Abstract
Nidoviruses (arteriviruses, coronaviruses, and roniviruses) are a phylogenetically compact but diverse group of positive-strand RNA viruses that includes important human and animal pathogens. Nidovirus RNA synthesis is mediated by a cytoplasmic membrane-associated replication/transcription complex that includes up to 16 viral nonstructural proteins (nsps), which carry common enzymatic activities, like the viral RNA polymerase, but also unusual and poorly understood RNA-processing functions. Of these, a conserved endoribonuclease (NendoU) is a major genetic marker that is unique to nidoviruses. NendoU activity was previously verified in vitro for the coronavirus nsp15, but not for any of its distantly related orthologs from other nidovirus lineages, like the arterivirus nsp11. Here, we show that the bacterially expressed nsp11 proteins of two arteriviruses, equine arteritis virus and porcine respiratory and reproductive syndrome virus, possess pyrimidine-specific endoribonuclease activity. RNA cleavage was independent of divalent cations in vitro and was greatly reduced by replacement of residues previously implicated in catalysis. Comparative characterization of the NendoU activity in arteriviruses and severe acute respiratory syndrome coronavirus revealed common and distinct features of their substrate requirements and reaction mechanism. Our data provide the first biochemical evidence of endoribonuclease activity associated with arterivirus nsp11 and support the conclusion that this remarkable RNA-processing enzyme, whose substrate in the infected cell remains to be identified, distinguishes nidoviruses from all other RNA viruses.
The replication of single-stranded RNA viruses with genomes of positive polarity (+RNA) relies on a process of RNA-templated RNA synthesis that takes place in the cytoplasm of the infected cell. Following translation of their genomes, +RNA viruses establish membrane-associated replication/transcription complexes that direct amplification of the viral genome and, in several cases, transcription of additional viral mRNAs. The key replication/transcription complex component in these processes is a virus-encoded RNA-dependent RNA polymerase (RdRp), but in addition, +RNA viruses generally encode multiple protein factors that support RdRp activity and/or are involved in the spatial and temporal regulation of viral RNA synthesis. Many of these enzymes were initially identified by comparative sequence analysis, subsequently supported by biochemical and molecular biological experiments. In nidoviruses, arguably the genetically most complex +RNA viruses (12), examples of such accessory enzymes are a second, low-fidelity RdRp (15), as well as an unusual set of “RNA-processing enzymes” (33), which are all genetically segregated in the viral replicase gene and have been implicated in various steps of the nidovirus life cycle (8, 16, 21, 24).
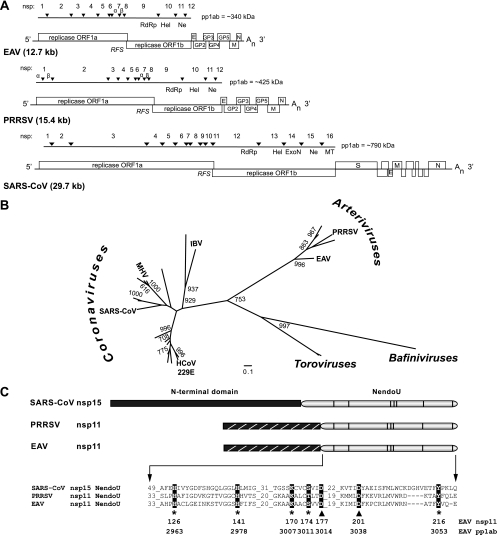
The order Nidovirales comprises the distantly related families Arteriviridae, Coronaviridae (genera Coronavirus and Torovirus), and Roniviridae and includes the virus groups with the largest RNA genomes known to date (12). The host range of Nidovirales extends from crustaceans to mammals, and the order includes pathogens associated with human respiratory infections, with diseases that can vary from the common cold to fatal pneumonia. Despite major differences in virion size and morphology, as well as pathogenesis, the evolutionary relationship among nidoviruses is evident from the organization and composition of the large replicase gene, which consists of the two 5′-most open reading frames (ORFs), ORF1a and ORF1b, of the polycistronic genome (Fig. 1A). Translation of the 5′-capped and 3′-polyadenylated nidovirus genomic RNA results in two large precursor polyproteins (pp1a and pp1ab) that are cleaved into 13 to 16 nonstructural proteins (nsps) by virus-encoded proteases. The relative abundances of nidovirus nsps are intricately regulated by the proteolytic processing of the pp1a and pp1ab precursors (references 35 and 39 and references therein), as well as the efficiency of the −1 ribosomal frameshift required for the translation of ORF1b (6). The nsps direct the formation of a membrane-anchored replication/transcription complex that mediates viral genome replication, as well as the synthesis of a 3′-coterminal nested set of subgenomic (sg) mRNAs (23, 31), from which the viral genes located downstream of the replicase gene are expressed.
FIG. 1.
Conservation of the NendoU endoribonuclease across the order Nidovirales. (A) Genome organization of the arteriviruses EAV and PRRSV, as well as SARS-CoV. The ORFs, as well as the ORF1a/1b ribosomal frameshift site (RFS) and the 3′poly(A) tail (An), are indicated. The ORFs encoding the viral structural proteins envelope (E), membrane (M), nucleocapsid (N), and glycoprotein 2 (GP2) to GP5 for EAV and PRRSV and E, M, N, and spike (S) for SARS-CoV are designated. The organization of the respective replicase gene products is depicted in the form of pp1ab. The arrowheads represent sites cleaved by virus-encoded proteases, and the resulting nsps are numbered. The locations of the viral RdRp, helicase (Hel), ExoN, NendoU (Ne), and ribose-2′-O-methyltransferase (MT) domains (see the text) are indicated. Note that the SARS-CoV genome is drawn to a different scale than the EAV and PRRSV genomes. (B) Phylogenetic tree of the NendoU domains of arteriviruses, coronaviruses, toroviruses, and bafiniviruses. For clarity, species names are given only for coronaviruses, for which NendoU activity was previously documented (HCoV 229E, human coronavirus 229E; SARS-CoV; MHV; IBV, infectious bronchitis virus), as well as the arteriviruses whose NendoU activities were analyzed in this study (EAV and PRRSV). The sequence alignment used for construction of the tree, as well as the complete version of the tree, is included as supplemental material (see Fig. S1 and S2 in the supplemental material). The numbers at branching points indicate the support of at least 600 out of 1,000 bootstraps, and the branch lengths of the tree represent the average numbers of substitutions per residue. (C) Domain organization of SARS-CoV nsp15, PRRSV nsp11, and EAV nsp11. Note the difference in size between the respective N-terminal domains. The homologous EAV and PRRSV nsp11 N-terminal domains (Gorbalenya, unpublished) are hatched. The partial sequence alignment shows the most conserved regions in the NendoU domains of the three viral proteins used in this study and was extracted from the alignment presented in Fig. S1 in the supplemental material. For EAV, the amino acid numbers given reflect the positions in EAV nsp11 (top row; used in this study) or in replicase polyprotein pp1ab (bottom row; used in previous studies). Conserved residues mutated in this study are highlighted. The asterisks indicate residues in EAV nsp11 NendoU that were replaced with Ala. The asterisks indicate proposed active-site residues, and nidovirus-wide conserved aspartic acid residues are indicated with triangles.
The core enzymatic activities commonly required for viral RNA synthesis, such as the viral RdRp and helicase functions, are encoded by ORF1b of the nidovirus genome, together with an additional conserved domain that has no counterpart in the RNA virus world and is thus considered a genetic marker of the order Nidovirales (12, 16). Bioinformatics has revealed a distant relationship between this domain and a small family of prokaryotic and eukaryotic proteins, of which XendoU, an endoribonuclease derived from Xenopus laevis that plays a role in the production of intron-encoded small nucleolar RNAs (19), is the prototype. XendoU cleaves U stretches in the RNA substrate in vitro in an Mn2+-dependent manner, releasing 2′-3′-cyclic phosphodiester products (19), and a similar endoribonuclease activity was recently shown for a mammalian homolog of XendoU, human placental protein 11 (PP11) (20). In coronaviruses, the proposed endoribonuclease activity maps to the C-terminal domain of nsp15, and uridylate-specific cleavage of RNA has been demonstrated in vitro for several nsp15 orthologs (3, 16, 38). The enzyme was termed NendoU (nidoviral endonuclease specific for U), and its crucial role in the coronavirus life cycle was established using recombinant coronaviruses that expressed mutant forms of nsp15 (16, 18). Biochemical studies have provided some insights into the catalytic activities of coronavirus NendoU proteins, such as their ability to process both single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) 3′ of uridylates and to release 2′-3′-cyclic phosphodiester products (5, 16). The crystal structures of nsp15 from mouse hepatitis virus (MHV) and severe acute respiratory syndrome coronavirus (SARS-CoV) were recently solved (29, 38), revealing an active site with two histidines and a lysine as likely catalytic residues. The spatial arrangement of the putative catalytic residues was strikingly similar to that found in RNase A, a feature also proposed for the catalytic site of XendoU (28). The critical roles of these three highly conserved residues (see Fig. S1 in the supplemental material) in catalysis was supported by the results of site-directed mutagenesis studies (11, 16, 38), and the importance of the two His residues for optimal viral RNA synthesis during MHV infection was established (18). Coronavirus nsp15 proteins form hexamers via intermolecular interactions that are mediated by their N-terminal domains (13, 29, 38). Hexamerization has been proposed to be required for optimal enzyme activity, most probably by stabilizing the catalytic site in its active conformation (17) and promoting substrate binding (38), and a recent study identified numerous residues of SARS-CoV nsp15, mapping both to the N terminus and to the catalytic domain of the protein, as contributing to RNA binding by the hexamer (4). Lastly, the presence of Mn2+ was shown to enhance coronavirus NendoU activity in vitro (16, 38), but its precise role in RNA cleavage remains unclear, since it appears to be dispensable for catalysis (38) and no metal-binding sites have been identified in any of the coronavirus enzymes characterized to date (29, 38).
The coronavirus NendoU domain has poorly characterized orthologs in other nidovirus lineages, like arteriviruses, toroviruses, and white bream virus, the prototype bafinivirus (32), which are all separated by large evolutionary distances (Fig. 1B; see Fig. S2 in the supplemental material). For example, the NendoU domains of coronavirus nsp15 and arterivirus nsp11 share only 26 to 27% identical residues and are fused to a family-specific N-terminal domain (Fig. 1C). Clearly, the biochemical characterization of arterivirus nsp11 and its counterparts in other nidoviruses is a prerequisite for understanding the functional consequences of the observed sequence variation. The prior analysis of arterivirus nsp11 was limited to a reverse genetics study of NendoU mutants using the arterivirus prototype, equine arteritis virus (EAV) (24), which revealed a key role in the EAV life cycle for the complete nsp11 subunit, as well as the specific importance of the NendoU domain and its catalytic residues for viral RNA synthesis. Characterization of the putative EAV nsp11 endoribonuclease activity in vitro, however, was previously hampered by the fact that recombinant nsp11 is extremely toxic to a variety of hosts (C. C. Posthuma, E. van den Born, and E. J. Snijder, unpublished observations). The use of a tightly regulated bacterial expression system has now enabled us to express and purify recombinant nsp11 for EAV and porcine reproductive and respiratory syndrome virus (PRRSV), two distantly related arteriviruses whose NendoU domains share ∼52% amino acid identity. This technical advance allowed us to initiate the protein's functional characterization, and here we provide the first direct evidence for the in vitro endoribonuclease activity of arterivirus nsp11. Furthermore, we have studied the enzyme's biochemical requirements for optimal RNA processing and the contributions of proposed active-site residues to catalysis. Our comparative approach allowed us to identify biochemical properties common to the arterivirus and coronavirus endoribonucleases, as well as distinct characteristics of the arterivirus NendoU, suggesting that it is able to process RNA efficiently in the absence of divalent cations by using a cleavage reaction mechanism resembling that of RNase A.
MATERIALS AND METHODS
Bioinformatics analyses.
Complete genome sequences of coronavirus, torovirus, bafinivirus, and arterivirus available on 16 April 2008 at the National Center for Biotechnology Information GenBank/RefSeq databases were downloaded into the Viralis platform (A. E. Gorbalenya, unpublished data). An amino acid alignment of the NendoU domain was produced using the MUSCLE program (9), and the alignment of poorly conserved columns was further manually refined to produce a final alignment (174 positions). A selection of 25 sequences representing virus species diversity (C. Lauber and A. E. Gorbalenya, unpublished data) was used for phylogenetic analyses. The following genome sequences were used: bovine torovirus Breda-1 (NC_007447), equine torovirus Berne (X52374), white bream virus (NC_008516), infectious bronchitis virus strain Beaudette (NC_001451), beluga whale coronavirus SW1 (EU111742), human coronavirus NL63 (NC_005831.2), human coronavirus 229E (NC_002645), Miniopterus bat coronavirus 1B (NC_010436), Miniopterus bat coronavirus HKU8 (NC_010438), Rhinolophus bat coronavirus HKU2 (NC_009988), Scotophilus bat coronavirus 512/05 (DQ648858), porcine epidemic diarrhea coronavirus (NC_003436), feline coronavirus (NC_007025), MHV A59 (NC_001846), human coronavirus HKU1 (NC_006577.2), human coronavirus OC43 (NC_005147), Tylonycteris bat coronavirus HKU4 (EF065505), Pipistrellus bat coronavirus HKU5 (EF065509), Rousettus bat coronavirus HKU9 (EF065513), SARS-CoV (AY291315), PRRSV strain VR-2332 (AY150564), PRRSV strain Lelystad (AY588319), lactate dehydrogenase-elevating virus neuro-virulent type C (L13298), simian hemorrhagic fever virus (NC_003092), and EAV (NC_002532.2). Utilizing the PhyML program (14), a maximum likelihood phylogenetic tree was compiled using the WAG model of amino acid substitution (37) and rate heterogeneity among sites (gamma distribution with eight categories). Support for internal nodes was measured by bootstrap analysis (1,000 replicates).
Plasmid construction.
Nucleotides 8735 to 9391 of the wild-type EAV infectious cDNA clone pEAV211 (36), encoding EAV nsp11, were PCR amplified with oligonucleotides E780 (5′-CATGCCATGGGCCATCACCATCACCATCACTCCAACAAAATTTCGTGCCTCCCG-3′), introducing a start codon followed by sequences specifying a His6 tag, and E584 (5′-CCGGAATTCGCATGCTAGCTCACTCTTGGACATAAAAGGTCGCG-3′), providing a stop codon. The fragments were cloned between the NcoI and EcoRI restriction sites of Gateway entry vector pENTR11 (Invitrogen, Carlsbad, CA) and, after sequence verification, placed downstream of the T7 promoter in Gateway expression vector pDEST14. Translation of the resulting construct, pDEST14-EAV nsp11HN, produced a protein corresponding to amino acids Ser-2838 to Glu-3056 of replicase pp1ab of the EAV Bucyrus strain (NCBI genome database accession number NC_002532), preceded by an N-terminal Met-Gly-His6 tag. Mutant pEAV211 plasmids encoding alanine substitutions in the EAV nsp11 region (24) (with the exception of Tyr-216 Ala) were used as templates for the generation of expression constructs encoding mutant derivatives of EAV nsp11. The Tyr-216 Ala substitution was newly engineered by using primers E780 and E1020 (5′-GGAATTCTCACTCTTGGACAGCAAAGGTCG-3′). The PRRSV nsp11 coding sequence (strain VR-2332; accession number AY150564) was PCR amplified from a full-length cDNA clone (22) using oligonucleotides HN-fwd (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATGCC ACCATGAAACATCACCATCACCATCACGGGTCGAGCTCTCCGCTCC C-3′) and HN-rev (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATTATTCAAGTTGGAAATAGGCTGTTTTG-3′). The corresponding PCR fragment was recombined into Gateway entry vector pDONR201, the entry clone was sequenced, and the fragment was transferred to pDEST14. Nsp11 expressed from the resulting construct, pDEST14-PRRSV nsp11HN, corresponded to amino acids Gly-3585 to Glu-3807 of PRRSV-VR2332 replicase pp1ab fused to an N-terminal Met-Lys-His6 tag. The pDEST14 vector encoding wild-type nsp15 from SARS-CoV (Frankfurt-1 strain) was described previously (29).
Protein expression and purification.
Escherichia coli BL21-AI cells (Invitrogen) were transformed with pDEST14-EAV nsp11HN, pDEST14-PRRSV nsp11HN, and pDEST14-SARS-CoV nsp15HN, respectively, and plated on LB agar containing 100 μg/ml ampicillin and 0.2% d-glucose. Subsequently, liquid cultures were inoculated by scraping colonies of freshly transformed cells into LB supplemented with 100 μg/ml ampicillin and 0.2% d-glucose and grown at 37°C until the optical density at 600 nm equaled 0.6. The growth medium was replaced with LB containing 100 μg/ml ampicillin, the culture temperature was lowered to 16°C, and protein expression was induced by addition of 0.2% l-arabinose (Sigma-Aldrich, St. Louis, MO) for 5 h. Cells were then harvested by centrifugation and lysed in 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 12.5 mM imidazole, 0.1% Triton X-100 supplemented with 50 U/ml DNase I (Invitrogen) and 1 mg/ml lysozyme (Sigma-Aldrich) for 1 h at 4°C. The Triton X-100 concentration was adjusted to 0.5%, and the lysates were incubated at 4°C for an additional 30 min and subsequently clarified by centrifugation at 20,000 × g for 45 min. Recombinant proteins were purified from supernatants using Talon metal affinity resin (Clontech, Mountain View, CA). The resin was washed extensively with 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5% glycerol, 12.5 mM imidazole, and the proteins were eluted in 20 mM HEPES, pH 7.4, 100 mM NaCl, 150 mM imidazole. The protein yield and purity of the preparations were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining using bovine serum albumin (Bio-Rad, Hercules, CA) as a concentration standard. The purified proteins were dialyzed against buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 50% glycerol and stored at −20°C.
RNA and DNA substrates.
RNA and DNA oligonucleotides were chemically synthesized by Eurogentec (Seraing, Belgium). Substrate RNA2 (5′-CGCAGUGAGCUCCUAAUCGCCC-3′) and markers RNAm2 (5′-CGCAGUU-3′), RNAm3 (5′-CGCAGU-3′), and RNAm4 (5′-CGCAG-3′) were described previously (16). dsRNA substrates were generated by annealing labeled RNA2 and unlabeled RNA13 (5′-GGGCGAUUAGGAGCUCACUGCG-3′) at equimolar concentrations. Oligoribonucleotides RNAsGUA (5′-AAAAAAAGUAAAAA-3′) and RNAsGCA (5′-AAAAAAAGCAAAAA-3′) were used to assess the preferences of the enzymes for cleavage after uridylate or cytidylate. DNA oligonucleotides MHV324 (5′-GATCCAATTGTCGACGCGTG-3′) and MHV325 (5′-AATTCACGCGTCGACAATTG-3′), which are partially complementary, were used to determine the activity of arterivirus nsp11 on ssDNA and dsDNA templates. RNA and DNA oligonucleotides were 5′ end labeled using T4 polynucleotide kinase (PNK) (Invitrogen) and [γ-32P]ATP (PerkinElmer, Waltham, MA) and were purified using RNase-free Micro Bio-spin 30 columns (Bio-Rad) prior to being used as substrates.
Endoribonuclease assays.
Proteins were incubated with the indicated concentrations of 5′-32P-labeled substrate in 25 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM dithiothreitol, 0.2 mM EDTA at 30°C. When MnCl2 was present, EDTA was omitted from the reaction buffer. EAV nsp11 and SARS-CoV nsp15 were used at a final concentration of 0.3 μM and PRRSV at 0.8 μM, unless otherwise indicated. Cleavage assays with SARS-CoV nsp15 were performed in the presence of 5 mM MnCl2. The reactions were stopped by the addition of an equal volume of Gel Loading Buffer II (95% formamide, 18 mM EDTA, 0.025% SDS; Ambion, Austin, TX) and incubation for 3 min at 95°C. The reaction products were separated by electrophoresis in 20% polyacrylamide/7 M urea/1× Tris-buffered EDTA gels, supplemented with 0.1% SDS for the analysis of double-stranded substrate processing. The gels were exposed to phosphorimager screens, which were subsequently scanned using a Typhoon Variable Mode Imager (GE Healthcare, Chalfont St Giles, United Kingdom). Images were analyzed and quantified using ImageQuant TL software (GE Healthcare).
Analysis of 3′ termini of cleavage products.
The analysis of the nature of the 3′ ends of 5′ cleavage products derived from NendoU-mediated RNA processing was performed essentially as previously described (16). Unlabeled RNAsGUA substrate was incubated with recombinant EAV nsp11 or SARS-CoV nsp15. The reaction was stopped by heat denaturation for 4 min at 95°C, and the cleavage products were incubated with T4 PNK buffer alone (70 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 100 mM KCl, 1 mM 2-mercaptoethanol), T4 PNK buffer in the presence of 5 U of T4 PNK, or calf intestine alkaline phosphatase (CIAP) buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2) in the presence of 0.5 U CIAP (Fermentas, Burlington, ON, Canada), for 30 min at 37°C. The phosphatases were subsequently heat inactivated at 95°C for 5 min. The RNAs were phenol extracted, ethanol precipitated using 10 μg glycogen (Roche, Basel, Switzerland) as a carrier, and ligated to 5′-[32P]pCp using T4 RNA ligase (Fermentas) as previously described (16). The reaction products were then resolved in denaturing polyacrylamide gels and visualized as described above.
RESULTS
Arterivirus nsp11 has Mn2+-independent endoribonuclease activity.
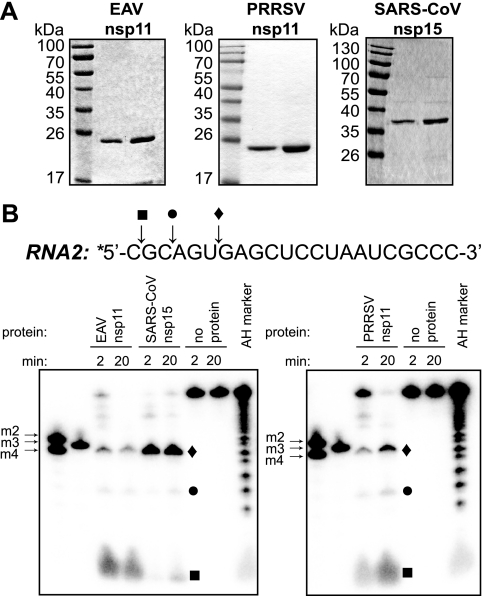
Previous attempts to express and purify recombinant EAV nsp11 had been unsuccessful due to the high toxicity that the wild-type protein displays in prokaryotic and eukaryotic cells (C. C. Posthuma and E. van den Born, unpublished data). We have now used the BL21-AI strain of E. coli as a host for the T7 RNA polymerase-driven expression of N-terminally His6-tagged nsp11. The regulation of T7 polymerase expression in this BL21-derived strain is controlled by the araBAD promoter, whose generally low basal expression was further repressed by the addition of glucose to the bacterial growth medium prior to induction of nsp11 expression. The combination of maximally repressed T7 RNA polymerase expression and induction of nsp11 expression at a low temperature allowed us to produce and purify wild-type nsp11 from EAV, as well as PRRSV (Fig. 2A). Nevertheless, the yield of wild-type nsp11 remained extremely low, and its toxicity was again illustrated by the fact that mutagenesis of putative catalytic residues dramatically improved expression levels under otherwise identical conditions (data not shown). We have used the same protocol for the expression and purification of SARS-CoV nsp15, whose endoribonuclease activity was previously characterized in some detail (3, 5, 13, 16). This allowed us to perform comparative functional studies of the arterivirus and coronavirus NendoU-containing nsps purified under identical conditions.
FIG. 2.
Purification and endoribonuclease activity of arterivirus nsp11. (A) Purified recombinant EAV nsp11 (0.5 μg and 1 μg), PRRSV nsp11 (2 μg and 4 μg), and SARS-CoV nsp15 (0.5 μg and 1 μg), each carrying an N-terminal His6 tag, were analyzed by SDS-PAGE and subsequent Coomassie brilliant blue staining. (B) Activity assays using recombinant protein and 2 μM of 5′-32P-labeled (indicated with an asterisk) RNA2 substrate were performed at 30°C for 2 or 20 min. The cleavage products were separated by electrophoresis in a 20% polyacrylamide/7 M urea gel and were visualized by phosphorimager analysis. A nucleotide ladder prepared by alkaline hydrolysis of RNA2 (AH marker), as well as 5′-32P-labeled synthetic short oligoribonucleotides (m2, m3, and m4), were included as size markers. Reactions without enzyme served as negative controls. The sequence of RNA2 is shown above the gel, and cleavage sites 3′ of the cytidylates present in the first (▪) and third (•) positions and 3′ of the uridylate in the sixth position (⧫), and the corresponding cleavage products, are indicated.
In order to assess their abilities to process RNA in vitro, recombinant EAV and PRRSV nsp11 proteins were incubated with a 5′-32P-labeled synthetic oligoribonucleotide, RNA2 (5′-CGCAGUGAGCUCCUAAUCGCCC-3′), that was shown to be efficiently cleaved by coronavirus NendoU proteins (16). Both arterivirus proteins were able to cleave the substrate in the absence of divalent cations, and the resulting products were similar to those derived from cleavage of RNA2 by SARS-CoV nsp15 (Fig. 2B). The size of the major cleavage product of the SARS-CoV enzyme was consistent with processing 3′ of the uridylate present at the sixth position, although its migration in denaturing PAGE was altered due to the presence of an additional phosphate group at the 3′ end that is lacking in the synthetic marker with the same nucleotide composition, RNA m3 (16, 29). A product of identical size was generated by cleavage of RNA2 by both EAV and PRRSV nsp11 (Fig. 2B). The arterivirus enzymes also generated products consistent with processing at the two cytidylates present in the first and third positions in RNA2, which were detectable in the SARS-CoV NendoU-driven reaction, as well, albeit to a lesser extent. An RNA ladder of fragments produced by alkaline hydrolysis of RNA2, and therefore containing 3′ phosphates, was used to verify the sizes of the cleavage products (Fig. 2B). These results indicated that arterivirus nsp11 possesses endonuclease activity and, in cleaving 3′ of pyrimidines, exhibits an in vitro substrate specificity similar to that of SARS-CoV nsp15. Interestingly, the arterivirus enzymes seem to cleave at cytidylates more efficiently than the SARS-CoV NendoU, as shown by the higher efficiency of processing at the cytidylate present in the first position in RNA2 (Fig. 2B), suggesting a less strict preference for uridylate than previously reported for the coronavirus endoribonuclease (4, 16).
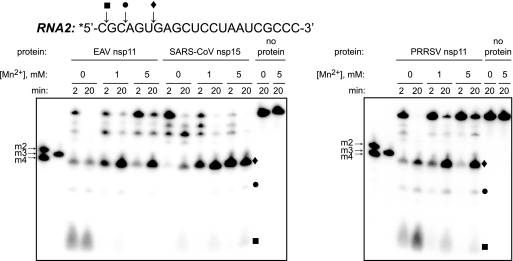
Prior studies had established that Mn2+ ions enhanced the NendoU activity of coronavirus nsp15 in vitro (5, 16, 38) and were strictly required for RNA cleavage by XendoU (11, 19). The stimulating effect of Mn2+ ions on SARS-CoV nsp15-mediated processing of RNA2 was confirmed in our assay (Fig. 3). In contrast, both EAV and PRRSV nsp11 proteins were able to process RNA2 efficiently in reaction buffers lacking divalent cations (Fig. 2B), suggesting that metal ions are not required for catalysis. To address this potential discrepancy, we investigated the role of Mn2+ ions for arterivirus NendoU activity. Addition of Mn2+ ions at concentrations previously shown to stimulate coronavirus NendoU activity in fact inhibited the cleavage of RNA2 by both EAV and PRRSV nsp11, as was evident from the increase of the remaining full-length substrate, as well as the accumulation of processing intermediates (Fig. 3). Inhibition of arterivirus nsp11-mediated RNA cleavage in the presence of Mn2+ ions was consistently reproduced with a range of substrates (data not shown). These results led us to conclude that Mn2+ ions are not required for optimal NendoU activity of arterivirus nsp11 in vitro, and consequently, the subsequent functional characterization of the arterivirus enzymes was performed in the absence of Mn2+.
FIG. 3.
Influence of Mn2+ ions on arterivirus endoribonuclease activity. Recombinant proteins were incubated with 5 μM of 5′-32P-labeled (*) RNA2 at 30°C in the absence of Mn2+ or in the presence of 1 mM or 5 mM of Mn2+, as indicated above the gel. The reactions were stopped after 2 or 20 min, and the cleavage products were separated by denaturing PAGE. For size markers and cleavage products, see the legend to Fig. 2.
Arterivirus NendoU cleavage: substrate requirements and preference for uridylate.
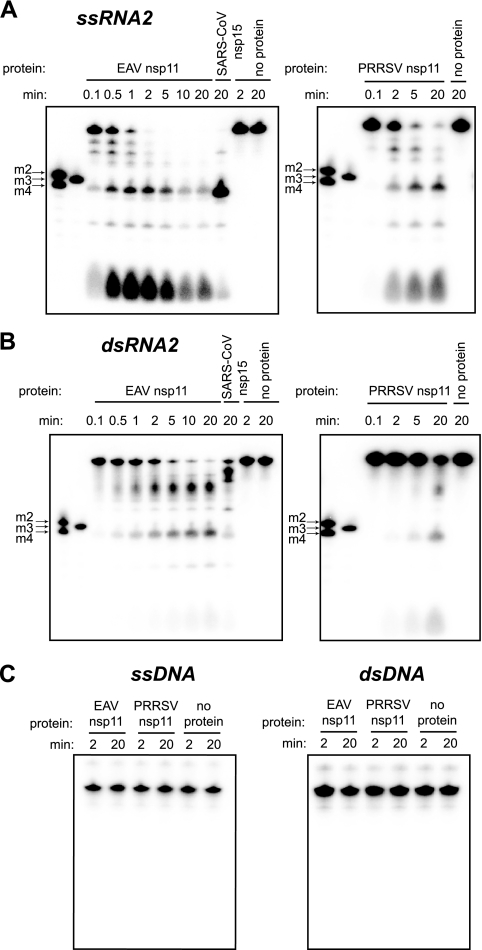
To gain more insight into the substrate requirements of the arterivirus NendoU, we examined its ability to cleave ssRNA and dsRNA molecules in vitro. Both ssRNA2 and dsRNA2 were cleaved by the arterivirus NendoU. ssRNA molecules were processed more efficiently by both arterivirus nsp11 and SARS-CoV nsp15 (Fig. 4A and B). Neither ssDNA nor dsDNA was hydrolyzed by the enzymes (Fig. 4C), confirming their specificity for RNA substrates.
FIG. 4.
Substrate requirements of the arterivirus endoribonuclease. Activity assays containing recombinant protein and 2 μM of 5′-32P-labeled ssRNA2 (A), dsRNA2 (B), or ss/dsDNA (C) were incubated at 30°C, and aliquots were taken at the time points indicated above the lanes. For size markers, see the legend to Fig. 2.
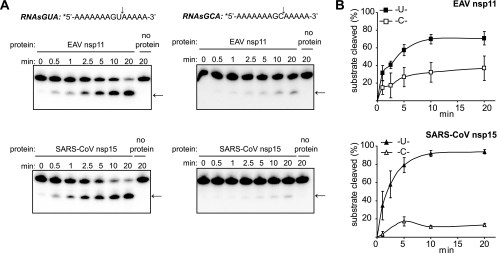
Recent studies of SARS-CoV and MHV nsp15 have shown that they can both process RNA at cytidylates, although much less efficiently than at uridylates (4, 18). The data we obtained using RNA2 as a substrate confirmed the strong preference of the SARS-CoV NendoU for uridylate and indicated that the EAV endoribonuclease is able to process RNA efficiently at pyrimidines (Fig. 2B). The large number of uridylates and cytidylates present in our initial substrate complicated the assessment of (potential) low-efficiency cleavage events. Therefore, in order to establish whether EAV NendoU prefers uridylate or cytidylate substrates, we used two additional oligoribonucleotides, both containing a single pyrimidine in an identical backbone: 5′-AAAAAAGUAAAAA-3′(RNAsGUA) and 5′-AAAAAAGCAAAAA-3′ (RNAsGCA). Incubation of these substrates with EAV nsp11 or SARS-CoV nsp15 yielded a single cleavage product, consistent with the pyrimidine specificity of the enzymes (Fig. 5A). Quantification of the processing efficiencies of these molecules under identical reaction conditions showed that both the EAV and the SARS-CoV NendoU proteins hydrolyzed the RNAsGUA substrate more efficiently than RNAsGCA (Fig. 5B). However, a larger fraction of the RNAsGCA substrate was cleaved by EAV nsp11 than by SARS-CoV nsp15 during the course of the reaction (Fig. 5B). These results suggest that EAV NendoU exhibits pyrimidine specificity with a clear preference for cleavage at uridylates, which nevertheless seems to be less pronounced than that of the SARS-CoV enzyme.
FIG. 5.
Arterivirus nsp11 cleaves RNA preferentially at uridylates. (A) One micromolar 5′-32P-labeled synthetic oligoribonucleotide containing a single uridylate (RNAsGUA) or a single cytidylate (RNAsGCA) was incubated with 0.3 μM SARS-CoV nsp15 or 0.6 μM EAV nsp11 at 30°C under standard reaction conditions. Aliquots were taken at different time points, and the reaction products were analyzed by denaturing PAGE and phosphorimager analysis. Cleavage sites and the corresponding products are indicated with arrows. (B) The amount of full-length substrate remaining was quantified, and the percentage of cleaved substrate was plotted as a function of the reaction time. The data points represent mean values from three independent experiments, and the error bars denote standard deviations.
Replacement of conserved amino acids in the EAV NendoU domain: effects on RNA cleavage and substrate specificity.
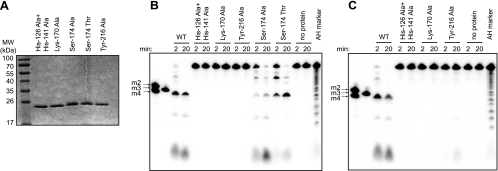
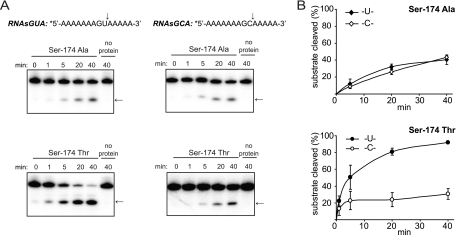
Functional studies of both XendoU and coronavirus NendoU proteins have confirmed the critical roles of two His residues and a Lys residue in catalysis (11, 16, 38). These three amino acids were initially proposed to form a catalytic center based on their conservation across the entire protein family (39). Structural studies substantiated this hypothesis, showing that these residues cluster in a positively charged groove of the SARS-CoV enzyme (29) and form a phosphate-binding site in XendoU (28). Superimposing the spatial arrangements of the catalytic triads of SARS-CoV NendoU and RNase A revealed that two additional residues in SARS-CoV NendoU, Tyr-342 and Ser-293, interact with the substrate and possibly confer uridylate specificity on the enzyme (29). To address the importance of the corresponding residues (indicated in the alignment in Fig. 1C) for the NendoU activity of EAV nsp11, they were replaced with Ala. The N-terminally His-tagged mutant proteins were expressed and purified under conditions identical to those used for purification of the wild-type nsp11 (Fig. 6A). A double replacement of His-126 and His-141, as well as single Lys-170- or Tyr-216-to-Ala substitutions reduced the cleavage of RNA2 to undetectable levels (Fig. 6B). Processing of this substrate was not observed, even at enzyme concentrations fivefold higher than those optimal for the wild-type protein (Fig. 6C), except for the Tyr-216-to-Ala mutant, for which trace amounts of cleavage products were observed upon longer reaction times. Taken together, these data confirmed the critical roles of the mutated residues in arterivirus NendoU-mediated RNA processing. The Ser-174-to-Ala substitution also impaired EAV NendoU activity, although not as dramatically, since cleavage of RNA2 was clearly detectable at a protein concentration similar to that used for wild-type EAV NendoU (Fig. 6B). Comparing the processing efficiencies of RNAsGUA and RNAsGCA revealed that the Ser-174-to-Ala substitution had rendered the EAV NendoU unable to discriminate between uridylates and cytidylates, since the mutant protein was able to cleave at both nucleotides with similar efficiencies (Fig. 7A and B). We also replaced Ser-174 with Thr, a residue that is present at this position of the NendoU alignment in all other nidovirus homologs, with the exception of SARS-CoV and closely related bat coronaviruses (see Fig. S1 in the supplemental material). This substitution resulted in somewhat impaired in vitro RNA processing in comparison with the wild-type EAV NendoU, as well (Fig. 6B), but it had no effect on the uridylate preference of the enzyme (Fig. 7A and B). These results strengthen the view that Ser-174 is important for substrate recognition and is involved in determining the substrate specificity of the EAV NendoU.
FIG. 6.
Catalytic activities of EAV nsp11 mutants. (A) Purified N-terminally His6-tagged EAV nsp11 proteins containing the indicated amino acid substitutions were electrophoresed on an SDS-polyacrylamide gel, which was subsequently stained with Coomassie brilliant blue. (B and C) Reaction mixtures containing 0.3 μM (B) or 1.5 μM (C) of mutant recombinant proteins and 2 μM of 5′-32P-labeled RNA2 were incubated at 30°C. The reactions were stopped at the indicated time points, and the cleavage products were separated in a 20% polyacrylamide/7 M urea gel. For size markers, see the legend to Fig. 2. WT, wild type.
FIG. 7.
Mutagenesis of Ser-174 influences EAV nsp11 substrate specificity. (A) One micromolar of RNAsGUA or RNAsGCA was incubated with 0.6 μM EAV nsp11 S3011A or 0.6 μM EAV nsp11 S3011T at 30°C under standard reaction conditions. Aliquots were taken at different time points, and the reaction products were analyzed by denaturing PAGE and phosphorimager analysis. (B) The amount of full-length substrate remaining was quantified, and the percentage of cleaved substrate was plotted as a function of the reaction time. The data points represent mean values from three independent experiments, and the error bars denote standard deviations.
Our previous analysis of the impact of NendoU mutagenesis on the EAV life cycle revealed that replacement of His-126, His-141, Lys-170, or Ser-174 with Ala caused major defects in viral RNA synthesis and production of infectious progeny (24). In addition, the critical roles of two nidovirus-wide conserved aspartic acid residues, Asp-177 and Asp-201 (Fig. 1C) in viral RNA synthesis was established. Nonconservative (to Ala) or conservative (to Glu or Asn) replacements of those residues rendered viral replication undetectable. Examining the effects of these mutations on the in vitro endonuclease activity of EAV NendoU could therefore provide valuable insight into the basis of this nonviable phenotype. Our attempts to purify recombinant EAV nsp11 containing an Asp-177-to-Ala or Asp-201-to-Ala substitution, however, were unsuccessful, since both mutations rendered the protein insoluble. Similar results were reported following mutagenesis of the Asp-177 counterpart in SARS-CoV NendoU (13) and the Asp-201 equivalent in the MHV enzyme (18). The crystal structure of SARS-CoV nsp15 revealed that these residues are part of an extensive hydrogen bond network in close proximity to the active site and that replacing them could have an indirect effect on the activity of the protein by disturbing its secondary structure (29), a notion supported by the insolubility of the recombinant mutant proteins.
EAV NendoU RNA cleavage: reaction mechanism.
A number of parallels have been drawn between the active sites of RNase A, XendoU (28), coronavirus NendoU (29), and, in this report, arterivirus NendoU. RNase A is a remarkably well-characterized enzyme that has been proposed to cleave the P-O5′ bond in RNA via general acid-base catalysis involving the side chains of the active-site His-12 and His-119 residues (reference 26 and references therein). In a first reaction, transphosphorylation of the RNA results in the accumulation of 2′,3′-cyclic phosphodiesters, which can be subsequently hydrolyzed to 3′-phosphomonoesters in a separate process (34) (Fig. 8A). 2′,3′-Cyclic phosphates have been detected at the 3′ ends of cleavage products generated by both XendoU (19) and coronavirus NendoU (16), suggesting similarities to RNase A in terms of cleavage chemistry. To investigate the chemical nature of the 3′ termini of cleavage products generated by the EAV NendoU, the products derived from cleavage of RNAsGUA after short (2-min) or prolonged (40-min) reaction times were treated with T4 PNK or CIAP. Following phenol extraction and ethanol precipitation, the cleavage products were ligated to 5′-[32P]pCp in a T4 ligase-mediated reaction. As expected, pCp could be readily ligated to the hydroxyl group present at the 3′ end of the unprocessed substrate (Fig. 8B), and this reaction was used as an internal control for determining the efficiency of pCp ligation. In contrast to the uncleaved substrate RNA, efficient transfer of the 32P label to the 3′ terminus of the 5′ cleavage product obtained after incubation with EAV nsp11 required phosphatase treatment prior to the pCp ligation reaction, suggesting the presence of a 3′ phosphate group. After a short EAV NendoU reaction, the 5′ cleavage products could be efficiently ligated only after dephosphorylation with T4 PNK (which is capable of removing both 2′,3′ and 3′ phosphates), but not after CIAP treatment (which can remove only 3′ phosphates), which suggests that the 5′ cleavage products contained predominantly 2′,3′-cyclic phosphates. Upon longer incubation of the substrate and enzyme, however, CIAP treatment was sufficient for efficient ligation of a portion of the 5′ cleavage products to 5′-[32P]pCp, indicating that nsp11-mediated hydrolysis of part of the cyclic phosphodiesters had occurred (Fig. 8B). In the SARS-CoV nsp15-mediated reaction, the fraction of 5′ cleavage products whose cyclic phosphate ends had been hydrolyzed was even larger, as illustrated by the higher efficiency of 5′-[32P]pCp ligation after dephosphorylation with CIAP (Fig. 8B). These results suggest that the EAV and SARS-CoV NendoU proteins are able to catalyze both transphosphorylation of RNA to form a 2′,3′-cyclic phosphodiester and its subsequent hydrolysis to a 3′-phosphomonoester, which would be consistent with an RNase A-like reaction mechanism.
FIG. 8.
Analysis of the 3′ termini of EAV and SARS-CoV NendoU products. (A) Proposed mechanism for the transphosphorylation (1) and hydrolysis (2) reactions catalyzed by RNase A. (B) Cleavage products obtained after incubation of 1 μM unlabeled RNAsGUA with 0.6 μM EAV nsp11 or SARS-CoV nsp15 for 2 min (1) or 40 min (2) at 30°C under standard reaction conditions were subsequently ligated to 5′-[32P]pCp without further treatment (lanes a) or after dephosphorylation with T4 PNK (lanes b) or CIAP (lanes c). The reaction products were analyzed by denaturing PAGE and phosphorimager analysis. The full-length substrate (*) and the 5′ cleavage product (▴) are indicated.
DISCUSSION
The NendoU activity is common to arteriviruses and coronaviruses, and likely all nidoviruses.
This report describes the in vitro endoribonuclease activity of nsp11 from two quite distantly related arteriviruses, EAV and PRRSV, and expands the short list of characterized XendoU/NendoU superfamily members with representatives of a new, deeply separated phylogenetic lineage. The sizes and domain compositions of the nidovirus replicase subunits harboring NendoU domains vary considerably among nidoviruses, which might be linked to different catalytic properties and functions of the enzymes in the life cycles of different nidoviruses. The biochemical characterization of arterivirus nsp11 documented its ability to hydrolyze RNA 3′ of pyrimidines, with a preference for uridylates, and revealed that this endoribonuclease activity is not dependent on the presence of divalent cations. Mutagenesis of predicted active-site residues abolished the protein's ability to cleave RNA and provided insights into the determinants of the enzyme's uridylate preference. Our comparative analysis of arterivirus nsp11 and coronavirus nsp15 purified under identical conditions established important parallels between the ribonucleolytic activities of these enzymes, such as the identities of key residues in their catalytic sites and their proposed reaction mechanisms, which are likely conserved across the entire order Nidovirales. Our data also revealed differences in their biochemical properties, such as the effects of Mn2+ on their in vitro activities, as well as their variable extents of preference for cleavage at uridylates, both probably owing to considerable divergence of arterivirus nsp11 and coronavirus nsp15, which carry distantly related NendoU domains fused to apparently unrelated N-terminal polypeptides (Fig. 1C).
The endoribonuclease activities of EAV and PRRSV nsp11 seem to be independent of the presence of metal ions as cofactors, since both proteins cleaved RNA very efficiently in reaction buffers lacking divalent cations and containing EDTA (Fig. 2B and 3). This finding stands in contrast to the previously documented stimulatory effect of Mn2+ on the NendoU activity of coronavirus nsp15 in vitro (16, 38), which was corroborated by the comparative enzyme activity studies included in this paper (Fig. 3). Our data show that arterivirus endoribonucleolytic activities are inhibited by Mn2+ ions at concentrations that have previously been shown to significantly enhance the activities of a number of coronavirus NendoU enzymes (3, 16, 38). Interestingly, similar Mn2+ concentrations have been reported to inhibit RNase A-mediated RNA hydrolysis as well, probably by binding to and blocking the catalytic histidine residues (1). The same study showed that low concentrations of Mn2+ may enhance RNase A activity, possibly through interactions between the metal ions and the phosphates present in the RNA substrate. Due to the high rate of RNA cleavage by the recombinant enzymes and the gel electrophoresis-based activity assays used in this study, we were unable to determine if arterivirus in vitro endoribonuclease activities can be further stimulated by low concentrations of Mn2+ or other divalent cations. It should be noted that the precise role of Mn2+ in RNA hydrolysis by the coronavirus NendoU and its cellular homologs, XendoU and human PP11, remains enigmatic. Although Mn2+ is essential for in vitro RNA processing by XendoU and human PP11, the interaction of the enzymes with a substrate does not require metal ions (11, 19, 20). The role of Mn2+ in coronavirus NendoU cleavage is even less clear. Previous studies have implicated Mn2+ ions in triggering conformational changes (3), as well as affecting the RNA binding properties of these proteins (5). Metal binding sites have not been identified in the currently available crystal structures of coronavirus nsp15 (4, 17, 29, 38) or XendoU (28). Furthermore, millimolar concentrations of Mn2+ ions are required for enhancing RNA cleavage by coronavirus nsp15 in vitro, while the intracellular free Mn2+ concentration is estimated to be in the micromolar range (references 5 and 27 and references therein), raising questions about the biological significance of this observation. Comparative analysis of both the structure and biochemistry of the arterivirus and coronavirus enzymes could therefore be used to gain more insight into the specific role(s) of divalent cations in coronavirus NendoU-mediated cleavage.
The EAV NendoU active site and mechanism of RNA cleavage.
Replacement of the predicted catalytic residues (His-126, His-141, and Lys-170) of EAV NendoU with alanine established their importance for in vitro RNA processing (Fig. 6B and C). This observation is consistent with the alignment in Fig. 1C and the results of similar experiments with related enzymes of other origins, as well as RNase A. Likewise, the replacement of Tyr-216, whose counterpart in the coronavirus NendoU was implicated in the proper positioning and orientation of the substrate in the coronavirus NendoU active site (29), also severely impaired EAV NendoU-mediated cleavage in vitro (Fig. 6B and C). Interestingly, replacement of Ser-174 with Ala only moderately affected cleavage by EAV NendoU (Fig. 6B) but rendered the enzyme unable to discriminate between uridylates and cytidylates (Fig. 7). The conservative replacement of Ser-174 with Thr largely restored efficient RNA processing, including the preference of EAV NendoU for cleavage at uridylates (Fig. 7). Ser-174 of EAV nsp11 is the counterpart of residue Ser-293 in SARS-CoV nsp15 (29), which has been proposed to be functionally equivalent to Thr-45 of RNase A, a major determinant of the enzyme's pyrimidine specificity (reference 26 and references therein). In a recent study (4), the effects of SARS-CoV nsp15 Ser-293 replacement by Ala or Thr were reported to be similar to those we describe here for EAV nsp11 Ser-174 replacements. Two additional residues of SARS-CoV nsp15, Pro-343 and Leu-345, which are highly conserved in the C-terminal regions of coronavirus NendoU domains (Fig. 1C), were also implicated in determining the substrate specificity of the endoribonuclease. Pro-343 was proposed to sterically hinder the access of cytosine to the active site, while the side chain of Leu-345 could prevent the access of purines. The extreme C-terminal residues of the EAV and SARS-CoV NendoU proteins differ substantially, with the counterpart residue of Pro-343 being Val-217 in EAV nsp11 while the bulky Leu-345 presumably has no counterpart (Fig. 1C). Structural information about the arterivirus endoribonuclease would shed more light on whether the protein's C-terminal tail is also involved in substrate binding and whether the limited conservation of this region could account for the more relaxed uridylate specificity we observed for EAV NendoU in comparison to the SARS-CoV enzyme (Fig. 5).
The similarities between the active sites of viral NendoU proteins and RNase A, which were previously suggested by others (29) and are also outlined in this report, prompted us to examine the mechanism of RNA cleavage by the EAV endoribonuclease. The reaction products of coronavirus NendoU were shown to possess 2′,3′-cyclic phosphate ends (16). Analysis of the 3′ ends of cleavage products generated by EAV NendoU indicated that this reaction also proceeds through the formation of 2′,3′-cyclic phosphodiesters, a fraction of which appear to be hydrolyzed to 3′ phosphomonoesters after longer reaction times (Fig. 8B). We obtained similar results upon analysis of SARS-CoV NendoU cleavage products. These data are consistent with the utilization of an RNase A-like mechanism of RNA cleavage by both the arterivirus and coronavirus endoribonucleases. The ability of the SARS-CoV enzyme to hydrolyze 2′,3′-cyclic phosphodiesters had not been detected previously, most likely due to the relatively low activity of the MBP-nsp15 fusion protein used in the earlier studies.
Role of NendoU in the life cycles of arteriviruses and coronaviruses.
Despite recent advances in our understanding of the biochemical properties of nidovirus RNA-processing enzymes, the characterization of their precise roles and mechanisms of action in the viral life cycle is still at an initial stage. Reverse-genetics-based mutagenesis studies of the coronavirus nsps that harbor the exoribonuclease (ExoN) and NendoU domains confirmed their importance for coronavirus RNA synthesis (7, 8, 16, 18, 21) and lent support to the involvement of ExoN in the fitness control of coronavirus RNA replication (8). A site-directed mutagenesis study of EAV nsp11 in the context of the complete EAV life cycle suggested that this replicase subunit is a multifunctional viral protein (24). Replacement of Asp-177 or Asp-201, which in this study were found to render bacterially expressed EAV nsp11 insoluble, resulted in a nonviable phenotype. A similar result was obtained by deleting the nsp11 coding sequence from the EAV genome. Most notably, however, replacement of any of the three putative catalytic residues (His-126, His-141, or Lys-170), which were shown in this study to reduce the in vitro EAV NendoU activity to background levels, yielded severely crippled but viable mutant viruses. These substitutions induced a specific defect in viral sg mRNA synthesis, which was accompanied by a dramatic reduction in the production of infectious progeny. We cannot formally exclude the possibility that a basal level of endoribonuclease activity, undetectable in our in vitro assay, may be retained by these catalytic-site mutants in vivo, which might be sufficient for viral replication and production of virus progeny, albeit at a low level. Examination of the role of the MHV NendoU in the viral life cycle revealed that mutagenesis of the catalytic His residues resulted in severely impaired but detectable RNA-processing activity in vitro and only modest defects in viral RNA synthesis and production of infectious progeny (18). Furthermore, no specific defects in sg mRNA synthesis were detected. This stands in contrast to the observations made upon EAV NendoU mutagenesis by reverse genetics (24) and results obtained by replacement of an NendoU active-site His in a SARS-CoV replicon. In the latter case, a severe decrease in sg mRNA levels was observed (2). Overall, these data suggest (partially) different roles of NendoU in the life cycles of arteriviruses and coronaviruses. Alternatively, a potentially higher basal activity retained by the MHV NendoU catalytic-site mutants, combined with differences in overall replication rates between MHV and EAV, might minimize the consequences of these substitutions for the MHV life cycle. Moreover, the significance of the uridylate preference observed for arterivirus and coronavirus endoribonucleases in vitro for viral RNA synthesis is difficult to assess; the in vivo substrates of these enzymes are still unknown, and identifying them will be a key step in linking the available in vitro and in vivo data mechanistically. In this respect, it remains unclear whether the reduction of EAV sg mRNA synthesis and the decrease in infectious-progeny production seen upon replacing Ser-174 of nsp11 with Ala (24) can be attributed to the loss of the mutant enzyme's ability to discriminate between uridylates and cytidylates (Fig. 7) or to its overall lower activity (Fig. 6B). In addition, the importance of viral determinants of pathogenesis can be difficult to assess in tissue culture, as illustrated by the coronavirus ADP-ribose 1″-phosphatase and the putative cyclic phosphodiesterase domains, which are dispensable for replication in cultured cells but are critically involved in viral pathogenicity in vivo (10, 25, 30).
Irrespective of the exact function of arterivirus and coronavirus NendoU proteins in the viral life cycle, the broad specificity that these endoribonucleases exhibit in vitro suggests that their activity in infected cells must be strictly regulated in order to protect viral (and possibly cellular) RNA species from rapid degradation. Indeed, expression of EAV nsp11 outside the context of the infected cell is extremely toxic to prokaryotic and eukaryotic cells (data not shown). For example, protein-protein interactions or modulation by cofactors might act as allosteric switches to regulate the ribonucleolytic activities of arterivirus nsp11 and coronavirus nsp15 and prevent nonspecific cleavage of viral and/or cellular RNAs. The hexamerization of coronavirus nsp15, mediated by its N-terminal domain, and the presence of Mn2+ have been implicated in ensuring optimal RNA-processing activity (5, 13, 17). In contrast, EAV NendoU activity is inhibited in the presence of Mn2+ (Fig. 3), and its quaternary structure may differ, as well. Although the yield of bacterially expressed EAV nsp11 was unfortunately too low for us to attempt to analyze its oligomeric state, preliminary gel filtration analyses of catalytic-site mutants suggest they are present as monomers in solution (data not shown). This observation may turn out to be linked to the properties of the N-terminal domain of arterivirus nsp11, which is relatively small and apparently unrelated to its coronavirus nsp15 counterpart (Fig. 1C) (Gorbalenya, unpublished). In conclusion, the spatial and temporal regulation of EAV NendoU activity is likely ensured by coordinated replicase polyprotein cleavage, interactions with other replicase subunits or cellular proteins, and/or its compartmentalization in a specifically organized membrane-bound replication/transcription complex. The continued biochemical and structural characterization of nidovirus NendoU proteins, in combination with the functional dissection of NendoU-containing ribonucleoprotein complexes in infected cells, should provide a basis for the identification of the in vivo target(s) of this replicative endoribonuclease that is unique in the RNA virus world.
Supplementary Material
Acknowledgments
We are grateful to our LUMC colleagues Linda Boomaars-van der Zanden and Inge Schaap and to Bruno Canard, Bruno Coutard, and their coworkers of the VIZIER expression platform (CNRS and University of Aix-Marseille, France) for generating the PRRSV nsp11 expression construct. A.E.G. thanks Michael Rozanov for sharing his observations. We thank Sjoerd van den Worm and Martijn van Hemert for helpful discussions and Igor Sidorov and Alexander Kravchenko for help with Viralis and its databases.
This work was supported by the Council for Chemical Sciences of The Netherlands Organization for Scientific Research (NWO-CW; grant 700.52.306), The Netherlands Bioinformatics Centre (NBIC; grant SP 3.2.2), and the VIZIER integrated project (LSHG-CT-2004-511960) of the European Union's Sixth Framework Programme.
Footnotes
Published ahead of print on 18 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alger, T. D. 1970. Copper(II) and manganese(II) effects on ribonuclease A activity. Biochemistry 93248-3255. [DOI] [PubMed] [Google Scholar]
- 2.Almazan, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 8010900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 7812218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhardwaj, K., S. Palaninathan, J. M. Alcantara, L. L. Yi, L. Guarino, J. C. Sacchettini, and C. C. Kao. 2008. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 2833655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhardwaj, K., J. Sun, A. Holzenburg, L. A. Guarino, and C. C. Kao. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decroly, E., I. Imbert, B. Coutard, M. Bouvet, B. Selisko, K. Alvarez, A. E. Gorbalenya, E. J. Snijder, and B. Canard. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 828071-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckerle, L. D., X. Lu, S. M. Sperry, L. Choi, and M. R. Denison. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 8112135-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson, K. K., L. Cervantes-Barragan, B. Ludewig, and V. Thiel. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose 1″-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 8212325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gioia, U., P. Laneve, M. Dlakic, M. Arceci, I. Bozzoni, and E. Caffarelli. 2005. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J. Biol. Chem. 28018996-19002. [DOI] [PubMed] [Google Scholar]
- 12.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 11717-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino, L. A., K. Bhardwaj, W. Dong, J. Sun, A. Holzenburg, and C. Kao. 2005. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 3531106-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 15.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 254933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 10112694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph, J. S., K. S. Saikatendu, V. Subramanian, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2007. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 816700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, H., K. Bhardwaj, Y. Li, S. Palaninathan, J. Sacchettini, L. Guarino, J. L. Leibowitz, and C. C. Kao. 2007. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 8113587-13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laneve, P., F. Altieri, M. E. Fiori, A. Scaloni, I. Bozzoni, and E. Caffarelli. 2003. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J. Biol. Chem. 27813026-13032. [DOI] [PubMed] [Google Scholar]
- 20.Laneve, P., U. Gioia, R. Ragno, F. Altieri, F. C. Di, T. Santini, M. Arceci, I. Bozzoni, and E. Caffarelli. 2008. The tumor marker human placental protein 11 is an endoribonuclease. J. Biol. Chem. 28334712-34719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 1035108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, H. S., G. Liu, J. Nielsen, M. B. Oleksiewicz, A. Botner, T. Storgaard, and K. S. Faaberg. 2003. Generation of an infectious clone of VR-2332, a highly virulent North American-type isolate of porcine reproductive and respiratory syndrome virus. J. Virol. 773702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense…? J. Gen. Virol. 871403-1421. [DOI] [PubMed] [Google Scholar]
- 24.Posthuma, C. C., D. D. Nedialkova, J. C. Zevenhoven-Dobbe, J. H. Blokhuis, A. E. Gorbalenya, and E. J. Snijder. 2006. Site-directed mutagenesis of the nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 801653-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 7912721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raines, R. T. 1998. Ribonuclease A. Chem. Rev. 981045-1066. [DOI] [PubMed] [Google Scholar]
- 27.Ranjith-Kumar, C. T., L. Gutshall, M. J. Kim, R. T. Sarisky, and C. C. Kao. 2002. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 7612526-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renzi, F., E. Caffarelli, P. Laneve, I. Bozzoni, M. Brunori, and B. Vallone. 2006. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc. Natl. Acad. Sci. USA 10312365-12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricagno, S., M. P. Egloff, R. Ulferts, B. Coutard, D. Nurizzo, V. Campanacci, C. Cambillau, J. Ziebuhr, and B. Canard. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA 10311892-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth-Cross, J. K., H. Stokes, G. Chang, M. M. Chua, V. Thiel, S. R. Weiss, A. E. Gorbalenya, and S. G. Siddell. 28 January 2009. Organ-specific attenuation of murine hepatitis virus (MHV-A59) by replacement of catalytic residues in the putative viral cyclic phosphodiesterase NS2. J. Virol. doi: 10.1128/JVI. 02203-08. [DOI] [PMC free article] [PubMed]
- 31.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 8120-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schutze, H., R. Ulferts, B. Schelle, S. Bayer, H. Granzow, B. Hoffmann, T. C. Mettenleiter, and J. Ziebuhr. 2006. Characterization of white bream virus reveals a novel genetic cluster of nidoviruses. J. Virol. 8011598-11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. M. Poon, Y. Guan, M. Rozanov, W. J. M. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. E., F. D. Venegas, and R. T. Raines. 1994. Energetics of catalysis by ribonucleases: fate of the 2′,3′-cyclic phosphodiester intermediate. Biochemistry 337408-7414. [DOI] [PubMed] [Google Scholar]
- 35.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 873473-3482. [DOI] [PubMed] [Google Scholar]
- 36.van den Born, E., A. P. Gultyaev, and E. J. Snijder. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10424-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whelan, S., and N. Goldman. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18691-699. [DOI] [PubMed] [Google Scholar]
- 38.Xu, X., Y. Zhai, F. Sun, Z. Lou, D. Su, Y. Xu, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 807909-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81853-879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.