Abstract
The propagation of autonomous parvoviruses is strongly dependent on the phosphorylation of the major nonstructural protein NS1 by members of the protein kinase C (PKC) family. Minute virus of mice (MVM) replication is accompanied by changes in the overall phosphorylation pattern of NS1, which is newly modified at consensus PKC sites. These changes result, at least in part, from the ability of MVM to modulate the PDK-1/PKC pathway, leading to activation and redistribution of both PDK-1 and PKCη. We show that proteins of the ezrin-radixin-moesin (ERM) family are essential for virus propagation and spreading through their functions as adaptors for PKCη. MVM infection led to redistribution of radixin and moesin in the cell, resulting in increased colocalization of these proteins with PKCη. Radixin was found to control the PKCη-driven phosphorylation of NS1 and newly synthesized capsids in vivo. Conversely, radixin phosphorylation and activation were driven by the NS1/CKIIα complex. Altogether, these data argue for ERM proteins being both targets and modulators of parvovirus infection.
Autonomous parvoviruses are small, icosahedric, nonenveloped particles with a 5.1-kb single-stranded linear DNA as a genome. This DNA encodes, besides two capsid proteins, at least four nonstructural proteins, of which only the large 83-kDa polypeptide NS1 is essential for progeny particle production in all cell types (for a review, see reference 8). NS1 is a multifunctional polypeptide with endonuclease and directional helicase activities that are important to drive rolling-circle-like replication of the viral DNA (5, 28). In addition to these enzymatic functions, NS1 serves as a transcription factor for the regulation of viral, as well as cellular, promoters (19). NS1 also interferes with the host cell physiology and morphology through its ability to directly interact with and/or induce posttranslational modifications in specific host cell proteins (3, 31-33). This involvement of NS1 in multiple facets of the parvoviral life cycle, in conjunction with host cell factories/proteins, implies that the various functions of the viral proteins are tightly regulated, in particular, through phosphorylation and subcellular (re)distribution (25).
In keeping with this view, NS1 becomes differentially phosphorylated during infection (7, 9). So far, we have identified two members of the protein kinase C (PKC) family, PKCλ and PKCη, as being able to activate NS1 for viral DNA amplification (10, 18, 26, 30). Interestingly, mutagenesis at consensus PKC phosphorylation sites dissociated NS1 functions necessary for virus amplifications, interactions with cellular proteins, and induction of host cell perturbations, death, and lysis (6, 9, 10, 26).
The activity of PKCs is also tightly regulated by phosphorylation (23). During activation, PKCs undergo a variety of conformational changes that allow their interaction with small ligands or regulatory proteins and facilitate translocation of the kinase to specific compartments in which substrates become available. In the course of this sequence of activation steps, PKCs undergo changes in their affinities for cellular scaffold and membrane structures (17). ERM (ezrin [Ez], radixin [Rdx], moesin [Moe]) family proteins are known to be mediators between cellular scaffold (actin) and membrane structures (14) and by analogy have also been proposed to serve as anchoring proteins for PKC (15). This is in agreement with reports showing an interaction of PKCα with ezrin in vivo (24) and with the identification of ERM proteins as anchoring molecules for cyclic AMP-dependent kinase (11).
ERM proteins contain an N-terminal FERM (4.1-ezrin- radixin-moesin) domain that binds to phosphatidylinositol(4,5)phosphate and cellular membrane proteins (37). Similarly to PKC, ERM proteins become phosphorylated at their conserved C-terminal threonines upon ligand binding, thereby undergoing conformational alterations. This leads to exposure of the actin binding site at the C terminus and allows ERM binding to the cellular cytoskeleton (35). The C-terminal phosphorylation of ERM proteins can be achieved by a variety of candidate kinases in vitro. Additional regulatory functions have been described for conserved tyrosine and S/T residues located in the N-terminal FERM and the central α-helical domains of ERM proteins (14).
During purification of NS1-activating kinases, we consistently found ERM proteins as “contaminants” in preparations of PKCη derived from HeLa cells. For the above-mentioned reasons, we hypothesized that these proteins might control PKC(η) in vivo through their adaptor function. To test this possibility, functional knockouts of Ez, Rdx, and Moe were designed and tested for their effects on minute virus of mice (MVM) DNA replication, NS1 phosphorylation, capsid phosphorylation, and virus spread. In addition, since ERM proteins themselves are subject to regulation, we analyzed the impact of MVM infection on ERM modification, intracellular distribution, activity, and colocalization with PKCη. The present results indicate that Rdx modulates the PKCη-driven phosphorylation, not only of NS1, but also of capsids in vivo, correlating with the stimulation of virus DNA replication and spreading, respectively. Furthermore, we show that Rdx is also a substrate for in vivo and in vitro phosphorylation by the recently described NS1/CKIIα complex (32). This regulation may contribute to retarget Rdx, thereby switching Rdx-dependent phosphorylation from an early (viral DNA replication) to a late (spreading) event of the MVM life cycle.
MATERIALS AND METHODS
Antibodies and reagents.
Antibodies specific for Ez (H-276), Rdx (C-15), and PKCη (C-15) were purchased from Santa Cruz; antibody for actin (C-4) from MP-Biomedicals; antibody for tubulin α (B-5-1-2) and Flag tag (M2) from Sigma; and antibody for BrdU from Biogenesis. Rdx P564 phosphospecific antibodies were kindly provided by S. Tsukita (37). The rabbit antisera αNS1C and αVP2 were generated using specific peptides from corresponding proteins (4). Monoclonal anti-capsid antibodies (B7) were kindly provided by J. M. Almendral. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse antibodies were purchased from Promega. Fluorescent-dye-labeled secondary antibodies were from Dianova and Alexis and protein G-Sepharose beads from Pharmacia Amersham. Specific short interfering RNAs (siRNAs) to knock down expression of ERM proteins (E1, MmVil2.1, no. si00185416; E2, MmVil2.2, no. si00185423; R1, Mm Rx5, no. si02668603; R2, MmRx6, no. si02688434; M1, MmMsn3, no. si0131945; M2, MmMsn4, no. si01319052) and control oligonucleotides (AllStarsNeg siRNA, no. 1027281) were purchased from Qiagen. Recombinant CKIIαβ was obtained from Roche.
Cells and viruses.
A9 fibroblasts and derivatives thereof were maintained as monolayers in Dulbecco's modified Eagle medium containing 10% fetal calf serum (FCS). HeLa-S3 cells were grown as suspension cultures in minimal Eagle's medium containing 5% FCS. MVM was propagated in adherent A9 cells, and virus stocks were prepared from infected cultures by repeated freezing and thawing in 10 mM Tris, pH 8.7, 1 mM EDTA. When indicated, full (DNA-containing) MVM particles were separated from empty capsids, on the basis of their buoyant density, by CsCl gradient centrifugation.
Plasmid constructs. (i) Isolation of mouse Ez, Rdx, and Moe cDNAs.
A9 cDNA libraries were generated from mRNA preparations using a Smart PCR cDNA synthesis kit. Full-length cDNAs were isolated essentially as described previously (18) in a single PCR from the mouse fibroblast library using the following primers corresponding to the published mouse sequences: Ez (NCBI NM 009510), N-terminal (5′-ATGCCCAAGCCAATCAACGTCCGGGTGACCACCATGGAT-3′) and C-terminal (5′-CTACATGGCCTCGAACTCGTCAATGCGTTGCTTGGTGTTG-3′); Rdx (NCBI NM 009041), N-terminal (5′-ATGCCGAAGCCAATCAATGTAAGAGTAACTACAATGGACG-3′) and C-terminal (5′-TCACATGGCTTCAAACTCATCGATGCGCTGCTTCGTATTG-3′); and Moe (NCBI NM 010833), N-terminal (5′-ATGGATGCAGAGCTGGAGTTTGC-3′) and C-terminal (5′-CTACATGGACTCAAACTCAT-3′). PCR fragments of the appropriate lengths were isolated from agarose gels, cloned directly into pCR2.1 (Invitrogen), and subjected to sequencing (Microsynth GmbH, Balgach, Switzerland).
(ii) Production of Flag-tagged wild-type or mutant ERM clones.
Site-directed mutagenesis was performed by single or chimeric PCR (33). PCRs were performed with an N-terminal primer (consisting of a unique restriction site generating blunt ends, followed by the Flag tag sequence and the first 40 nucleotides of Ez, Rdx, or Moe) and a C-terminal reverse primer (consisting of a unique NotI site followed by the respective coding sequences). Mutations altering the ERM C-terminal threonine phosphorylation sites in the actin-binding domain were performed by single PCR using C-terminal primers harboring the appropriate mutations, followed by ∼15 wild-type nucleotides: EzT566A, 5′-…CAGCGCCTT…-3′, and EzT566E, 5′-…CAGCTCCTT…-3′; RdxT564A, 5′-… AAGCCCCTT…-3′, and RdxT564E, 5′-…AAGGTCCTT…-3′; MoeT547A, 5′-…CAGGGCCTT…-3′, and MoeT547E, 5′-…CAGCTCCTT…-3′. Internal substitutions and deletions were performed by chimeric PCR using two complementary internal primers harboring the mutations: RdxY146F, 5′-AGTCTGTCATTAGCCAGAAAGCCTGGTTTTCTGGCTAATG-3′ and 5′-CATTAGCCAGAAAACCAGGCTTTCTGGCTAATGACAGACT-3′; Rdxdl[P], 5′-TAAAAACTGTGATGTCTGCGdlGTCATCCTCCAACGGAGAA-3′ and 5′-TTCTCCGTTGGAGGATAGACdlCGCAGACATCACAGTTTTTA-3′, where mutated codons are indicated by italics, with the changed nucleotides in boldface; deletions are indicated by dl; ellipses indicate variable numbers of wild-type nucleotides which are not shown in the sequence. The Flag-tagged mutants were subcloned into pCR2.1, sequenced, and transferred as PmeI/NotI restriction fragments into HpaI/NotI-cleaved pP38 (18), yielding pP38-FlagEzT566A, pP38-FlagEzT566E, pP38-Flag-RdxT564A, pP38-FlagRdxT564E, pP38FlagRdxY146F, pP38-FlagRdxdl[P], pP38-FlagMoeT547A, and pP38-FlagMoeT547E, respectively.
Expression constructs for the bacterial production of recombinant proteins.
The sequence encoding Rdx was transferred after SmaI and HindIII digestion in frame into SmaI- and HindIII-cleaved pQE-32 (Qiagen), generating pQE-Rdx.
Purification of HA-2 fractions by affinity column chromatography.
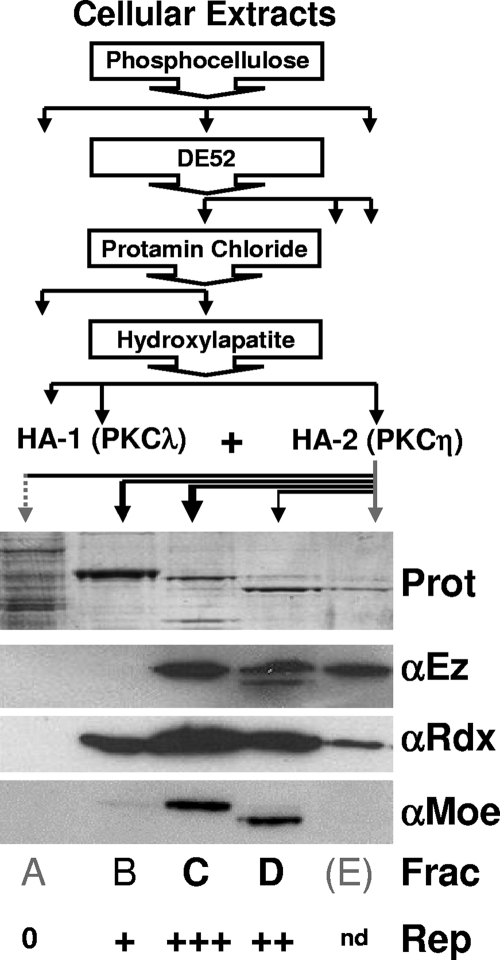
The purification and identification of NS1-activating PKCλ and PKCη from HeLa cellular extracts by consecutive column chromatography (phosphocellulose, DE52, protamine chloride, and hydroxyl apatite [HA]) have been described previously (18, 26). The presence of ERM proteins at the final purification step (i.e., in the PKCη-containing, NS1-activating fractions from HA-2) was determined by analyzing HA-bound proteins eluting at 116 mM (Fig. 1A), 164 mM (Fig. 1B), 212 mM (Fig. 1C), 260 mM (Fig. 1D), and 500 mM (Fig. 1E) KPO4, pH 7.5.
FIG. 1.
Detection of ERM proteins in cell fractions activating dephosphorylated NS1 for viral DNA amplification. (Top) Purification scheme used to identify PKCλ/PKCη as NS1-activating protein kinases to drive viral DNA amplification (18, 26). The presence of PKCλ and PKCη in low-affinity (HA-1) and high-affinity (HA-2) fractions from HA chromatography, respectively, is indicated. (Bottom) Analysis of HA-2 fractions by Coomassie blue staining (Prot) and Western blot detection of Ez, Rdx, and Moe. Activation of dephosphorylated NS1 (NS1O) in the presence of recombinant PKCλ was achieved to a significant extent using HA-2 fractions C (+++) and D (++), while only minor (+) or no (0) replication activity (Rep) was measured with fractions B and A, respectively. nd, not determined.
Knockdown of ERM expression using siRNA treatment.
A9 cells grown on spot slides were transfected with 300 ng of the respective siRNA with 1 μl Lipofectamine 2000 (Gibco BRL). After 24 h, the treated cells were infected with 30 PFU/cell CsCl-purified MVM and analyzed for the presence of Ez, Rdx, Moe, and NS1 by immunofluorescence. Viral DNA replication was measured upon addition of 1 μM bromodeoxyuridine (BrdU) for 2 h, followed by immunofluorescence determination of BrdU incorporation using specific antibodies (18). Statistical evaluation was performed based on three individual experiments, each analyzing >200 cells.
Generation of stably transfected A9 cell lines.
Stable transfectants were generated with pP38-Flag ERM (using the above-mentioned mutants of Ez, Rdx, and Moe) and the selection plasmid pSV2neo at a molar ratio of 25:1 (18). Colonies were pooled after growth under selection, and frozen stocks were prepared. Experiments were performed in the absence of G418. Transfectants were kept in culture for less than 25 passages. Use was also made of previously established A9 cell lines stably containing P38-FlagPKCηT512A (18), P38-FlagCKII:E81A, or P38-FlagCKIImATP (32).
Biochemical fractionation of cell extracts.
The association of proteins with cellular scaffolds and membrane structures was determined as previously described (31). Briefly, extracts were prepared, and the insoluble material was separated from the soluble fraction by low-speed centrifugation. (i) The insoluble pellet was extracted with 1% Triton X-100 and separated by centrifugation into insoluble scaffold proteins (iS fraction; pellet) and nuclear-membrane-associated proteins (nM fraction; supernatant). (ii) The soluble components were further fractionated by high-speed centrifugation, yielding the cytosolic constituents (C fraction) in the supernatant and a pellet that was extracted with Triton X-100 and further separated by centrifugation into soluble scaffold proteins (sS fraction) and postnuclear membrane proteins (pM fraction). All volumes were adjusted to the equivalent original cell numbers, making it possible to compare the relative amounts and distributions of selected proteins in differently treated cells.
Immunofluorescence microscopy.
Cells were grown on spot slides (Roth), mock or MVM infected, and further incubated for the appropriate times. The cultures were fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. Specimens were preadsorbed with 20% FCS, incubated with primary antibodies, and stained with specific Alexa Fluor 594-, CY2-, CY3-, or rhodamine-conjugated anti-species antibodies. After being mounted with Elvanol, the cells were analyzed by laser scanning microscopy with a Leica DMIRBE apparatus (63× lens; red laser, 543 nm; green laser, 488 nm) and Powerscan software or by spinning-disk confocal microscopy with a Perkin-Elmer ERS 6Line microscope (100× lens; red laser, 568 nm; green laser, 488 nm) presenting a single slice of a stack. Quantitative analyses were performed on all slices of a stack, and mean colocalizations were calculated with ImageJ software (4).
Western blotting analyses.
Protein extracts were fractionated by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto nitrocellulose membranes. Proteins of interest were detected by incubation with appropriate primary antibodies in 10% dry milk/phosphate-buffered saline or with phosphospecific antibodies in 2% bovine serum albumin, 10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 for 18 h and staining with horseradish peroxidase-conjugated secondary antibodies for 1 h, followed by chemiluminescence detection (Amersham).
MVM DNA replication in infected cells.
Accumulation of MVM DNA was determined by Southern blotting (7). Cells were harvested in Tris-EDTA buffer and digested with proteinase K, and total DNA was sheared by passage through a syringe. Viral DNA was analyzed by agarose gel electrophoresis and detected, after being blotted onto nitrocellulose membranes, with a 32P-labeled probe corresponding to nucleotides 385 to 1885 of the NS1-encoding region of MVM DNA.
Metabolic labeling, purification, and phosphopeptide analyses.
Metabolic labeling and tryptic phosphopeptide analyses were essentially performed as previously described (27). A9 cell cultures were infected with MVMp, kept for 24 h before being incubated for 4 h in labeling medium (complete medium lacking phosphate and supplemented with [32P]orthophosphate). The labeled cells were harvested directly in 800 μl of immunoprecipitation buffer (20 mM HEPES-KOH, pH 7.5, 300 mM NaCl, 1 mM EDTA, 0.2% NP-40). Immunoprecipitations were carried out using 10 μl of antiserum (NS1, αNS1C; capsids, αVP2+B7) and protein G-Sepharose. 32P-labeled proteins were separated by 10% SDS-PAGE and blotted on polyvinylidene difluoride membranes, and the bands corresponding to NS1 or VP protein were excised. Membrane-bound proteins were digested with trypsin and analyzed by two-dimensional thin-layer electrophoresis/chromatography.
In vitro kinase reactions.
In vitro kinase reactions and tryptic phosphopeptide analyses were performed as described previously (4) using recombinant CKIIαβ (Roche) and, when indicated, purified glutathione S-transferase-tagged wild-type NS1 protein (33). Rdx used as substrates was produced in bacteria and purified as described previously (32). Assays were performed for 40 min at 37°C with 30 μCi [γ-32P]ATP in 50 μl of 20 mM HEPES-KOH, pH 7.5, 7 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol. The reactions were stopped, and the reaction products were analyzed after immunoprecipitation by 10% SDS-PAGE and semidry transfer onto polyvinylidene difluoride membranes (Millipore). The phospholabeled proteins were then digested with trypsin and analyzed by two-dimensional thin-layer electrophoresis/chromatography (electrophoresis at pH 1.9/phosphochromatography).
Plaque assays.
To determine the formation and release of progeny virions, A9 cultures or derivatives thereof were infected with serial dilutions of CsCl-purified MVMp. The number and morphology of the lysis plaques were determined as described previously (9). Briefly, cell monolayer cultures were seeded at a concentration of 105 cells per 60-mm2 dish, infected 24 h later, and covered with a Bacto agar overlay. After incubation for 6 days, the cultures were stained for 18 h by addition of neutral-red-containing Bacto agar. The stained cells were fixed on the plates with formaldehyde after the agar overlay was removed.
RESULTS
Rdx is required for MVM DNA replication.
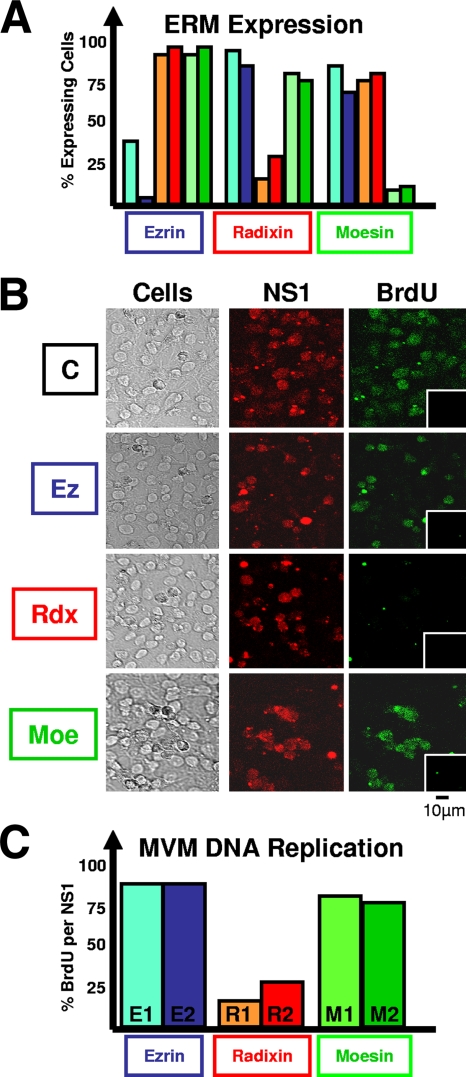
Reconstitution experiments have identified PKCλ (10) and PKCη (18) as being sufficient to activate dephosphorylated NS1 polypeptides for MVM DNA amplification in a kinase-free in vitro replication system (29). As illustrated in Fig. 1, ERM proteins were consistently found to become copurified with NS1-activating PKCη during chromatographic fractionation. To investigate whether these cellular proteins, known for their mediator functions, play a role during parvovirus replication, we performed functional knockdown experiments using siRNAs specific for Ez, Rdx, or Moe. As determined by confocal laser scanning microscopy after immunolabeling with appropriate antibodies, transfection of A9 cultures with selected siRNAs led to a significant reduction of the fraction of cells expressing the targeted ERM protein without affecting the levels of the other proteins (Fig. 2A). Single cells were then analyzed for the ability to sustain viral DNA replication by measuring BrdU incorporation 24 h after infection with MVM (Fig. 2B and C). While MVM replication in cells lacking Ez or Moe was similar to that observed in the scrambled siRNA-treated control, knockdown of Rdx led to a significant reduction of BrdU incorporation into viral DNA, suggesting a specific requirement for the protein in viral DNA amplification in A9 cells.
FIG. 2.
MVM replication after depletion of individual ERM proteins by siRNA treatment. A9 cells grown on spot slides were transfected with siRNAs designed to specifically target mRNAs of Ez, Rdx, and Moe. Treated and mock-treated cells were infected (or not) after 24 h with CsCl-purified MVM (30 PFU/cell), fixed 24 h p.i., and analyzed by confocal laser scanning microscopy. (A) Knockdown of ERM protein production through siRNA treatment. Individual ERM proteins were detected after being labeled with specific antisera. The numbers of fluorescent cells are expressed relative to mock-treated (control oligonucleotide) cultures. (B and C) Impact of siRNA treatment on MVM DNA replication. MVM infection was revealed through NS1 expression and detected by αNS1C antiserum. Viral DNA amplification was detected by measuring BrdU incorporation for 2 h under conditions in which cellular DNA was not labeled (insets, noninfected cell populations). (C) Statistical evaluation of the proportion of MVM-infected (NS1-positive) cells sustaining viral DNA amplification (BrdU incorporation), as determined from three independent experiments (>200 cells each). Blue, E1 and E2, treatment with siRNA specific for Ez; red, R1 and R2, treatment with Rdx siRNAs; green, M1 and M2, Moe.
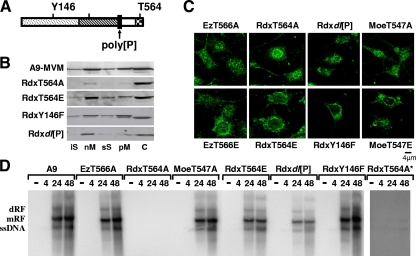
These data raised the possibility that Rdx is an essential component in the DNA replication complex and/or acts as a mediator for phosphorylation and activation of the viral replicator protein NS1. This was tested by generating stably transfected cell lines in which the activities of ERM proteins were selectively modulated in the presence of NS1, due to overexpression of (dominant) negative mutants under the control of the parvoviral P38 promoter (18). Figure 3A depicts the domain structure of Rdx (14) with the motifs that were targeted for mutagenesis in our approach highlighted: (i) the conserved threonine in the actin binding domain (Ez, T566; Rdx, T564; Moe, T547), (ii) the conserved tyrosine phosphorylation site at position Y146 in the FERM domain, and (iii) the polyproline region, which is rather heterogeneous among ERM proteins. Phosphorylation of the conserved C-terminal threonine controls the overall activity of ERM proteins through conformational alterations of the polypeptide (35). This threonine was mutated to either alanine or glutamic acid in all three polypeptides. While replacement of the (phospho)threonine by inert alanine should give rise to a dominant-negative phenotype, the glutamic acid substitution at this position was expected to generate a constitutively active protein. Substitution of Y146F and the deletion of the polyproline region were sought to generate an intermediate inhibitory effect on endogenous Rdx, due to a partial functional knockout and/or aberrant subcellular distribution caused by this mutagenesis. This was confirmed by fractionation experiments monitoring endogenous Rdx in MVM-infected cell lines (Fig. 3B). As previously reported for active ERM proteins (35), besides its soluble cytosolic (C) form, endogenous Rdx was found to be associated with membranes (nM and pM) and scaffold structures (iS and sS) in both MVM-infected parental A9 cells and derivatives expressing RdxT564E. In contrast, expression of the inactive RdxT564A mutant led to reduction of membrane-associated Rdx and to an almost complete disappearance of scaffold-associated Rdx. Intermediate phenotypes were observed upon expression of RdxY146F or Rdxdl[P]. While only a little reduction of “active” Rdx was caused by RdxY146F, Rdxdl[P] exerted a strong dominant-negative effect, leaving only a low proportion of “active” Rdx associated with membrane/scaffold structures.
FIG. 3.
MVM replication in cell lines expressing ERM variant proteins. (A) Schematic representation showing the various domains of ERM proteins: N-ERMAD/FERM (dotted); α-helical (hatched); polyproline (poly[P]), aa 470 to 477 (black); C-ERMAD (white); and F-actin binding (checkered). The amino acids targeted for mutagenesis in Rdx are indicated. The threonine residues T564 (Rdx), T566 (Ez), and T547 (Moe) are involved in ERM protein activation through phosphorylation. (B) Impact of mutant Rdx expression on the subcellular distribution of endogenous Rdx. A9 cells or derivatives expressing RdxT564A (dominant negative), RdxT564E (constitutively active), RdxY146F, or Rdxdl[P] under the control of the NS1-induced P38 promoter were infected with MVM (30 PFU/cell). Cells were harvested 24 h p.i., and the affinity of endogenous Rdx for cellular membranes and scaffold structures was measured through biochemical fractionation. The distribution of Rdx between the individual fractions was determined by Western blotting using Rdx-specific antiserum. (C) Subcellular localization of ERM mutant proteins in MVM-infected cells. A9 cells or derivatives thereof expressing ERM variants were infected with MVM (30 PFU/cell) and fixed 24 h p.i. Recombinant proteins were specifically detected by means of their Flag epitopes using αM2 antibodies and were analyzed by confocal laser scanning microscopy. Endogenous proteins are not recognized by αM2 (data not shown). (D) MVM replication in the presence of ERM variants. A9 cells or derivatives thereof were infected with MVM (30 PFU/cell) and harvested at the indicated times p.i. (in hours, shown above the lanes). Monomeric and dimeric RF (mRF and dRF) and ssDNA were detected by Southern blotting using a 32P-labeled probe corresponding to the NS1 coding region. RdxT564A* shows a prolonged exposure.
The expression and subcellular localization of the variant proteins were first examined by confocal laser scanning microscopy using monoclonal αM2 antibodies, specifically recognizing the recombinant proteins due to their N-terminal Flag epitopes. As shown in Fig. 3C, all variant proteins were expressed to comparable levels upon MVM infection, while no signal was detected in the parental A9 cells (data not shown). Interestingly, while active polypeptides seemed to accumulate in the perinuclear area with only minute amounts entering the nuclear compartment, the inactive polypeptides were found in both nuclear and cytoplasmic compartments. We then tested these cell lines for the ability to sustain viral DNA amplification and production of progeny virions, as evidenced by the accumulation of replicative forms (RF) and the displacement of progeny single-stranded genomes, respectively. A9 cells or derivatives thereof expressing variant ERM proteins were infected with MVMp, harvested at the indicated times postinfection (p.i.), and analyzed for the production of monomeric and dimeric RF, as well as single-stranded virion DNA (ssDNA), by Southern blotting (Fig. 3D). As expected from the BrdU incorporation experiments with cells depleted of ERM proteins (cf. Fig. 2), inactivation of endogenous Ez and Moe only marginally affected the production of RF and ssDNA. In contrast, expression of RdxT564A dramatically impaired viral DNA synthesis, which could hardly be detected after prolonged exposure. Partial inhibition of viral DNA replication was also seen upon expression of Rdxdl[P], while the RdxY146F mutant failed to interfere with endogenous Rdx-dependent MVM DNA production. Unexpectedly, overexpression of constitutively active RdxT564E did not further promote viral DNA amplification, which underwent, instead, a slight but significant reduction. This inhibition may possibly result from side effects of disregulated Rdx activation, e.g., on cell survival.
Rdx mediates phosphorylation of NS1 by PKCη.
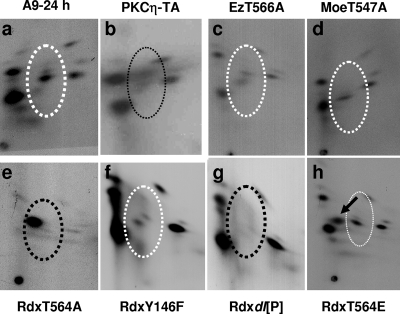
Parvoviral DNA amplification is strongly dependent on specific phosphorylation of NS1 (18, 29, 30). As mentioned above, ERM proteins ended up in PKCη preparations during purification, raising the possibility of ERM also becoming associated with and controlling the activity of PKCη in the cellular context. To investigate this possibility, we first compared the phosphorylation patterns of NS1 extracted from parental A9 cells and derivatives that either expressed mutant ERM proteins or were impaired for PKCη activity. A9 and A9:P38-PKCηT512A, -EzT566A, -MoeT547A, -RdxT564A, -RdxY146F, -Rdxdl[P], and -RdxT564E were infected with MVMp and metabolically labeled with [32P]orthophosphate at 24 h p.i. Radiolabeled NS1 was then isolated from cellular extracts by immunoprecipitation, purified by SDS-PAGE, and subjected to tryptic phosphopeptide analysis. As illustrated in Fig. 4a and b, presenting wild-type A9 versus A9 with inactivated PKCη, and in agreement with our previous finding (18), PKCη drove the phosphorylation of several peptides migrating in the center of the NS1 phosphopeptide map, characteristic of the replicative phase of infection (7). Upon expression of EzT566A, MoeT547A, and RdxY146F, no significant differences in the NS1 phosphopeptide patterns were seen compared to A9 cells (Fig. 4c, d, and f versus a), in keeping with wild-type levels of MVM DNA amplification (Fig. 3D). In contrast, there were significant differences in NS1 phosphorylation upon expression of the Rdx mutants RdxT564A and Rdxdl[P], correlating with an impairment of viral DNA synthesis in these cell lines (Fig. 3D). In A9-RdxT564A cells (Fig. 4e), NS1 phosphorylation was almost completely inhibited, being restricted to a single prominent phosphopeptide that most likely does not represent a genuine NS1 phosphorylation site and may reflect aberrant processing of the protein. On the other hand, the selective loss of the PKCη-specific phosphopeptides in Rdx[P] cells is more relevant, as all other NS1 phosphorylations were retained (Fig. 4g). The loss of this specific subset of phosphopeptides points to the involvement of Rdx in the control of PKCη-driven phosphorylation. Interestingly, expression of constitutively active RdxT564E led to the generation of a new NS1 phosphopeptide (Fig. 4h). In agreement with previously published data, this phosphopeptide may reveal a “late” phosphorylation event taking place during the postreplicative phase of MVM infection. This observation suggests that ERM activation may contribute to the alteration of the NS1 phosphorylation pattern, which is known to occur in the course of a parvovirus infection (7, 9).
FIG. 4.
Impacts of ERM proteins on NS1 phosphorylation. A9 cells or derivatives thereof were infected with MVM (30 PFU/cell), metabolically labeled 24 h p.i., and processed further for determination of the NS1 phosphorylation pattern by two-dimensional tryptic phosphopeptide analysis. The dotted ovals mark peptides whose phosphorylation is PKCη dependent. White ovals, phosphorylation pattern of NS1 produced in parental A9 cells; black ovals, patterns lacking specific phosphopeptides; black arrow, additional phosphopeptide. PKCη-TA, PKCηT512A.
MVM infection induces distribution of ERM proteins.
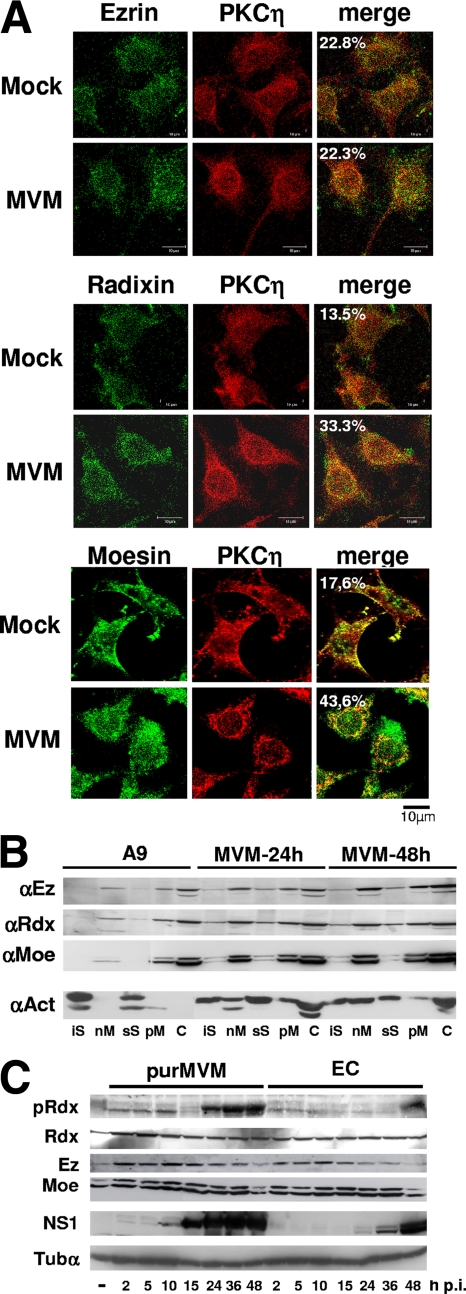
The above observations indicate that ERM proteins, and in particular Rdx, participate in the interplay between parvoviral NS proteins and regulation of PKCs. Ongoing MVM infection is associated with changes in the subcellular distribution of both the NS (25) and PKC (17) proteins, raising the question of whether ERM trafficking may also become altered during the viral life cycle. The recruitment of ERM proteins to distinct cell compartments was investigated in MVM- versus mock-infected A9 cells using confocal laser scanning microscopy, biochemical fractionation (for the measurement of associations with cellular scaffold and membrane structures), and Western blotting (for the detection of proteins activated through specific phosphorylation). As illustrated in Fig. 5A, the distributions of ERM and PKCη were visualized by double-immunofluorescence labeling of mock- and MVM-infected A9 cells at 24 h p.i. Few changes in Ez localization were observed between mock-treated and MVM-infected A9 cells. In contrast, as previously reported for PKCη (17), MVM induced redistribution of Rdx and Moe, which accumulated in the perinuclear area, showing a strong colocalization with PKCη.
FIG. 5.
Impact of MVM infection on the subcellular distribution of ERM proteins. (A) A9 cells grown on spot slides were infected (or not [Mock]) with CsCl-purified MVM (30 PFU/cell) and analyzed for the localization of individual ERMs (green) and PKCη (red) by confocal laser scanning microscopy at 24 h p.i. Colocalization appears yellow in the merge and was quantified using Image J software. (B) A9 cells were infected (or not [A9]) with MVM (30 PFU/cell) and harvested at the indicated times p.i. Association of ERMs with scaffold and membrane structures was determined by fractionating cell extracts and measuring the protein contents of individual fractions by Western blotting using specific antisera. Actin served as a control. (C) Asynchronously grown A9 cells were infected (or not) with CsCl-purified MVM (30 PFU/cell) or an equivalent amount of empty capsids (EC). Total cellular extracts were prepared at the indicated times p.i. and analyzed for their indicated protein contents by Western blotting. pRdx denotes the Rdx species activated through phosphorylation at residue T564 and detected by means of phosphospecific antibodies (37). α-Tubulin (Tubα) served as a loading control.
As previously reported (35), and modulated through the expression of dominant-negative versus constitutively active Rdx mutants (Fig. 3B), ERM proteins show affinity for (cellular) scaffold and membrane structures. To test whether MVM infection interferes with the ability of ERM proteins to bind their ligands and partner proteins, we performed biochemical fractionation of MVM- versus mock-treated A9 cell extracts. The proportions of ERM proteins associated with cellular scaffold structures (iS and sS) and membranes (nM and pM), relative to the soluble cytosolic fraction (C), were determined by Western blotting using actin as a loading control. As shown in Fig. 5B, MVM infection resulted in an increase in both the total amounts of all three ERM species and their binding to membranes and cellular scaffold structures. Besides confirming the ERM redistribution induced by MVM, these results argued for a (in)direct effect of viral products on the properties of ERM proteins.
To further substantiate this hypothesis, we measured the activity of Rdx, as evidenced by its threonine 564 phosphorylation (35), in time course experiments using a phosphorylation-specific antibody (37). As shown in Fig. 5C, the amount of phosphorylated Rdx increased early after infection, indicating that the protein became activated upon MVM infection. Surprisingly, this activation was biphasic, with a more slowly migrating phosphorylated species becoming prominent from 16 h p.i. onward. In contrast, after an early increase at 2 h p.i., the total amount of Rdx remained constant over the time course. Therefore the late phosphorylation-dependent activation and further modification of Rdx were likely induced by viral products. The viral NS1 protein was a good candidate for triggering these effects, since its production started slightly before and persisted during the second wave of Rdx activation. Furthermore, infection of A9 cells with empty capsids resulted in an initial increase in the (activated) Rdx level but failed to induce major T564 phosphorylation-dependent activation of Rdx at 16 to 48 h p.i. The marked activation and further modification of Rdx at late stages of infection led us to speculate that, aside from facilitating viral DNA amplification (see above), ERM proteins may control subsequent steps in the MVM life cycle, such as packaging of ssDNA, virus egress, or induction of cell death and lysis.
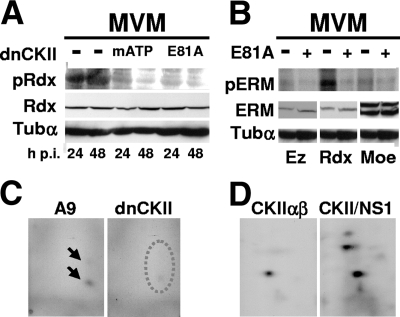
Rdx is phosphorylated by the NS1/CKIIα complex in vivo and in vitro.
It was recently shown that NS1 is able to physically interact with CKIIα (32), thereby modulating the substrate specificity of the cellular kinase (33). To investigate whether this complex of a parvoviral protein with a cellular protein kinase could mediate the activation of ERM proteins during MVM infection of A9 cells, we made use of A9 derivative cell lines expressing dominant-negative mutant forms of CKIIα under the control of the NS1-inducible P38 promoter. To this end, MVM-infected A9 cells and derivatives containing P38-CKIImATP or P38-CKII-E81A, were analyzed at the indicated times p.i. for (i) Rdx phosphorylation at T564 (Western blotting) and (ii) overall ERM protein phosphorylation (metabolic 32P labeling followed by tryptic phosphopeptide analyses). As shown in Fig. 6A, Rdx-pT564 was readily detected in A9 cells at 24 and 48 h p.i. In contrast, the species was hardly detectable in the derivative cell lines expressing the dominant-negative CKII mutants mATP and E81A, while the total amount of Rdx protein was not reduced in comparison with control cells. Likewise, as was apparent from incorporated [32P]orthophosphate, the overall phosphorylation of Rdx was much reduced in the presence of dominant-negative CKII (Fig. 6B). This result was confirmed by 2D tryptic phosphopeptide analysis of Rdx in which only one of the two resolved phosphopeptides was detected in samples from dominant-negative CKII-expressing cells (Fig. 6C). No differences could be seen for Ez and Moe, whose overall phosphorylation was reduced to the level of that of Rdx in the absence of CKII activity (Fig. 6B). Therefore, these two polypeptides did not appear to be targets for NS1/CKIIα in vivo. To confirm that NS1/CKIIα were able, on the other hand, to target Rdx for phosphorylation, we subjected recombinant, bacterially expressed Rdx to in vitro phosphorylation by either CKIIαβ alone or NS1/CKIIα. As shown in Fig. 6D, in the presence of wild-type NS1, Rdx became strongly phosphorylated at two additional locations compared to Rdx treated with CKIIαβ alone, becoming resolved in three distinct phosphopeptides. These data argue for the ability of the NS1/CKIIα complex to directly phosphorylate and eventually modulate Rdx in MVM-infected A9 cells.
FIG. 6.
Phosphorylation and activation of Rdx by the NS1/CKIIα complex. (A to C) A9 cells or derivatives thereof expressing dominant-negative CKIIα (dnCKII) under the control of the NS1-inducible P38 promoter were infected with MVM (30 PFU/cell) and analyzed for Rdx phosphorylation. (A) At the indicated times, cellular extracts were prepared and analyzed by Western blotting for the amounts of total Rdx (Rdx) or Rdx activated through phosphorylation at residue T564 (pRdx). α-Tubulin (Tubα) served as a loading control. Dominant-negative CKIIα was mutated either in the ATP-binding pocket (mATP) or at the conserved glutamic acid in position 81 (E81A). (B) Metabolic 32P labeling was performed at 24 h p.i. in A9 cells expressing (+) or not expressing (−) CKIIαE81A. Individual 32P-labeled ERM proteins were isolated by immunoprecipitation and SDS-PAGE (pERM). The total amounts of the respective proteins in cellular extracts were determined by Western blotting (ERM). α-Tubulin served as a loading control. (C) Tryptic phosphopeptide analysis of 32P-labeled Rdx produced in the presence (A9) or absence (dnCKII) of active CKII. The arrows indicate labeled peptides that are underrepresented in the absence of active CKII (dotted oval). (D) Tryptic phosphopeptide analysis of Rdx phosphorylated in vitro with either CKIIαβ or CKIIαβ supplemented with wild-type glutathione S-transferase-NS1 (CKII/NS1).
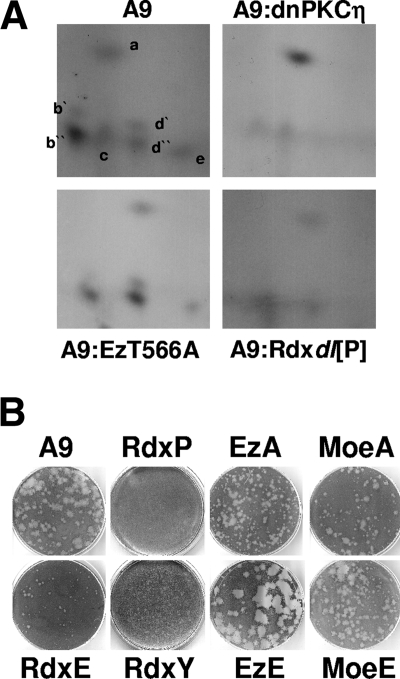
Impacts of ERM proteins on MVM maturation and spread.
The virus-induced modulation of Rdx properties and, more particularly, the biphasic activation of this polypeptide during MVM infection of A9 cells suggested that ERM proteins may have an additional function(s) besides regulation of viral DNA amplification. As a modulator of PKCη-driven phosphorylation, Rdx may conceivably affect the modification of parvoviral capsids. This possibility was tested by metabolic 32P labeling and tryptic phosphopeptide analyses of MVM capsids. To this end, A9 cells or derivatives expressing dominant-negative PKCηT512A, Rdxdl[P], or EzT566A were infected with MVMp and metabolically labeled at 24 h p.i. MVM virions were isolated by immunoprecipitations, and 32P-labeled VP2 was purified by SDS-PAGE. Tryptic phosphopeptide analyses were then performed as previously described for NS1. Despite the reduction of overall production and phosphorylation of MVM virions in the cell lines expressing dominant-negative PKCη (PKCηT512A) (18) or Rdx (Rdxdl[P]) (Fig. 3D), it is possible to monitor qualitative differences in the two-dimensional phosphopeptide patterns derived from different cell lines by comparing the relative intensity of the individual spots to each other. As shown in Fig. 7A, trypsin-digested VP2 derived from virions produced in either EzT566A (which was shown in our preceding experiments to have no impact on MVM propagation) or A9 cells resolved into five to seven distinct phosphopeptides. The phosphorylation of five of these peptides (b′, b″, d′, d″, and e) appeared to depend on PKCη activity, since it was drastically reduced in the presence of PKCηT512A. In addition, four of these peptides (b′, d′, d″, and e) were also missing in the phosphopeptide pattern of Rdxdl[P]-treated cells, further supporting the role of a functional Rdx/PKCη complex in the phosphorylation of parvoviral proteins.
FIG. 7.
Impacts of ERM proteins on late stages of MVM infection. (A) A9 cells or derivatives thereof expressing dominant-negative PKCηT512A (A9:dnPKCη), dominant-negative Ez (A9:EzT566A), or dominant-negative Rdx (A9:Rdxdl[P]) were infected with MVM (30 PFU/cell) and 32P labeled with orthophosphate at 24 h p.i. Newly synthesized capsids were isolated by immunoprecipitation with αB7/αVP2 to enrich for DNA-containing virions and analyzed for the tryptic phosphopeptide pattern of VP2. (B) Impacts of ERM proteins on the capacity of MVM to form lysis plaques in parental A9 cells (A9) or derivatives expressing variant ERM proteins: Rdxdl[P] (RdxP), RdxT564E (RdxE), RdxY146F (RdxY), EzT566A (EzA), EzT566E (EzE), MoeT547A (MoeA), and MoeT547E (MoeE). No lysis plaques were detected in the presence of dominant-negative Rdxdl[P], while RdxY147F reduced plaque numbers 50-fold; in the remaining cell lines, plaque numbers were comparable to those of parental A9 cells.
Although little is known about the impact of capsid phosphorylation on virus egress and spreading, this result prompted us to investigate whether ERM proteins are involved in the control of second-round infections. To this end, cell lines expressing variant ERM proteins were compared to parental A9 cells for the ability to sustain plaque formation after MVM inoculation. Readouts consisted of plaque numbers and morphology. As shown in Fig. 7B, Rdx activity had a major effect on the development of MVM lysis plaques. No plaques could be seen after inoculation of ERM-deficient A9:P38-Rdxdl[P] cells with MVMp. The cell line expressing RdxY146F allowed the formation of pinpoint plaques at a only 50-fold-reduced efficiency compared to parental A9 cells. While overexpression of constitutively active RdxT564E resulted in a normal number but much reduced size of plaques, activated EzT566E promoted virus spread, giving rise to a large-plaque phenotype. No significant effects were seen with constitutively active MoeT547E and dominant-negative EzT566A or MoeT547A, although plaque sizes were somewhat reduced in cultures expressing the last mutant compared to those in A9 cells. Altogether, these results indicated that, besides controlling viral DNA amplification in vivo, ERM proteins exerted additional functions involved in virus maturation and/or spreading.
DISCUSSION
The multiple functions of the MVM nonstructural protein NS1 are tightly regulated by its expression, modification, and subcellular localization (25). In the present study, we showed that the ERM family of proteins and, more particularly, Rdx are essential for both the replication and spreading of parvoviruses. This could be traced back to a new function of ERM proteins, in addition to their known roles as mediators between the actin cytoskeleton and cellular membranes. Besides their general impact on viral fitness (as is apparent from MVM's ability to form lysis plaques), Rdx, a member of this protein family, was found to modulate PKCη activity in A9 cells, resulting in changes in the phosphorylation patterns of both the replicator viral protein NS1 and progeny virions. Interestingly, this modulator is itself subject to a feedback control consisting of phosphorylation by the NS1/CKIIα complex.
Since the PKC canonical recognition motif is rather nonspecific (22), it was proposed that adaptor proteins might serve to target these kinases selectively to their substrates (15, 21). Increasing numbers of PKC interaction partners have been identified. In keeping with this view, it was shown that atypical PKCs achieve their regulatory functions in a complex with multiple cellular proteins (34). In this study, we showed that the family of ERM proteins is involved in the modulation of PKCη activity in the cellular context. Upon MVM infection, Rdx not only is modified through phosphorylation, but also colocalizes with PKCη, correlating with the PKCη-driven phosphorylation of both NS1 and progeny virions. Considering that ERM proteins function as mediators between the (actin) cytoskeleton and membrane structures (35), this finding is not surprising, since activation of PKCs is associated with changes in their affinity from cellular scaffold to membrane association (17).
NS1 was shown to be regulated in its various functions during productive virus infection by selected phosphorylation events (6, 10, 27). Indeed, some of the changes in the NS1 phosphorylation pattern observed during infection (7) are thought to contribute to the dissociation of NS1 functions necessary for progeny particle production from activities involved in cytolysis and virus release (9). The current finding that Rdx is involved in PKCη-driven phosphorylations of viral proteins provides a possible clue to the mechanism of NS1 modulation during infection. As ongoing parvovirus infection has an impact on ERM protein modification and colocalization with PKCη, ERM proteins may be instrumental in regulating the substrate targeting or specificity of PKCη, resulting more particularly in changes in the phosphorylation patterns of viral proteins. This would be in agreement with the reported control of ERM proteins over PKCη-driven phosphorylation of both NS1 and VP proteins.
PKCη was found to participate in multiple processes involved in cell proliferation (2, 12), differentiation (16), survival, and death (1). It was shown that the activity of PKCη is regulated by phosphorylation/dephosphorylation events, interaction with small molecules (diacylglycerol and acid lipids), and ubiquitin-dependent degradation (13). The present study revealed an additional mode of PKCη regulation, through direct or indirect interaction with ERM proteins. In addition to targeting PKCη to a distinct microenvironment in the cell, this interaction may contribute to the activation and/or substrate specificity of the kinase. This possibility is supported by the known function of ERMs as adaptor proteins between the actin cytoskeleton and membrane structures (35). On one hand, such interaction between ERMs and PKCs might induce conformational changes to the kinase necessary for the activation process (13). On the other hand, local changes in the substrate recognition site of PKCs might influence the substrate specificity. It should be stated in this regard that ERMs are a family of similar but functionally distinct proteins. This diversity further increases the potential of these adaptor proteins for fine tuning of cellular kinases.
ERM proteins, and particularly Rdx, are subject to regulation by posttranslational modifications (14, 35). This was substantiated by the present study, which showed that Rdx is a substrate of cellular kinases in MVM-infected cells, resulting in Rdx (and consequently PKCη [17]) activation. Moreover, our data indicate that the main phosphorylation of Rdx at later stages of infection depends on newly synthesized parvoviral proteins, particularly arguing for the involvement of the recently described NS1/CKIIα complex (32). The time lag between NS1-dependent MVM DNA amplification and the major part of Rdx activation strongly suggests a contribution of Rdx to postreplicative steps of the viral life cycle. Although an effect of ERM proteins on the assembly and/or maturation of infectious virions cannot be ruled out, it is tempting to speculate that Rdx/PKCη activity may control virus egress and/or cytolysis at the end of infection. Capsid phosphorylation at the VP2 N terminus has been implicated in controlling virus egress (20), a possibility that is facilitated by cellular vesicles shuttling newly produced virions to the cellular periphery (4). It is worth noting in this respect that ERM proteins, such as Ez, have been shown to be involved in vesicle transport (36) and that MVM capsid phosphorylation was found in the present study to depend on both PKCη and Rdx. The possible role of ERM proteins in virion release was further supported by the present demonstration of a marked impact of ERM activity on the capacity of MVM-infected cells to sustain the formation of lysis plaques. Alternatively, Rdx-driven PKCη activity might also be involved in NS1-mediated induction of cytopathic effects leading to cell lysis.
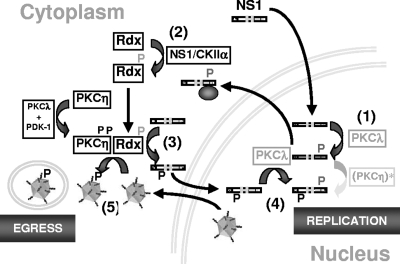
Figure 8 depicts a hypothetical model of the involvement of ERM proteins in the MVM replication cycle and of the regulation of these proteins through NS1/CKIIα according to our current knowledge. At early stages of infection, newly synthesized NS1 is phosphorylated in the nucleus by PKCλ at residues T435 and S473 (26), two modifications necessary for helicase function (10) and interaction with the catalytic subunit of CKII, CKIIα (32). In order to promote viral DNA amplification, NS1 requires additional modifications driven by the novel PKCη (18). Early during infection, active PKCη is found in limited amounts in the nucleus (17) to drive the initial replication steps. As reflected by the lag period observed in the beginning of the MVM infection cycle, this PKCη-driven phosphorylation of NS1 might, however, be rather inefficient. Upon synthesis of viral (NS) proteins, PKCη translocates toward the perinuclear area and becomes increasingly activated through phosphoinositol-dependent kinase 1 (PDK-1) in a PKCλ-dependent manner (17). This boost of PKCη activation correlates with the activation and translocation of Rdx to the perinuclear area, where it colocalizes with PKCη (Fig. 5). This MVM-induced activation of Rdx is dependent on active CKIIα and appears to be mediated through phosphorylation by the NS1/CKIIα complex (Fig. 6). Cooperation of Rdx with PKCη seems to be essential for efficient phosphorylation of both NS1 (on its way to the nucleus in order to drive viral DNA replication) and newly synthesized MVM virions (whose active export from the nucleus to the outside of cells is phosphorylation dependent [20] and vesicle associated [4]) (Fig. 7A). Additional roles may be played by ERM family proteins (e.g., Rdx or Moe) in the loading of newly synthesized virions into vesicles, thereby controlling virus egress and spread (Fig. 7B). Our current investigations aim to determine the molecular mechanisms underlying the modulation of parvovirus propagation by ERM proteins and the control of PKCη activity by these proteins in more detail.
FIG. 8.
Schematic presentation of the impacts of ERM proteins on the MVM replication cycle and of their regulation through NS1/CKIIα. Newly synthesized NS1 is transported to the nucleus, where it becomes phosphorylated by PKCλ (step 1), activating the viral polypeptide for helicase function and making it able to physically interact with CKIIα (step 2). In addition to PKCλ, NS1 requires PKCη-driven phosphorylations to promote viral DNA amplification. These modifications may occur rather inefficiently at early stages due to the presence of active PKCη in the nucleus. Upon synthesis of viral proteins, PKCη becomes activated through PKCλ and PDK-1 and translocates to the perinuclear region, colocalizing with activated Rdx, which might serve as an adaptor protein for PKCη. In MVM-infected cells, Rdx activation is dependent on CKII and is mediated through the NS1/CKIIα complex (step 2). This association of Rdx and PKCη in the perinuclear area leads to efficient phosphorylation and activation of newly synthesized NS1 on its way to the nucleus (step 3), where NS1 is also phosphorylated by PKCλ and becomes competent to drive viral DNA replication (step 4). The PKCη/Rdx connection also appears to be important for the phosphorylation of progeny virions during their transport from the nucleus to the cell periphery (step 5).
Acknowledgments
Special thanks are due to Claudia Plotzky for excellent technical assistance. We are also grateful to Michèle Vogel, Nathalie Salomé, José Almendral, and Sachiko Tsukita for providing us with monoclonal antibodies and antisera. Spinning-disc laser confocal microscopy was performed at the Nikon Imaging Center in Heidelberg, Germany (Bioquant BQ 0004, INF 267, and D-69120 Heidelberg).
Séverine Bär was supported by von Humboldt Foundation and EMBO fellowships.
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Abu-Ghanem, S., G. Oberkovitz, D. Benharroch, J. Gopas, and E. Livneh. 2007. PKCη expression contributes to the resistance of Hodgkin's lymphoma cell lines to apoptosis. Cancer Biol. Ther. 61375-1380. [DOI] [PubMed] [Google Scholar]
- 2.Aeder, S. E., P. M. Martin, J. W. Soh, and I. M. Hussaini. 2004. PKC-eta mediates glioblastoma cell proliferation through the Akt and mTOR signaling pathways. Oncogene 239062-9069. [DOI] [PubMed] [Google Scholar]
- 3.Anouja, F., R. Wattiez, S. Mousset, and P. Caillet-Fauquet. 1997. The cytotoxicity of the parvovirus minute virus of mice nonstructural protein NS1 is related to changes in the synthesis and phosphorylation of cell proteins. J. Virol. 714671-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar, S., L. Daeffler, J. Rommelaere, and J. P. Nuesch. 2008. Vesicular egress of non-enveloped lytic parvoviruses depends on gelsolin functioning. PLoS Pathog. 4e1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 766518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbau, R., V. Duverger, J. Rommelaere, and J. P. Nuesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278151-167. [DOI] [PubMed] [Google Scholar]
- 7.Corbau, R., N. Salom, J. Rommelaere, and J. P. Nuesch. 1999. Phosphorylation of the viral nonstructural protein NS1 during MVMp infection of A9 cells. Virology 259402-415. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 3391-174. [DOI] [PubMed] [Google Scholar]
- 9.Daeffler, L., R. Horlein, J. Rommelaere, and J. P. Nuesch. 2003. Modulation of minute virus of mice cytotoxic activities through site-directed mutagenesis within the NS coding region. J. Virol. 7712466-12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dettwiler, S., J. Rommelaere, and J. P. Nuesch. 1999. DNA unwinding functions of minute virus of mice NS1 protein are modulated specifically by the lambda isoform of protein kinase C. J. Virol. 737410-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dransfield, D. T., A. J. Bradford, J. Smith, M. Martin, C. Roy, P. H. Mangeat, and J. R. Goldenring. 1997. Ezrin is a cyclic AMP-dependent protein kinase anchoring protein. EMBO J. 1635-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fima, E., M. Shtutman, P. Libros, A. Missel, G. Shahaf, G. Kahana, and E. Livneh. 2001. PKCη enhances cell cycle progression, the expression of G1 cyclins and p21 in MCF-7 cells. Oncogene 206794-6804. [DOI] [PubMed] [Google Scholar]
- 13.Gould, C. M., and A. C. Newton. 2008. The life and death of protein kinase C. Curr. Drug Targets 9614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeflich, K. P., and M. Ikura. 2004. Radixin: cytoskeletal adopter and signaling protein. Int. J. Biochem. Cell Biol. 362131-2136. [DOI] [PubMed] [Google Scholar]
- 15.Jaken, S., and P. J. Parker. 2000. Protein kinase C binding partners. Bioessays 22245-254. [DOI] [PubMed] [Google Scholar]
- 16.Kashiwagi, M., M. Ohba, K. Chida, and T. Kuroki. 2002. Protein kinase C eta (PKC eta): its involvement in keratinocyte differentiation. J. Biochem. 132853-857. [DOI] [PubMed] [Google Scholar]
- 17.Lachmann, S., S. Bar, J. Rommelaere, and J. P. Nuesch. 2008. Parvovirus interference with intracellular signalling: mechanism of PKCη activation in MVM-infected A9 fibroblasts. Cell Microbiol. 10755-769. [DOI] [PubMed] [Google Scholar]
- 18.Lachmann, S., J. Rommeleare, and J. P. Nuesch. 2003. Novel PKCη is required to activate replicative functions of the major nonstructural protein NS1 of minute virus of mice. J. Virol. 778048-8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Legendre, D., and J. Rommelaere. 1994. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J. Virol. 687974-7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maroto, B., J. C. Ramirez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 7410892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mochly-Rosen, D., and A. S. Gordon. 1998. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1235-42. [PubMed] [Google Scholar]
- 22.Newton, A. C. 1997. Regulation of protein kinase C. Curr. Opin. Cell Biol. 9161-167. [DOI] [PubMed] [Google Scholar]
- 23.Newton, A. C. 2003. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem. J. 370361-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng, T., M. Parsons, W. E. Hughes, J. Monypenny, D. Zicha, A. Gautreau, M. Arpin, S. Gschmeissner, P. J. Verveer, P. I. Bastiaens, and P. J. Parker. 2001. Ezrin is a downstream effector of trafficking PKC-integrin complexes involved in the control of cell motility. EMBO J. 202723-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nüesch, J. 2006. Regulation of non-structural protein functions by differential synthesis, modification and trafficking, p. 275-286. In C. S. Bloom, R. M. Linden, C. R. Parrish, and J. R. Kerr (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 26.Nuesch, J. P., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 755730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuesch, J. P., R. Corbau, P. Tattersall, and J. Rommelaere. 1998. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J. Virol. 728002-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nuesch, J. P., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209122-135. [DOI] [PubMed] [Google Scholar]
- 29.Nuesch, J. P., S. Dettwiler, R. Corbau, and J. Rommelaere. 1998. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J. Virol. 729966-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuesch, J. P., S. Lachmann, R. Corbau, and J. Rommelaere. 2003. Regulation of minute virus of mice NS1 replicative functions by atypical PKCλ in vivo. J. Virol. 77433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuesch, J. P., S. Lachmann, and J. Rommelaere. 2005. Selective alterations of the host cell architecture upon infection with parvovirus minute virus of mice. Virology 331159-174. [DOI] [PubMed] [Google Scholar]
- 32.Nuesch, J. P., and J. Rommelaere. 2006. NS1 interaction with CKII alpha: novel protein complex mediating parvovirus-induced cytotoxicity. J. Virol. 804729-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nuesch, J. P., and J. Rommelaere. 2007. A viral adaptor protein modulating casein kinase II activity induces cytopathic effects in permissive cells. Proc. Natl. Acad. Sci. USA 10412482-12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno, S. 2001. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 13641-648. [DOI] [PubMed] [Google Scholar]
- 35.Simons, P. C., S. F. Pietromonaco, D. Reczek, A. Bretscher, and L. Elias. 1998. C-terminal threonine phosphorylation activates ERM proteins to link the cell's cortical lipid bilayer to the cytoskeleton. Biochem. Biophys. Res. Commun. 253561-565. [DOI] [PubMed] [Google Scholar]
- 36.Stanasila, L., L. Abuin, D. Diviani, and S. Cotecchia. 2006. Ezrin directly interacts with the alpha1b-adrenergic receptor and plays a role in receptor recycling. J. Biol. Chem. 2814354-4363. [DOI] [PubMed] [Google Scholar]
- 37.Tsukita, S., and S. Yonemura. 1999. Cortical actin organization: lessons from ERM proteins. J. Biol. Chem. 27434507-34510. [DOI] [PubMed] [Google Scholar]