds. Four of the remaining five birds either died soon after arrival at the clinic or were euthanized due to severe disease symptoms. Available organs of these animals were screened for the presence of ABV-derived nucleic acids by RT-PCR. High concentrations of viral RNA were present in most organs of the diseased birds including brain, crop, liver, kidney, heart, and lung (Table 1). Feces samples were available from two birds. They also contained readily detectable concentrations of ABV-specific nucleic acids (Table 1).
Isolation of ABV from organs of infected parrots using avian cell lines.
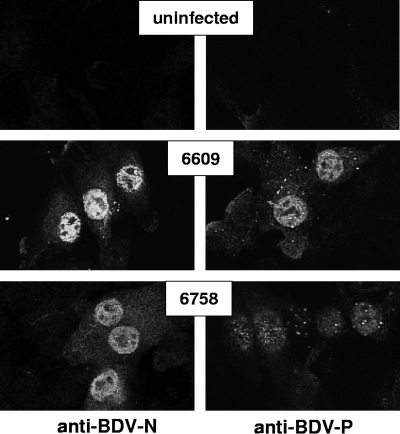
A crop sample of bird 6609 (containing ABV genotype 2) and a brain sample of bird 6758 (containing ABV genotype 4) were available for virus isolation experiments. Homogenates of these organs were added to various avian and mammalian cell cultures. At days 7, 10, and 14 postinfection, the cultures were inspected for the presence of ABV-infected cells by indirect immunofluorescence analysis using cross-reactive rabbit antiserum raised against either the N or the P protein of BDV. Many cells of the quail CEC32 cultures contained viral antigen even at very early times postexposure to organ extracts (Table 2). The N and P antigens of ABV strains 6609 and 6758 were present in both the cytoplasm and the nucleus of infected cells (Fig. 2). The isolated viruses spread fast through the CEC32 cultures and eventually reached all cells (Table 2). No obvious destruction of infected cells was observed, and the persistently infected cells grew at seemingly normal speed.
TABLE 2.
Virus isolation from organ extracts using avian and mammalian cell lines
| Sample source | Virus detection ina:
|
|||||
|---|---|---|---|---|---|---|
| Avian cells
|
Mammalian cells
|
|||||
| CEC32 (quail) | QM7 (quail) | LMH (chicken) | Vero (monkey) | MDCK (dog) | C6 (rat) | |
| Crop 6609b | ++ | + | − | − | − | − |
| Brain 6758 | +++ | ++ | + | − | − | − |
Cultures were analyzed for the presence of virus antigen-positive cells at 7, 10, and 14 days postinfection with samples of organ extracts. Scoring based on immunofluorescence assay (IFA) was as follows: +++, many IFA-positive cells on day 7 and rapid subsequent viral spread; ,++, few IFA-positive cells on day 7 and many positive cells at later analysis; +, no IFA-positive cells on day 7, but few positive cells on days 10 or 14; −, no IFA-positive cells on day 7, 10, or 14.
Titration experiments on CEC32 cells showed that the crop sample of animal 6609 contained about 100-fold less infectious virus than the brain sample of animal 6758.
FIG. 2.
Visualization of ABV in infected CEC32 cells. Cells persistently infected with either ABV strain 6609 or strain 6758 were stained for viral antigens using cross-reactive antibodies raised against the N and P antigens of BDV.
The efficacy of virus isolation was slightly reduced if the quail cell line QM7 was employed instead of CEC32. If the chicken cell line LMH was used, ABV strain 6758 but not strain 6609 could be isolated. The spread of virus 6758 in LMH cultures was much slower than in CEC32 or QM7 cultures (Table 2). All our attempts to isolate virus from parrot brain or crop samples were unsuccessful if Vero monkey kidney cells, C6 rat glioma cells, or MDCK canine kidney cells were employed. Since these cell types are routinely used for propagating BDV, our results indicate a high preference of ABV for cells of avian origin (Table 2).
Conserved features of the ABV genome.
The complete genome of ABV isolate 6609 (genotype 2) was determined and compared to available sequences of other ABV isolates. Although originating from a bird in Austria, the nucleotide sequence of isolate 6609 is 97% identical to the ABV genotype 2 strain bil (accession number EU781967) from a bird that lived in the United States (19). The proteins of the two viruses showed identities between 96.5% (X protein) and 100% (N protein). As predicted from the remote positions in the phylogenetic tree (Fig. 1), isolate 6609 is only 81.4% identical to the sequenced regions of genotype 3 strain 1367 (accession number FJ169440) and 78.8% identical to genotype 4 strain 1034_1322 (accession number FJ169441) (17).
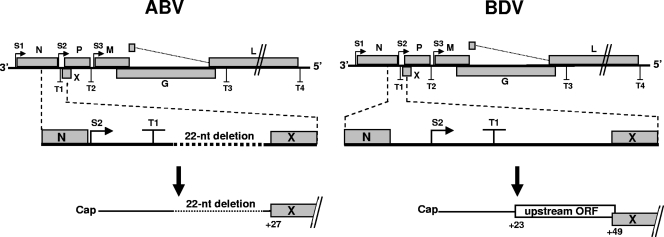
As noted previously (17, 19), ABV strongly resembles classical BDV in that the number and order of genes on the linear viral genome, the structure of transcription initiation and termination signals, and the location of splice donor and acceptor sites are highly conserved. However, we noted one remarkable difference between avian and mammalian bornaviruses. In ABV, the region between the N and X genes is shorter than in BDV (Fig. 3). As in BDV, the S2 start site for X/P mRNA synthesis is positioned upstream of the T1 termination signal, but unlike BDV, the S2 site in ABV is located immediately downstream of the stop codon of the N open reading frame (ORF). Furthermore, a 22-nucleotide fragment found between the T1 signal and the X ORF in BDV is missing in ABV. This deletion removes a short ORF located upstream of the X coding region in all known strains of BDV. This motif was demonstrated to serve regulatory functions (24, 25).
FIG. 3.
Bornaviruses of avian and mammalian origin seem to employ different strategies for controlling expression of the second viral transcription unit. Schematic drawing shows transcriptional start (S1 to S3) and stop (T1 to T4) sites in both viral genomes. The enlarged regions show the position of regulatory elements present between transcription units 1 and 2 in ABV and BDV. In all known ABV strains, the S2 site is located immediately downstream of the ORF for N. The ABV strains further lack a regulatory out-of-frame ORF located upstream of the X coding region. nt, nucleotide.
ABV replicates in neuronal and nonneuronal cells of parrot tissue.
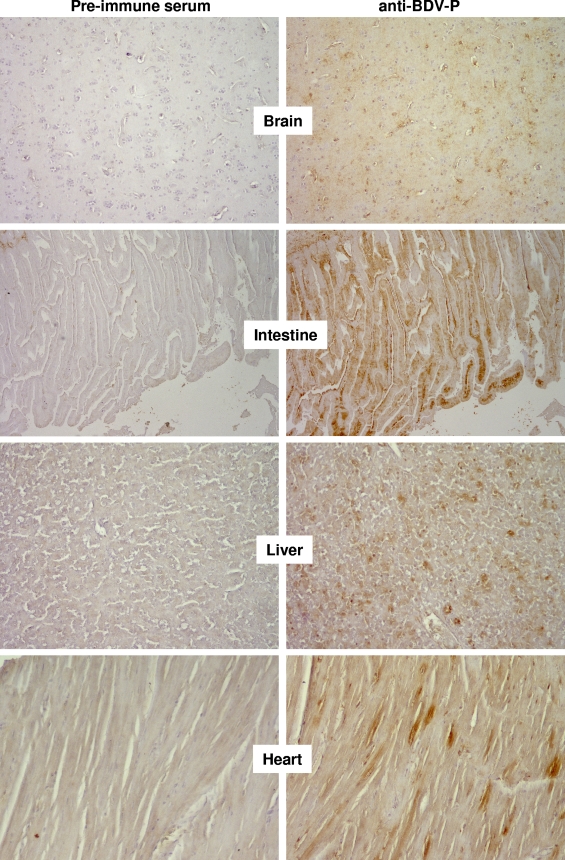
Formalin-fixed and paraffin-embedded samples from various tissues of parrots 6609 and 6758 were processed for immunohistochemical analyses. None of the available cross-reactive antisera detected antigen of ABV strain 6758 in tissue sections. However, a cross-reactive antiserum directed against the P protein of BDV recognized ABV strain 6609 with sufficient sensitivity and specificity. It strongly stained a large number of cells in the brain of diseased parrot 6609, while the preimmune serum did not (Fig. 4). The P-specific antiserum further strongly stained the villi of the intestine from the diseased animal. A close inspection revealed that epithelial cells as well as cells of the lamina propria contained viral antigen (Fig. 4). A large number of virus antigen-positive cells were also present in the liver of the diseased animal. Although the identity of these cells remains unclear at present, from their abundance and their location in the tissue it seems unlikely that they represent infected neurons (Fig. 4). The view that ABV can productively infect nonneuronal cells is supported by the viral antigen distribution pattern in the heart of parrot 6609. In this tissue, viral antigen was mainly present in myocytes (Fig. 4).
FIG. 4.
Detection of ABV antigens in various organs of a parrot with PDD. Consecutive sections of paraffin-embedded organs of diseased parrot 6609 were stained with either a rabbit antiserum against the P protein of BDV or preimmune serum, as indicated. Brown stain indicates specific staining of ABV antigens by the cross-reactive antiserum.
DISCUSSION
We identified ABV strains of genotypes 2 and 4 in parrots from central Europe that showed typical clinical signs of PDD, and we report the successful isolation of one representative member of each ABV genotype. Infection experiments with avian and mammalian cell cultures revealed a high preference of ABV for avian host cells. We observed that a surprisingly broad spectrum of organs and cell types of diseased birds contained replicating virus, demonstrating that ABV does not exhibit a high preference for cells of the central and peripheral nervous system like classical BDV in mammals.
As reported in a previous study (19), we also failed to detect ABV in 100% of birds with typical clinical signs of PDD. At present we cannot distinguish between the possibilities that (i) the clinical diagnosis of the negative birds was incorrect, (ii) other ABV genotypes exist that are not readily detected by the current diagnostic PCR assay, and (iii) unknown viral agents others than ABV cause a similar disease. In the six ABV-positive birds described in this study, we identified viruses of genotype 2 in two animals and genotype 4 in four others, indicating that these two genotypes represent the dominant ABV types in central Europe. However, the fact that we did not identify viruses of genotypes 1, 3, and 5 might be explained by a sampling error as the number of diseased birds in our study was rather small.
In comparing the viral genome structures, we found that the ABV and BDV genomes are highly similar, as reported previously (17, 19). However, we noted that the genomes differed with respect to recently discovered regulatory elements that determine the expression of virus-encoded regulatory factor X (24, 25). A critical sequence motif, namely a short ORF located in the 5′ noncoding region of the bicistronic mRNA for X and P, is missing in ABV (Fig. 3). This short upstream ORF in BDV appears to decrease the probability that X protein is translated from the bicistronic X/P mRNA, thus favoring expression of the P protein from the downstream ORF (24). In addition, a poorly defined sequence element located in the same region of the BDV genome seems to control the frequency by which the viral polymerase recognizes the transcriptional termination signal T1 (25). The absence of these regulatory elements indicates that ABV may use a different strategy to synthesize optimal amounts of X and P proteins from its bicistronic 0.8-kb mRNA.
Infection experiments showed that ABV has a high preference for avian cells in tissue culture. The quail cell line CEC32 turned out to be highly suitable for the isolation of ABV strains of genotypes 2 and 4 from clinical specimens. Virus isolation was also possible using other avian cell lines, but virus isolation was not successful if various mammalian cells lines were used. These results strongly suggest that ABV is a viral pathogen that selectively infects birds. It will be of great interest to determine in future experiments which bird species are susceptible to experimental infection with ABV and which species develop ABV-induced disease.
Although formal proof is lacking at present, our results collectively suggest that natural transmission of ABV may be from bird to bird and that such transmission may occur by the fecal-oronasal route. First, we noted a surprisingly high sequence similarity between ABV strains from the United States and from Europe, which argues against infection of parrots by an unknown resident reservoir species. If local virus reservoirs existed, birds from distinct regions should carry viruses with distinct signatures. The latter scenario may apply for classical BDV, where farm animals seem to get the virus from persistently infected rodents or insectivores (7, 8, 16, 27). Second, in our study we observed that viruses 7308 and 7225, which originated from two partner birds that lived in the same household, were almost identical. Third, our histological analysis revealed many ABV-infected cells in the villi of the intestines of a bird with PDD (Fig. 4), suggesting that infected cells and virus particles are shed from this organ and excreted by the feces. Supporting this view, we found that the feces of two birds with PDD from which samples were available did contain ABV as measured by RT-PCR. If our data interpretation is correct, transmission of ABV among parrots would resemble the transmission of avian influenza viruses among water fowl.
Our RT-PCR data and the results of the immunohistochemical analysis of organs from ABV-infected birds were in good agreement. They both showed that ABV is present in many if not all organs of birds with PDD. By contrast, BDV was reported to exhibit a high preference for the central nervous system in naturally infected horses and sheep as well as in experimentally infected laboratory animals. Thus, ABV and BDV exhibit strikingly different tissue distributions. Our immunohistological study also showed that ABV replication is not restricted to neuronal cells but, rather, takes place in myocytes and other as yet unidentified nonneuronal cell types in liver and gut. It should be noted that Hilbe and coworkers (16) reported the abundant presence of BDV antigen in myocytes of a wild shrew, the hypothetical natural reservoir of BDV. It is intriguing to speculate that neurotropism is not an intrinsic feature of BDV but rather represents a property that BDV has adopted in dead-end hosts that play no role in the ecology of this virus.
Supplementary Material
Acknowledgments
We thank Annette Ohnemus, Rosita Frank, Benjamin Meyer, and Reiner Schmider for excellent technical assistance and Kirsi S. Honkavuori and Thomas Briese for sharing positive control samples as well as sequences of primers and probes for real-time RT-PCR.
Footnotes
Published ahead of print on 18 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Berhane, Y., A. D. Smith, S. Newman, M. Taylor, E. Nagy, B. Binnington, and B. Hunter. 2001. Peripheral neuritis in psittacine birds with proventricular dilatation disease. Avian Pathol. 30653-670. [DOI] [PubMed] [Google Scholar]
- 2.Boutette, J. B., and M. Taylor. 2004. Proventricular dilation disease: a review of research, literature, species differences, diagnostics, prognosis, and treatment, p. 175-181. In Proceedings of the Association of Avian Veterinarians, New Orleans, LA.
- 3.Carbone, K. M., B. D. Trapp, J. W. Griffin, C. S. Duchala, and O. Narayan. 1989. Astrocytes and Schwann cells are virus-host cells in the nervous system of rats with Borna disease. J. Neuropathol. Exp. Neurol. 48631-644. [DOI] [PubMed] [Google Scholar]
- 4.Daoust, P. Y., R. J. Julian, C. V. Yason, and H. Artsob. 1991. Proventricular impaction associated with nonsuppurative encephalomyelitis and ganglioneuritis in two Canada geese. J. Wildl. Dis. 27513-517. [DOI] [PubMed] [Google Scholar]
- 5.de la Torre, J. C. 2002. Bornavirus and the brain. J. Infect. Dis. 186(Suppl. 2)S241-S247. [DOI] [PubMed] [Google Scholar]
- 6.Doneley, R. J., R. I. Miller, and T. E. Fanning. 2007. Proventricular dilatation disease: an emerging exotic disease of parrots in Australia. Aust. Vet. J. 85119-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durrwald, R., J. Kolodziejek, S. Herzog, and N. Nowotny. 2007. Meta-analysis of putative human bornavirus sequences fails to provide evidence implicating Borna disease virus in mental illness. Rev. Med. Virol. 17181-203. [DOI] [PubMed] [Google Scholar]
- 8.Durrwald, R., J. Kolodziejek, A. Muluneh, S. Herzog, and N. Nowotny. 2006. Epidemiological pattern of classical Borna disease and regional genetic clustering of Borna disease viruses point towards the existence of to-date unknown endemic reservoir host populations. Microbes Infect. 8917-929. [DOI] [PubMed] [Google Scholar]
- 9.Geib, T., C. Sauder, S. Venturelli, C. Hassler, P. Staeheli, and M. Schwemmle. 2003. Selective virus resistance conferred by expression of Borna disease virus nucleocapsid components. J. Virol. 774283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosztonyi, G., and H. Ludwig. 1995. Borna disease—neuropathology and pathogenesis. Curr. Top. Microbiol. Immunol. 19039-73. [PubMed] [Google Scholar]
- 11.Gregory, C. R., K. S. Latimer, R. P. Campagnoli, and B. W. Ritchie. 1996. Histologic evaluation of the crop for diagnosis of proventricular dilatation syndrome in psittacine birds. J. Vet. Diagn. Investig. 876-78. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, C. R., K. S. Latimer, F. Niagro, B. W. Ritchie, R. P. Campagnoli, T. M. Norton, and C. B. Greenacre. 1994. A review of proventricular dilatation syndrome. J. Assoc. Avian. Vet. 869-75. [Google Scholar]
- 13.Hallensleben, W., M. Schwemmle, J. Hausmann, L. Stitz, B. Volk, A. Pagenstecher, and P. Staeheli. 1998. Borna disease virus-induced neurological disorder in mice: infection of neonates results in immunopathology. J. Virol. 724379-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallensleben, W., and P. Staeheli. 1999. Inhibition of Borna disease virus multiplication by interferon: cell line differences in susceptibility. Arch. Virol. 1441209-1216. [DOI] [PubMed] [Google Scholar]
- 15.Herzog, S., and R. Rott. 1980. Replication of Borna disease virus in cell cultures. Med. Microbiol. Immunol. 168153-158. [DOI] [PubMed] [Google Scholar]
- 16.Hilbe, M., R. Herrsche, J. Kolodziejek, N. Nowotny, K. Zlinszky, and F. Ehrensperger. 2006. Shrews as reservoir hosts of Borna disease virus. Emerg. Infect. Dis. 12675-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honkavuori, K. S., H. L. Shivaprasad, B. L. Williams, P. L. Quan, M. Hornig, C. Street, G. Palacios, S. K. Hutchison, M. Franca, M. Egholm, T. Briese, and W. I. Lipkin. 2008. Novel Borna virus in psittacine birds with proventricular dilatation disease. Emerg. Infect. Dis. 141883-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper, D. C., C. Sauder, G. S. Scott, B. Dietzschold, and J. A. Richt. 2002. Immunopathology and immunoprotection in CNS virus infections: mechanisms of virus clearance from the CNS. Curr. Top. Microbiol. Immunol. 265163-182. [DOI] [PubMed] [Google Scholar]
- 19.Kistler, A. L., A. Gancz, S. Clubb, P. Skewes-Cox, K. Fischer, K. Sorber, C. Y. Chiu, A. Lublin, S. Mechani, Y. Farnoushi, A. Greninger, C. C. Wen, S. B. Karlene, D. Ganem, and J. L. DeRisi. 2008. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol. J. 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolodziejek, J., R. Durrwald, S. Herzog, F. Ehrensperger, H. Lussy, and N. Nowotny. 2005. Genetic clustering of Borna disease virus natural animal isolates, laboratory and vaccine strains strongly reflects their regional geographical origin. J. Gen. Virol. 86385-398. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig, H., L. Bode, and G. Gosztonyi. 1988. Borna disease: a persistent virus infection of the central nervous system. Prog. Med. Virol. 35107-151. [PubMed] [Google Scholar]
- 22.Mannl, A., H. Gerlach, and R. Leipold. 1987. Neuropathic gastric dilatation in psittaciformes. Avian Dis. 31214-221. [PubMed] [Google Scholar]
- 23.Pauli, G., and H. Ludwig. 1985. Increase of virus yields and releases of Borna disease virus from persistently infected cells. Virus Res. 229-33. [DOI] [PubMed] [Google Scholar]
- 24.Poenisch, M., P. Staeheli, and U. Schneider. 2008. Viral accessory protein X stimulates the assembly of functional Borna disease virus polymerase complexes. J. Gen. Virol. 891442-1445. [DOI] [PubMed] [Google Scholar]
- 25.Poenisch, M., S. Wille, P. Staeheli, and U. Schneider. 2008. Polymerase read-through at the first transcription termination site contributes to regulation of Borna disease virus gene expression. J. Virol. 829537-9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richt, J. A., L. Stitz, H. Wekerle, and R. Rott. 1989. Borna disease, a progressive meningoencephalomyelitis as a model for CD4+ T-cell-mediated immunopathology in the brain. J. Exp. Med. 1701045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staeheli, P., C. Sauder, J. Hausmann, F. Ehrensperger, and M. Schwemmle. 2000. Epidemiology of Borna disease virus. J. Gen. Virol. 812123-2135. [DOI] [PubMed] [Google Scholar]
- 28.Stitz, L., T. Bilzer, and O. Planz. 2002. The immunopathogenesis of Borna disease virus infection. Front. Biosci. 7d541-555. [DOI] [PubMed] [Google Scholar]
- 29.Stitz, L., T. Bilzer, J. A. Richt, and R. Rott. 1993. Pathogenesis of Borna disease. Arch. Virol. Suppl. 7135-151. [DOI] [PubMed] [Google Scholar]
- 30.Tomonaga, K., T. Kobayashi, and K. Ikuta. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect. 4491-500. [DOI] [PubMed] [Google Scholar]
- 31.Zoller, B., I. Redman-Muller, I. Nanda, M. Guttenbach, E. Dosch, M. Schmid, R. Zoorob, and C. Jungwirth. 2000. Sequence comparison of avian interferon regulatory factors and identification of the avian CEC-32 cell as a quail cell line. J. Interferon Cytokine Res. 20711-717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.