Abstract
Human cytomegalovirus (HCMV) infection results in the formation of nuclear viral transcriptosomes, which are sites dedicated to viral immediate-early (IE) transcription. At IE times of the infection, viral and cellular factors, including several components of transcription such as cyclin-dependent kinase 9 (cdk9), localize at these sites. To determine the mechanism and requirements of specific recruitment of cdk9 to the viral transcriptosomes, infection in the presence of inhibitor drugs and infection of cell lines expressing exogenous mutant cdk9 were performed. We found that cdk9 localization to the viral transcriptosomes requires de novo protein synthesis. In addition, active transcription is required for recruitment and maintenance of cdk9 at the viral transcriptosomes. In cells infected with a recombinant IE2 HCMV (IE2 86 ΔSX virus) in which IE2 gene expression is greatly reduced, cdk9 localization at the transcriptosome is delayed and corresponds to the kinetics of accumulation of the IE2 protein at these sites. Infection in the presence of the cdk9 inhibitors Flavopiridol and DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) allowed cdk9 localization to the viral transcriptosomes. A kinase-inactive cdk9 (D167N) expressed during the infection also localizes to the viral transcriptosomes, indicating that kinase activity of cdk9 is not a requirement for its localization to the sites of IE transcription. Exogenous expression of additional cdk9 mutants indicates that binding of Brd4 to the cdk9 complex is not required but that efficient binding to cyclin T1 is essential.
Human cytomegalovirus (HCMV) is a member of the Herpesviridae family and is of clinical concern in immunocompromised patients, organ transplant recipients, and the developing fetus (for a review, see reference 34). Congenital HCMV is the major viral cause of birth defects and can lead to permanent disabilities such as hearing and vision loss, mental disabilities, and even death. At present, there is no cure or available vaccine for treatment of HCMV.
Immediately after the viral particles contact the cellular plasma membrane, many host functions are altered. It is a combination of the interactions between the virus and host that are established and the disruption of cellular functions that creates an optimal environment for viral replication (for a review, see reference 17). Viral gene expression is temporally regulated, beginning with the immediate-early (IE) genes. The IE genes do not require de novo cellular or viral protein synthesis for expression and can be classified as the set of viral transcripts that accumulate in the presence of cycloheximide (CHX). The IE gene products activate the expression of viral early genes, which in turn initiate and regulate viral DNA synthesis. After the onset of viral DNA synthesis, the late viral genes, which primarily encode structural proteins, are expressed, and that expression leads to the eventual release of virus from the cell.
HCMV utilizes cellular RNA polymerase II (RNAP II) and the accompanying host machinery for transcription of viral genes. In humans, the C-terminal domain (CTD) of the largest subunit of RNAP II is composed of 52 repeats of the consensus heptapeptide sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser and is susceptible to high levels of phosphorylation during the transcription cycle (for reviews, see references 29, 33, and 40). A hypophosphorylated form of RNAP II (RNAP IIa) is recruited to the preinitiation complex at the gene promoter by the general transcription factors. Initiation proceeds when the cyclin-dependent kinase 7 (cdk7) complex phosphorylates the CTD at the serine 5 residues, hyperphosphorylating RNAP II (RNAP IIo). The CTD is further phosphorylated by the cdk9 complex at the serine 2 residues, which promotes transcription elongation by weakening the association of negative elongation factors with the paused RNAP II complex. Brd4 has been shown to enhance transcription elongation by recruiting cdk9 via cyclin T1 to paused RNAP II at acetylated promoter regions and possibly stimulating cdk9 phosphorylation of RNAP II (52). At this time, RNA processing factors are also recruited to the transcription complex. During the infection, both the cdk9 and cdk7 active complexes are upregulated in terms of RNA and protein levels and activity (49). This contributes to an increase in hyperphosphorylation of RNAP II to levels greater than in uninfected cells. HCMV also encodes a kinase, UL97, which can phosphorylate RNAP II CTD in vitro, although UL97 does not significantly contribute to CTD phosphorylation in vivo (4).
Viral IE transcription must be robust for initiation of a productive infection, and a key step in this process is the formation of the viral transcriptosomes (1, 3, 23, 24, 27, 49). Viral transcriptosomes are subnuclear foci that consist of several viral and cellular components that localize adjacent to cellular promyelocytic leukemia (PML) oncogenic domains (also known as ND10 structures) and function as the sites of viral IE transcription. To date, these sites have been shown to consist of the input viral genome, IE2-86 (IE2), UL112-113, UL69, and several cellular transcription regulators and chromatin-modifying proteins, including RNAP II (IIa and IIo) and its kinases, cdk9 and cdk7, cyclin T1, Brd4, histone deacetylase 1 (HDAC1), and HDAC2 (1, 3, 23, 24, 27, 39, 49). The input viral genomes serve as the templates for viral IE transcription, and the IE RNAs are found at high concentrations at these sites (3, 24). The newly synthesized major IE proteins IE1-72 (IE1) and IE2 also localize to the growing viral transcriptosomes. At between 3 and 6 h postinfection (p.i.), IE1 protein and several PML oncogenic domain-associated proteins (including PML protein) become dispersed throughout the nucleus due to IE1 activity (2, 3, 36, 51). IE2 protein, however, persists at the viral transcription sites and serves as a suitable marker for the viral transcriptosomes as the infection progresses (27, 39, 49). Eventually, the viral transcriptosomes develop into replication compartments where viral DNA synthesis occurs (1, 47).
Through a series of studies, we have shown that treatment with Roscovitine, the cdk inhibitor, during an HCMV infection significantly alters viral gene expression (43). This alteration results in the differential processing of several IE transcripts, inhibition of viral early and late gene expression and DNA replication, and decreased viral titers. Additionally, in the presence of Roscovitine, virus-specific alterations in the host transcription machinery, such as decreases in the levels of hyperphosphorylated RNAP II, decreases in cdk9 and cdk7 RNA levels, decreases in cdk9 protein levels, and impaired localization of cdk9 and cdk7 to the viral transcriptosomes, are observed (27, 49). The cyclin partner for cdk9, cyclin T1 (known as positive transcription elongation factor b [P-TEFb] when the two are paired in a complex), is still maintained at the viral transcriptosomes with IE2 in the presence of Roscovitine. The localization at the viral transcriptosomes of Brd4, which binds to P-TEFb, is also unaffected in the presence of Roscovitine. It was subsequently shown that the impaired recruitment of cdk9 to the viral transcriptosomes in the presence of Roscovitine is not due to the observed decrease in cdk9 protein levels. Many of the effects of Roscovitine on the infection, including alterations in viral gene expression and the impairments in viral transcriptosome formation, are limited to treatment within the first 8 h of infection. This suggests that the requirements for cdk9 and cdk7 during the infection in terms of viral gene expression and viral transcriptosome formation are fulfilled within the first few hours of infection.
Although much information regarding the composition of the viral transcriptosomes has been gained in the past decade, very little is known about how the viral transcriptosomes are established. The combination of cellular and viral signals and the kinetics of this dynamic process have not been elucidated. In an attempt to begin addressing this question, we have focused on the requirements for recruitment of a single component of the viral transcriptosome, cdk9. We have found that viral entry into the nucleus, the presence of the input viral genome, and initiation of viral IE RNA synthesis are not sufficient for recruitment of cdk9 to the viral transcriptosomes. Once cdk9 localizes to the viral transcriptosomes, continuous active transcription is required to maintain localization at these sites. During infection with an IE2 mutant virus (IE2 86 ΔSX) in which IE2 gene expression is reduced (42), cdk9 is still recruited to the viral transcriptosomes but with a delay compared to a wild-type (wt) viral infection. The results of exogenous expression of various cdk9 mutants indicate that kinase activity of cdk9 and binding of Brd4 to the cdk9 active complex are not required for its recruitment to the viral transcriptosomes but that binding to cyclin T1 is essential.
MATERIALS AND METHODS
Cell culture and virus.
Human foreskin fibroblasts (HFF) were obtained from the Medical Center of the University of California, San Diego, and cultured in Earle's minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum, 1.5 μg/ml amphotericin B, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. All reagents were from Invitrogen (Carlsbad, CA). Cells were kept in incubators maintained at 37°C with 7% CO2. The constitutively expressing wt, S175D-, E55A-, E57A-, and R65A-cdk9HA cell lines were maintained in HFF media supplemented with 15 to 30 μg/ml hygromycin B (Invitrogen). The doxycycline (Dox)-inducible S175A- and E55A/E57A/R65A-cdk9HA cell lines were maintained in HFF media supplemented with 400 μg/ml G418 (Invitrogen). The Towne strain of HCMV (HCMV Towne; VR 977) was obtained from the American Type Culture Collection and propagated as previously described (48). Construction and propagation of wt IE2 86-enhanced green fluorescent protein (EGFP) HCMV and ΔSX-EGFP HCMV were described previously (42). Construction and propagation of wt-cdk9HA HCMV and D167N-cdk9HA HCMV are described below.
cdk9 mutagenesis and lentivector cloning.
The pRC-CMV-cdk9HA plasmid was obtained from X. Graña (Temple University School of Medicine, Philadelphia, PA) and has been described previously (27). The forward primers used for site-directed mutagenesis of cdk9 were as follows: for the S175D mutant, 5′-CTGGCCCGGGCCTTCGACCTGGCCAAGAACAGC-3′; for the S175A mutant, 5′-CTGGCCCGGGCCTTCGCCCTGGCCAAGAACAGC-3′; for the E55A mutant, 5′-TGCTGATGGAAAACGCGAAGGAGGGGTTCCC-3′; for the E57A mutant, 5′-TGGAAAACGAGAAGGCGGGGTTCCCCATTAC-3′; for the R65A mutant, 5′-CCCATTACAGCCTTGGCGGAGATCAAGATCCT-3′; and for the E55A/E57A/R65A mutant, 5′-AAGGATCTTGATCTCCGCCAAGGCTGTAATGGGGAACCCCGCCTTCGCGTTTTCCATCAGCAC-3′. The reverse primers were the reverse complements of the forward primers. Mutagenesis was performed following the protocol for a QuikChange site-directed mutagenesis kit (Stratagene, San Diego, CA).
Each cdk9HA mutant was introduced into the constitutively expressing lentivector, pLV-EF1α-TKHygro (27), or into the Dox-inducible lentivector, pSLIK-neo (gift of M. Simon, California Institute of Technology, Pasadena, CA) (44), when the possibility of a dominant-negative effect was a concern. pLV-EF1α-S175D-cdk9HA-TKHygro, pLV-EF1α-E55A-cdk9HA-TKHygro, pLV-EF1α-E57A-cdk9HA-TKHygro, and pLV-EF1α-R65A-cdk9HA-TKHygro were cloned exactly as described previously (27) into the pLV-EF1α-TKHygro lentivector. The Dox-inducible pSLIK-S175A-cdk9HA-neo and pSLIK-E55A/E57A/R65A-cdk9HA-neo plasmids were cloned as follows. The pRC-S175A-cdk9HA and pRC-E55A/E57A/R65A-cdk9HA plasmids were linearized with EcoRI, and the blunt ends were filled in using Klenow T4 DNA polymerase. The mutant cdk9HA-containing DNA fragments were released by digestion with XbaI. A plasmid containing the Dox-inducible tetracycline response element (TRE) promoter, pEN-TRmiRc2 (gift of M. Simon), was prepared for insert ligation by digestion with SpeI, followed by a Klenow T4 DNA polymerase reaction and then an XbaI digestion. The mutant cdk9HA inserts were ligated into pEN-TRmiRc2. The TRE mutant cdk9HA cassettes were flanked by attL1 and attL2 recombination sites, and an LR Clonase kit (Invitrogen) was used to recombine the TRE mutant cdk9HA cassettes into the pSLIK-neo lentivector, which contains attR1 and attR2 recombination sites flanking the ccdB gene.
cdk9HA cell lines.
293FT cells were cultured and transfected with the lentiviral vector and packaging mix (ViraPower lentiviral expression system; Invitrogen) as described previously (27) with the following modifications. At 48 and 72 h posttransfection, the lentivirus-containing supernatant was collected, spun down to remove cellular debris, and filtered. It was immediately used to transduce subconfluent flasks of HFF cells with a 1:1 or 3:1 dilution of lentivirus and HFF media in the presence of 3 μg/ml Polybrene. At 48 h after the second transduction, the HFF cells were subjected to hygromycin B selection (15 to 30 μg/ml) or G418 selection (400 μg/ml). After 10 days under selection, cdk9HA expression was confirmed by Western blot and immunofluorescence analysis (IFA).
Recombinant cdk9 HCMV BAC cloning and virus production.
Construction of a recombinant cdk9 HCMV bacterial artificial chromosome (BAC) was performed using a method for the expression of exogenous genes in the HCMV BAC that has been previously described in detail (9, 45). Recombinant cdk9HA BAC constructs (wt and D167N mutant), under the control of HCMV IE promoter, were inserted between US11 and US12 by the Tn7-mediated transposition method (20). Briefly, pRC-wt-cdk9HA and pRC-D167N-cdk9HA were digested with XbaI and EcoRI and were cloned between the Tn7 arms of XbaI- and EcoRI-cut pFastBac1:IE vector (45). The pFB11:IE-wt-cdk9HA and pFB11:IE-D167N-cdk9HA constructs were transformed into DH10B bacteria containing the pHB5:LacZ/att BAC and the transposase plasmid, pMON7124 (20). Replacement of lacZ/att with a recombinant cdk9HA gene was selected for with chloramphenicol, tetracycline, and gentamicin on plates with blue-white colony screening (Invitrogen). White colonies, indicative of a transposition event, were screened for insertion of the recombinant IE-cdk9HA cassette by PCR. Positive clones were confirmed by DNA sequencing and detailed restriction enzyme analysis using a field inversion gel electrophoresis mapper (Bio-Rad) (data not shown). The resulting BACs were named HB5:IE-wt-cdk9HA and HB5:IE-D167N-cdk9HA.
Recombinant HB5:IE-cdk9HA BAC viruses were reconstituted by electroporation into HFF cells as described previously (45, 35). HB5:IE-wt-cdk9HA virus was collected from the cell supernatant at day 16 postelectroporation, and HB5:IE-D167N-cdk9HA virus was collected at day 20, after the infection had spread throughout the entire monolayer of cells. The virus was checked by sequencing of the altered region of the viral DNA to confirm that it had not reverted to the wt HB5 virus. Stocks of recombinant cdk9HA viruses were prepared and were subjected to titration by a plaque assay as previously described (48).
Infections and drug treatments.
HFF cells (passages 15 to 21) and constitutively expressing wt-, S175D-, E55A-, E57A-, and R65A-cdk9HA cells were synchronized in G0 phase by allowing them to grow to confluence as previously described (41). Three days after confluence, the cells were trypsinized, replated at a lower density (without selective media) to allow progression into the cell cycle, and infected at a multiplicity of infection (MOI) of 5 with HCMV Towne or were mock infected with tissue culture supernatants. The Dox-inducible S175A- and E55A/E57A/R65A-cdk9HA cell lines were trypsinized and induced with 5 μg/ml Dox for 3 to 4 days prior to infection. Infections were performed in the presence of 1 μg/ml Dox (Clontech, Palo Alto, CA) and done in parallel with infection of untransduced HFF cells in the presence of 1 μg/ml Dox to confirm that Dox had no effect on the progression of the infection for up to 8 h p.i. At various times p.i., cells were harvested and stored at −80°C for Western blot analysis or fixed in 2% formaldehyde (FA) solution for IFA. For recombinant HCMV infections with wt-cdk9HA and D167N-cdk9HA viruses and with wt IE2 86-EGFP and IE2 86 ΔSX-EGFP viruses, cells were infected on coverslips at an MOI of 5 as described above.
For infections in which CHX (Sigma-Aldrich, St. Louis, MO) was added at the beginning of the infection, HFF cells were infected at an MOI of 3 with viral or mock supernatant at 1 h post-G0 release and seeding. At 30 min p.i., the medium was replaced with CHX (100 μg/ml) or control medium. At 8 h p.i., cells were fixed with 2% FA. For infections in which the addition of CHX was delayed, Go-synchronized HFF cells were released into G1 and infected at an MOI of 5 with HCMV Towne supernatant or mock supernatant. At 8 h p.i., 100 μg/ml CHX or control medium was added, and cells were fixed for IFA at 14 h p.i. For infections with actinomycin D (Act D; Calbiochem, San Diego, CA), Go-released HFF cells were seeded onto coverslips and infected at an MOI of 5 with viral or mock supernatant. At 5 or 10 h p.i., the medium was replaced with Act D (10 μg/ml) or control medium. Cells were fixed at 10 and 14 h p.i. with 2% FA. For infections in the presence of Roscovitine (Calbiochem), Flavopiridol (gift from J. Brady, National Institutes of Health), or DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole; Sigma-Aldrich), G0-synchronized HFF cells were infected upon release into G1 in the presence of 16 μM Roscovitine, 50 nM Flavopiridol, 15 μM DRB, or control medium and seeded onto glass coverslips or 3-cm-diameter dishes. Cells were harvested for Western blot analysis or fixed for IFA at 8 h p.i.
Antibodies.
CH16.0 monoclonal antibody (MAb) and UL44 MAb (Virusys Corporation, Sykesville, MD); cdk9 sc-484 polyclonal antibody, hemagglutinin (HA) sc-805 polyclonal antibody, and HA sc-7392 MAb (Santa Cruz Biotechnology, Santa Cruz, CA); β-actin MAb (Sigma-Aldrich); IE2 MAb (Chemicon, Temecula, CA); cyclin T1 NCL MAb (Novocastra, Newcastle on Tyne, United Kingdom); H5 MAb (Covance, Berkeley, CA); pp65 MAb (gift from W. Britt); goat anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) and goat anti-rabbit IgG-HRP (Calbiochem); goat anti-mouse IgM-HRP, goat anti-rabbit IgG-Cy3, fluorescein-conjugated isotype-specific secondary antibodies, and normal rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA); and mouse isotype-specific Ig controls (Zymed, San Francisco, CA) were used for the experiments.
Western blot analysis.
Cells were lysed in Laemmli reducing sample buffer (50 mM Tris [pH 6.8], 0.2% sodium dodecyl sulfate [SDS], 10% glycerol, 5% 2-β-mercaptoethanol, 50 mM NaF, 0.5 mM Na3VO4, 5 mM β-glycerophosphate). The lysates were then sonicated briefly and boiled for 5 min. Proteins from an equivalent number of cells were separated on a 6% or 8% SDS-polyacrylamide gel and transferred to nitrocellulose. Membranes were stained with amido black to assess protein loading in each lane. The blots were incubated with 5% nonfat dried milk in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, and 0.1% Tween 20) and incubated with primary antibodies diluted in 5% milk-TBST as follows: rabbit anti-cdk9, 1:200; cyclin T1 MAb, 1:250; β-actin MAb, 1:10,000; H5 MAb, 1:250; CH16.0 MAb, 1:10,000. Following washes with TBST, blots were incubated with anti-mouse IgG-HRP, anti-rabbit IgG-HRP diluted 1:2,000, or anti-mouse IgM-HRP diluted 1:8,000. After being washed in TBST, proteins were detected using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) according to the manufacturer's instructions. For detection of cyclin T1, SuperSignal Femto (Pierce) diluted 1:5 in double-distilled water was used.
Immunofluorescence.
Cells were plated onto sterile glass coverslips and infected as described above. At specified time points, the coverslips were washed in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and fixed with 2% FA solution in PBS for 10 min. Formaldehyde-fixed cells were permeabilized in 0.2% Triton X-100 and incubated in 10% normal goat serum. Cells were subsequently incubated with one or more of the following specific antibodies diluted in 10% normal goat serum: IE2 MAb, 1:500; rabbit anti-cdk9, 1:50; HA MAb, 1:20; pp65 MAb, 1:500; or CH16.0, 1:1,000. Nonspecific IgG antibodies were used as controls. After PBS washes, cells were incubated with Hoechst stain and the appropriate fluorescein-conjugated secondary antibodies diluted in blocking solution, namely, isotype-specific mouse IgG (1:50) or goat anti-rabbit IgG-Cy3 (1:600). After PBS washes, the slips were mounted onto glass slides with SlowFade Gold (Molecular Probes, Eugene, OR). Confocal images were captured at the Cancer Center Digital Imaging Shared Resource of the University of California, San Diego, by using a Deltavision microscope and were deconvolved with SoftWorx software. All images represent 0.2-μm sections and ×1,000 magnification under conditions of oil immersion.
Immunoprecipitations.
Uninfected cdk9HA-expressing cells were harvested by trypsinization, and 4 × 105 cells were lysed in NETN buffer (100 mM NaCl, 20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5% IGEPAL CA-630 [Sigma-Aldrich], 1× protease inhibitor cocktail [Roche Chemicals, Indianapolis, IN], 50 mM NaF, 0.5 mM Na3VO4, and 5 mM β-glycerophosphate) by rotation at 4°C for 30 min. Cell lysates were immunoprecipitated with 15 μg of polyclonal rabbit anti-HA, polyclonal rabbit anti-cdk9, or a nonspecific rabbit IgG (RbIgG) control coupled to 30 μl of protein G plus agarose beads (Santa Cruz Biotechnology) with dimethyl pimelimidate dihydrochloride (Sigma-Aldrich). Lysates and antibody-coupled beads were incubated overnight with rotation at 4°C. Immunocomplexes were washed three times in NETN buffer, and each sample was boiled in 2× Laemmli buffer for Western blotting. For Western blot analysis, approximately 6 × 104 cells were loaded for the pre-immunoprecipitation (pre-IP) and post-IP samples, and 3.4 × 105 cells were loaded for the IP samples.
RESULTS
Formation of the viral transcriptosomes requires de novo protein synthesis at the beginning of the infection.
The HCMV transcriptosomes are the designated regions of viral IE transcription and the sites to which viral and cellular proteins are actively recruited. In addition to the input viral genome, these sites contain IE2, UL112-113, UL69, and several cellular transcription regulators and chromatin-modifying proteins, including RNAP II (RNAP IIa and IIo) and its kinases cdk9 and cdk7, cyclin T1, Brd4, HDAC1, and HDAC2 (1, 3, 23, 24, 27, 39, 49). The size and abundance of the viral transcriptosomes increase as the infection progresses, until they eventually develop into viral replication compartments during the early stage of the infection (1, 47).
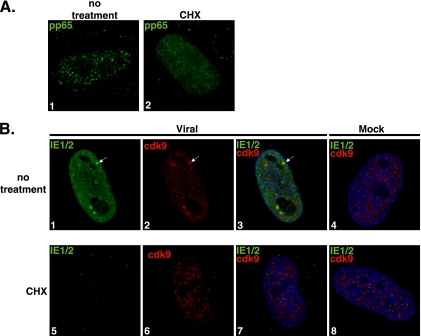
The exact factors and processes required for nucleating the viral transcriptosome and recruiting cellular proteins are not known. Since cdk9 is a major positive regulator of transcription, we were interested in whether viral IE RNA synthesis was sufficient for efficient recruitment of cdk9 to the viral genome. We sought to address this possibility by using conditions under which cellular and viral transcription can still occur, but protein synthesis was inhibited. Synchronized HFF cells were released into G1 and infected at an MOI of 5 with virus or mock supernatant. At 30 min p.i., the medium was replaced with CHX protein synthesis inhibitor (100 μg/ml) or with control medium. At 8 h p.i., cells were fixed for IFA. Figure 1A shows that the results of pp65 entry into the host cell and its localization to the nucleus were comparable for infections in both the absence (panel 1) and the presence (panel 2) of CHX, and no pp65 staining was detected in any of the mock samples (data not shown). Costaining with CH16.0 MAb, which detects both IE1 and IE2, and with anti-cdk9 showed that in the absence of CHX, there was expression of IE1 and IE2 protein (Fig. 1B, panels 1 to 3). As expected, although active transcription was taking place in the presence of CHX, no protein synthesis was occurring and therefore no viral IE protein could be detected at 8 h p.i. in the treated cells (Fig. 1B, panel 5). In the presence of the protein synthesis inhibitor, there was also no development of cdk9 aggregates (panels 5 to 7). CHX treatment did not affect the diffuse cdk9 localization in mock-treated infections (panel 8). These data show that viral entry, translocation to the nucleus, and ongoing RNA synthesis are not sufficient for viral transcriptosome formation.
FIG. 1.
Formation of viral transcriptosomes requires de novo protein synthesis. G0-synchronized cells were released into G1, infected with HCMV Towne (MOI of 5), and seeded onto glass coverslips. At 30 min p.i.,100 μg/ml CHX or control medium was added. Mock infections were performed under the same conditions. At 8 h p.i., cells were washed with PBS, fixed with 2% FA, permeabilized, and stained with antibodies specific for pp65 (A) and for IE1/2 and cdk9 (B). Fluorescein isothiocyanate- and Cy3-conjugated isotype-specific secondary antibodies were used. Nuclei were stained with Hoechst dye. White arrows show examples of viral transcriptosomes, although more are present. All of the images represent confocal optical 0.2-μm sections at a magnification of ×1,000 under conditions of oil immersion.
Viral IE RNA and protein synthesis are not sufficient for cdk9 accumulation at viral transcriptosomes.
In order to address whether the proteins synthesized during IE and early stages of the infection are sufficient for viral transcriptosome formation and cdk9 recruitment or whether continued transcription into early stages of the infection is also required, the following experiment was performed. Synchronized HFF cells were released into G1 and seeded onto coverslips. At 1 h postseeding, cells were infected at an MOI of 5. At 5 h p.i., Act D or control medium was added, and cells were fixed for IFA at 10 and 14 h p.i. (Fig. 2A and B). The addition of Act D at 5 h p.i. prevented any further viral gene transcription but protein synthesis was able to continue. Under these conditions, IE proteins were present, but very little early transcription would have occurred and accumulation of early proteins would have been limited. In a subset of samples, the addition of Act D or control medium was delayed until 10 h p.i., and the cells were fixed at 14 h p.i. (Fig. 2B). In the drug-treated cells, IE and some early proteins would have been present and the IE2 and cdk9 would already have colocalized at the transcriptosomes, but no further transcription could have occurred. The presence of Act D during these intervals has no significant effect on the steady-state levels of the IE proteins or cdk9 (data not shown).
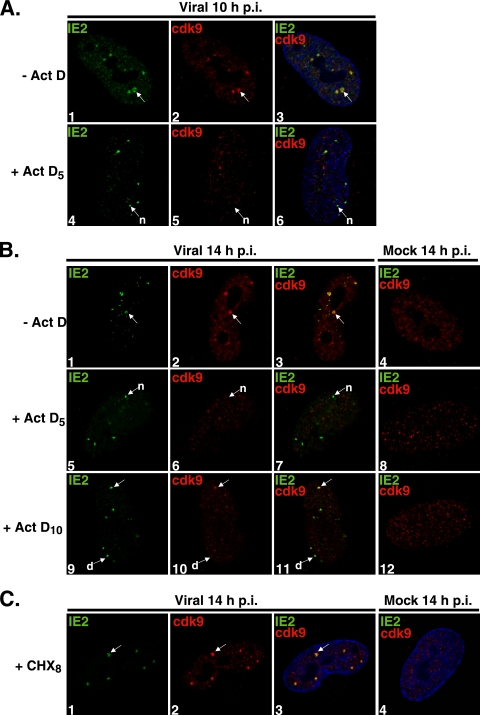
FIG. 2.
Cdk9 localization at the viral transcriptosomes requires active transcription. G0-synchronized cells were released into G1 and seeded onto glass coverslips. One hour later, cells were infected with HCMV Towne (MOI of 5) or with mock infection supernatant. (A) At 5 h p.i., Act D (10 μg/ml) (+ Act D5) or control medium (− Act D) was added; at 10 h p.i., cells were fixed for IFA. (B) Act D or control medium (− Act D) was added at 5 h p.i. (+ Act D5) or at 10 h p.i. (+ Act D10), and cells were fixed at 14 h p.i. for IFA. (C) At 8 h p.i., cells were treated with CHX (100 μg/ml) (+ CHX8) or control medium; cells were fixed at 14 h p.i. for IFA. Cells were stained with antibodies specific for IE2 and cdk9, followed by fluorescein isothiocyanate- and Cy3-conjugated isotype-specific secondary antibodies. Nuclei were stained with Hoechst dye. The white arrows marked “n” indicate viral transcriptosomes that do not show colocalization between stained proteins, the arrows marked “d” show decreased cdk9 staining at viral transcriptosomes, and the unmarked white arrows point to examples of colocalization. All of the images represent confocal optical 0.2-μm sections at a magnification of ×1,000 under conditions of oil immersion.
Figure 2A shows the localization of cdk9 with respect to IE2 localization at 10 h p.i. In the absence of any drug treatment, IE2 protein expression could be detected and viral transcriptosomes with accumulation of both IE2 and cdk9 were observed (panels 1 to 3). However, the addition of Act D at 5 h p.i. prevented the accumulation of cdk9 at viral transcriptosomes, although foci of IE2 were still present (panels 4 to 6). These results show that the viral transcripts were synthesized before 5 h p.i. and that the proteins they had encoded were not sufficient for the accumulation of cdk9 at the viral transcriptosomes.
To determine whether the recruitment of cdk9 at the viral transcriptosomes was inhibited or merely delayed, samples fixed at 14 h p.i. were examined (Fig. 2B). As expected, at 14 h p.i., the viral transcriptosomes were still detected in the absence of Act D by staining for IE2 and cdk9 (panels 1 to 3). The absence of cdk9 recruitment to the viral transcriptosomes during Act D treatment, however, is not the result of a delayed process. Introducing Act D at 5 h p.i. and waiting until 14 h p.i. still did not allow any cdk9 aggregates to develop and colocalize with IE2 (panels 5 to 7).
As shown in Fig. 2A, IE2 and cdk9 proteins are localized to the viral transcriptosomes by 10 h p.i. Therefore, delaying the addition of Act D until 10 h p.i. would effectively inhibit transcription after IE2 and cdk9 have colocalized at the viral transcriptosomes. The late addition of Act D should have also permitted some early gene expression. Surprisingly, when the addition of Act D was delayed until 10 h p.i. and the cells were fixed at 14 h p.i., foci of IE2 were still observed, but the concentration of cdk9 at these sites was markedly decreased or absent (Fig. 2B, panels 9 to 11). Figure 2B (panels 9 to 11) also shows an example of a cell in which one of the transcriptosomes in the nucleus contained both IE2 and cdk9. The loss of cdk9 at the viral transcriptosomes in the presence of Act D was not an immediate event, because treatment with Act D for 1 h at 10 h p.i. did not result in a complete loss of cdk9 from the viral transcriptosomes (data not shown).
Interestingly, inhibition of protein synthesis at 8 h p.i., after viral transcriptosomes have been formed, did not result in a loss of IE2 or cdk9 from those sites (Fig. 2C). Together, these data show that transcription and de novo protein synthesis are both required for establishment of viral transcriptosomes and that continuous transcription is required in order to maintain cdk9 already localized at viral transcriptosomes.
Cdk9 recruitment to the viral transcriptosomes is delayed during infection with an IE2 86 ΔSX-EGFP virus.
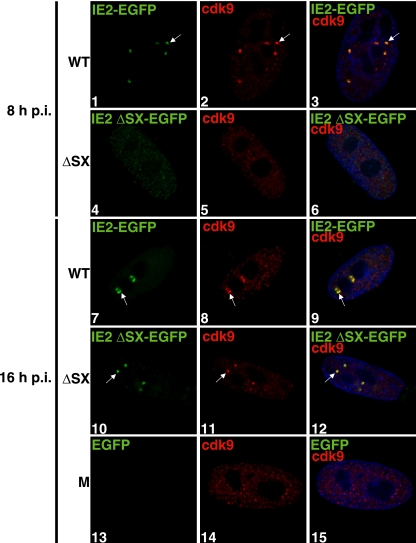
As has already been reported, several viral factors are present at the viral transcriptosomes, including IE2. During infection at a high MOI, IE2 can be detected by immunofluorescence at the viral transcriptosomes as early as 2 h p.i. (27, 49). IE2 has been implicated in interacting with many cellular proteins, including transcription factors such as TBP and TFIIB, by in vitro binding assays (19, 46). We wanted to determine whether IE2 plays a role in the recruitment of cdk9 to the viral transcriptosomes in vivo during infection. We addressed this question by examining cdk9 localization during infection with the IE2 86 ΔSX-EGFP virus, which has a deletion between amino acids 136 and 290 in exon 5 and is fused to EGFP at the C terminus of IE2 (42). This virus produces greatly reduced levels of the IE2 protein. Synchronized HFF cells were released into G1 and infected at an MOI of 5 with wt IE2 86-EGFP virus, IE2 86 ΔSX-EGFP virus, or an equivalent volume of mock supernatant. The cells were fixed at 8 and 16 h p.i. for IFA. Figure 3 shows that at 8 h p.i., wt IE2 86-EGFP and cdk9 colocalize at the viral transcriptosomes in cells infected with the wt IE2 86-EGFP virus (panels 1 to 3). However, at 8 h p.i., IE2 86 ΔSX-EGFP expression as determined by immunofluorescence was still very low, and no cells contained any obvious cdk9 aggregates (panels 4 to 6; the brightness of the image elements representing the intensity of EGFP was increased in those panels for visualization purposes).
FIG. 3.
Cdk9 recruitment to the viral transcriptosomes is delayed during infection at a high MOI with the IE2 86 ΔSX-EGFP virus. G0-synchronized cells were released into G1, infected with HCMV IE2 86 ΔSX-EGFP or IE2 86 wt-EGFP virus at an MOI of 5, or treated with mock supernatant and were seeded onto glass coverslips. At 8 (panels 1 to 6) and 16 (panels 7 to 15) h p.i., cells were washed with PBS, fixed with 2% FA, permeabilized, and stained with anti-cdk9 and Cy3-conjugated secondary antibodies. Nuclei were stained with Hoechst dye. The brightness of the image elements representing the intensity of EGFP expression in panels 4 to 6 was increased by use of the Softworx program for visualization purposes. Mock (M) infected samples at 16 h p.i. are represented by panels 13 to 15. The white arrows point to examples of colocalization. All of the images represent confocal optical 0.2-μm sections at a magnification of ×1,000 under conditions of oil immersion. wt, IE2 86 wt-EGFP virus; ΔSX, IE2 86 ΔSX-EGFP virus.
At 16 h p.i., wt IE2 86-EGFP and cdk9 were maintained at the viral transcriptosomes (Fig. 3, panels 7 to 9). By this time, the mutant IE2 86 ΔSX-EGFP protein also formed visible aggregates in the nucleus (panel 10). These IE2 86 ΔSX-EGFP proteins colocalized with readily detectable cdk9 aggregates (panels 10 to 12). Mock-treated cells did not show any EGFP fluorescence and did not develop cdk9 aggregates (panels 13 to 15). These data indicate that although full-length IE2 86 is not essential for cdk9 localization to the viral transcriptosomes during infection at a high MOI, a threshold level of IE2 is required.
Cdk9 kinase activity and binding to Brd4 are not required for cdk9 recruitment to the viral transcriptosomes.
We previously reported that in the presence of the cdk inhibitor, Roscovitine, cdk9 and cdk7 localization to the viral transcriptosomes is impaired but IE2 localization to those sites is not (27). Although Roscovitine treatment leads to a decrease in cdk9 protein and RNA levels compared to control sample results, we showed that the decrease in protein levels does not account for the impaired localization to the viral transcriptosomes. It has also been reported that the status of cdk9 kinase activity can affect cdk9 localization in uninfected cells (37).
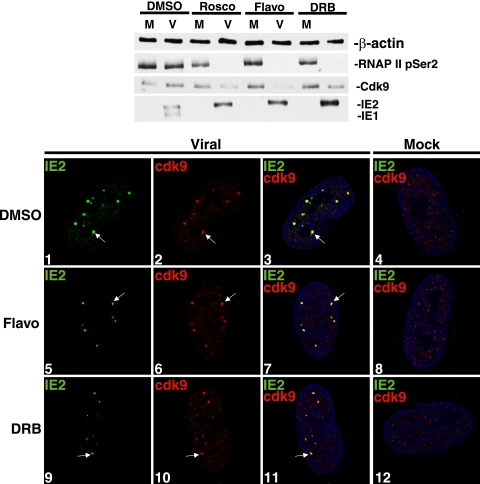
In order to assess further whether inhibition of cdk9 activity prevents cdk9 localization to the viral transcriptosomes, we compared the effects of Roscovitine with those of Flavopiridol and DRB. The latter two drugs have a different inhibitory profile and greater specificity for cdk9 compared to other kinases. Cells were released into G1 and infected at an MOI of 5 with viral or mock supernatant in the presence of an inhibitor drug or control medium. At 8 h p.i., cells were harvested for Western blot analysis (Fig. 4, top panel). The protein levels for cdk9 were lower in the presence of all three inhibitors compared to the levels seen with the control samples in infected cells at 8 h p.i. Virus-specific decreases in the level of hyperphosphorylated RNAP IIo were also observed in the presence of each of the inhibitors compared to control sample results for phosphorylation of serine 2 residues of the RNAP II CTD.
FIG. 4.
Cdk9 kinase activity is not a requirement for its recruitment to the viral transcriptosomes. G0-synchronized cells were released into G1 and infected with HCMV Towne (V; MOI of 5) or mock (M) supernatant. Infections took place in the presence of Roscovitine (Rosco; 16 μM), Flavopiridol (Flavo; 50 nM), DRB (15 μM), or control dimethylsulfoxide (DMSO). Cells were harvested at 8 h p.i. Total cell lysates from equal numbers of cells were subjected to Western blotting using H5 (which detects pSer2-RNAP IIo), anti-cdk9, and CH16.0 (which detects IE1/2). β-actin protein levels were checked as a loading control. Cells were fixed for IFA at 8 h p.i. and stained with antibodies specific for IE2 and cdk9, followed by fluorescein isothiocyanate- and Cy3-conjugated isotype-specific secondary antibodies, which were used for IFA. Nuclei were stained with Hoechst dye. The white arrow in each panel indicates a region of colocalization, although more are present. All of the images represent confocal optical 0.2-μm sections at a magnification of ×1,000 under conditions of oil immersion.
The progression of the infection was greatly altered by treatment with each of these inhibitors. We have already reported that, in similarity to infection in the presence of Roscovitine treatment, treatment with Flavopiridol alters the ratio of the IE1 and IE2 transcripts during the infection and inhibits viral early protein expression (43). Figure 4 shows that treatment with DRB also led to an altered accumulation of IE1 and IE2 protein such that IE2 was present at greater levels than IE1 protein. When the viral early proteins were examined at later time points, comparable decreases in viral UL44 were also observed (data not shown). These results show that the addition of several inhibitors of cdk9 at the beginning of the infection alters the expression of viral proteins.
To determine the effect of the drugs on localization of cdk9, cells were infected at an MOI of 5 in the presence of Flavopiridol, DRB, or control medium and were fixed at 8 h p.i. for immunostaining (Fig. 4, bottom). As expected, the control-treated infected samples displayed colocalization between IE2 and cdk9 at the viral transcriptosomes (panels 1 to 3). However, in contrast to what was observed when cells were treated with Roscovitine, cdk9 colocalized with IE2 at nuclear aggregates in the presence of both Flavopiridol (panels 5 to 7) and DRB (panels 9 to 11). Treatment with Flavopiridol and DRB did not affect cdk9 localization in uninfected cells (panels 8 and 12). These data suggest that cdk9 kinase activity is not essential for accumulation at the viral transcriptosomes.
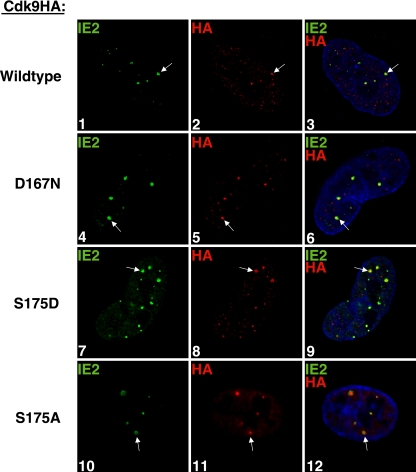
Although the results described above indicate that cdk9 kinase activity is not required for recruitment of cdk9, the fact that these kinase inhibitors may differentially affect the activities of other kinases complicates the interpretation. To further investigate whether cdk9 activity is required for recruitment to the viral transcriptosomes, we constructed two recombinant viruses that expressed at IE times a kinase-inactive cdk9 mutant (D167N) that is C-terminally tagged with HA and a wt HA-tagged cdk9. This was accomplished by cloning the wt or kinase-inactive cdk9 driven by the major IE promoter into a nonessential region of the AD169 HCMV HB5 BAC. The recombinant BAC DNAs were electroporated into HFF cells, and the titered viral supernatants were then used to infect HFF cells at an MOI of 5.
The wt-cdk9HA colocalized with IE2 at the viral transcriptosomes at 8 h p.i., as detected by an IFA with antibodies against HA and IE2 (Fig. 5, panels 1 to 3). The kinase-inactive D167N-cdk9HA, detected by HA, also colocalized with IE2 at the viral transcriptosomes (panels 4 to 6). Together, these data suggest that the kinase activity of cdk9 alone is not the determining factor for its recruitment to the sites of viral IE transcription. However, these experiments do not exclude the possibility that another property of cdk9 in conjunction with cdk9 activity may regulate cdk9 localization to the viral transcriptosomes during the infection.
FIG. 5.
Neither cdk9 kinase activity nor binding to Brd4 is required for cdk9 localization at the viral transcriptosomes. G0-synchronized cells were released into G1, infected at an MOI of 5 with wt-cdk9HA HCMV (panels 1 to 3) or D167N-cdk9HA HCMV (panels 4 to 7), and seeded onto glass coverslips. The S175A-cdk9HA-expressing cells were induced with Dox as described in Materials and Methods. S175D- and S175A-cdk9HA cells were infected with HCMV Towne (MOI of 5) and seeded onto glass coverslips (panels 7 to 9 and 10 to 12, respectively). At 8 h p.i., cells were washed with PBS, fixed with 2% FA, permeabilized, and costained with antibodies specific for IE2 and HA (to detect exogenous cdk9HA). Fluorescein isothiocyanate- and tetramethylrhodamine isothiocyanate-conjugated isotype-specific secondary antibodies were used. Nuclei were stained with Hoechst dye. The white arrows point to examples of colocalization. All of the images represent confocal optical 0.2-μm sections at a magnification of ×1,000 under conditions of oil immersion.
Since cdk9 activity is not a requirement for cdk9 localization to the viral transcriptosomes, we sought to determine what other properties of cdk9 might contribute to its localization at these sites. It is known that cdk9 forms an active complex with cyclin T1 called the P-TEFb complex. P-TEFb has also been reported to bind with Brd4, enhancing transcription elongation efficiency (26, 52). We have already shown that both cyclin T1 and Brd4 are found at the viral transcriptosomes, and, unlike the effects on cdk9 and cdk7, treatment with Roscovitine does not impair their recruitment to those sites (27). In an attempt to determine whether binding to Brd4 may be required for cdk9 recruitment to the viral transcriptosomes, site-directed mutagenesis was used to create two C-terminally HA-tagged cdk9 mutants that have previously been shown to have altered kinase activity and binding to Brd4 (6, 26, 28, 31, 52, 53). S175D-cdk9HA was kinase active but no longer bound to Brd4, and S175A-cdk9HA lacked both kinase activity and binding to Brd4. These mutants were then cloned into lentiviral constructs and used to create stable HFF cell lines that express the cdk9HA mutants, either constitutively (S175D-cdk9HA) or by Dox induction (S175A-cdk9HA), as described in Materials and Methods.
Each of these recombinant cdk9HA cell lines was infected with HCMV at an MOI of 5, and at 8 h p.i., they were processed for IFA by staining with anti-IE2 antibody and an anti-HA antibody to detect the cdk9HA mutant (Fig. 5). We previously showed that exogenous wt-cdk9HA in recombinant cells infected with HCMV localized at the viral transcriptosomes with IE2 (27). In Fig. 5, we show that HCMV infection of cells expressing either S175D- or S175A-cdk9HA also resulted in the mutant cdk9HA colocalizing with IE2 at the viral transcriptosomes (panels 7 to 9 or 10 to 12, respectively). Since both S175D-cdk9HA and S175A-cdk9HA are unable to bind to Brd4 but S175D-cdk9HA is active with respect to kinase, these data indicate that neither cdk9 binding to Brd4 nor cdk9 kinase activity is required for cdk9 localization at the sites of IE transcription.
Efficient binding of cdk9 to cyclin T1 is required for recruitment of cdk9 to the transcriptosome.
To determine whether binding to cyclin T1 is necessary for the accumulation of cdk9 at the transcriptosomes, we constructed HFF cell lines that expressed cdk9HA mutants in which the residues E55, E57, and R65 on cdk9 were individually changed to alanine. These sites were chosen before the structure of the cdk9-cyclin T1 complex had been solved (6), and their selection was based on structural analysis of the cdk2-cyclin A complex that demonstrated that the corresponding residues on cdk2 (E40, E42, and R50) interacted directly with cyclin A (10). The recently published structure of the cdk9-cyclin T1 complex now shows that E57 interacts directly with cyclin T1 and that R65 both contributes to the positively charged surface of cdk9 and is adjacent to the residue L64 that does contact cyclin T1.
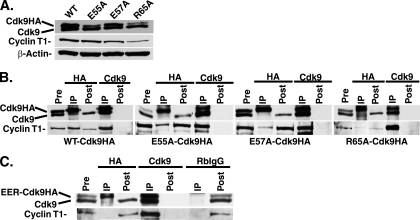
Figure 6A shows that the mutant cell lines express differing amounts of the exogenous mutant cdk9HA but very similar levels of endogenous cdk9. It should be noted that in cells that constitutively expressed R65A-cdk9HA, the level of cyclin T1 in the cell lysates was slightly lower. The ability of each cdk9HA mutant to bind to cyclin T1 was analyzed in co-IP experiments (Fig. 6B). For each cell line, we immunoprecipitated lysates with either an anti-HA antibody (immunoprecipitating only those complexes containing exogenous cdk9HA) or an anti-cdk9 antibody (immunoprecipitating all complexes containing endogenous cdk9 and exogenous cdk9HA). The amount of cyclin T1 that was present in the co-IP was determined by Western blot analysis. Again, the differences in exogenous mutant cdk9HA compared to endogenous cdk9 results are reflected in the pre-IP lanes.
FIG. 6.
Amino acid changes in the N terminus of cdk9HA result in differential cyclin T1 binding. (A) The cell lines constitutively expressing wt-, E55A-, E57A-, and R65A-cdk9HA were lysed in 1× NETN buffer, prepared using 8% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and checked by Western blot analysis for the presence of cdk9 (endogenous and exogenous) and cyclin T1 protein. β-actin served as a loading control. (B) The cell lines constitutively expressing wt-, E55A-, E57A-, and R65A-cdk9HA were lysed in 1× NETN buffer and subjected to IP with antibodies for HA or cdk9. The IP, pre-IP, and post-IP lysates were prepared using 8% SDS-PAGE and checked by Western blot analysis for cdk9 and cyclin T1 protein. The E55A-cdk9HA and E57A-cdk9HA IP experiments were run on the same gel. The pre- and post-IP lanes represent 20% cell equivalents of the IP lanes. (C) The cell line expressing E55A/E57A/R65A-cdk9HA (EER-cdk9HA) was induced with Dox as described in Materials and Methods, lysed in 1× NETN buffer, and subjected to IP with antibodies for HA, cdk9, and an RbIgG control. The immunocomplexes (IP), pre-IP, and post-IP lysates were prepared using 8% SDS-PAGE and checked by Western blot analysis for the presence of cdk9 (endogenous and exogenous) and cyclin T1 protein.
For each cell line, the total cdk9 IP resulted in isolation of both the endogenous cdk9 and slower migrating wt or mutant cdk9HA. The lysates were immunodepleted of cdk9, as neither form could be detected in the post-IP lanes. Analysis of the Western blots with the antibody to cyclin T1 also showed that all of the cyclin T1 was in the cdk9 immunoprecipitate.
To assess the relative amount of cyclin T1 that was in a complex with the exogenous cdk9, we immunoprecipitated the lysates with the anti-HA antibody. When the lysates from each of the cell lines were immunoprecipitated with the anti-HA antibody, only the exogenous cdk9HA form was present in the immunoprecipitate. The post-IP samples show that the exogenous form was immunodepleted whereas the endogenous form remained. For the lysates from the wt-cdk9HA cell line, approximately 50% of total cyclin T1 was present in the HA co-IP, which was expected, since the exogenous cdk9HA comprises approximately 50% of the total cdk9 in the cell. When similar co-IP analyses with the anti-HA antibody were done with lysates from the E55A-cdk9HA, E57A-cdk9HA, and R65A-cdk9HA cell lines, significantly reduced levels of cyclin T1 were immunoprecipitated compared to the levels seen with wt-cdk9HA co-IP. The relative amount of cyclin T1 in complex with the exogenous cdk9 can be visualized by comparing the cyclin T1 present in the HA co-IP to the amount present in total cdk9 co-IP. It should be noted that the E55A-cdk9HA cell line expresses less exogenous cdk9HA than endogenous cdk9, while the E57A-cdk9HA cell line shows comparable levels of exogenous and endogenous cdk9. The fact that the amount of cyclin T1 in complex with E55A-cdk9HA relative to that in complex with total cdk9 is greater than that in complex with the E57A-cdk9HA suggests that E55A-cdk9HA binds to cyclin T1 more efficiently than E57A-cdk9HA. It also appears that R65A-cdk9HA binds less efficiently to cyclin T1 than E55A-cdk9HA. Only with long exposures of the blot can the cyclin T1 be visualized. Although there is a slightly lower level of total cyclin T1 in the R65A-cdk9HA cell line, the relative amount of cyclin T1 that can co-IP with the R65A-cdk9HA, versus the total cdk9 amount, is still substantially lower than the amounts seen with the wt- and E55A-cdk9HA cell lines.
Since each of the single mutants was able to form a complex with a small amount of cyclin T1, we constructed a Dox-inducible cell line that expressed a cdk9HA mutant with all three substitutions. In Fig. 6C, we show that IP of lysates from the cell line expressing the cdk9 triple mutant (E55A/E57A/R65A-cdk9HA) with an antibody directed against HA did not yield any detectable cyclin T1 (even with long exposures of the blot), whereas the cyclin T1 did co-IP when the lysates were immunoprecipitated with a cdk9 antibody directed against total cdk9 (both endogenous and exogenous). As a control, lysates were also immunoprecipitated with nonspecific RbIgG, and neither cdk9 nor cyclin T1 was present in the immunoprecipitate.
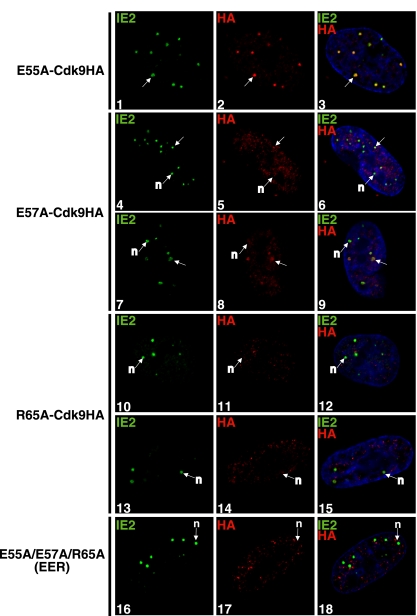
To determine whether the binding to cyclin T1 correlates with the recruitment of the mutant cdk9 to the viral transcriptosomes, these cell lines were then infected at an MOI of 5 and processed at 8 h p.i. for IFA by staining with an anti-IE2 antibody and an anti-HA antibody to detect the exogenous cdk9HA (Fig. 7). E55A-cdk9HA still colocalized with IE2 (panels 1 to 3). When E57, which contacts cyclin T1, was mutated to alanine, an impairment in E57A-cdk9HA localization to the viral transcriptosomes was observed, and several IE2 spots in each cell did not have any detectable cdk9 (panels 4 to 9). The R65A-cdk9HA mutant, which complexes with cyclin T1 less efficiently than the wt- and E55A-cdk9HA mutants, also did not localize to the viral transcriptosomes with IE2, even in well-developed transcriptosomes (panels 10 to 15). Finally, in the cells expressing the cdk9HA with three mutations (E55A/E57A/R65A-cdk9HA), which is unable to bind to cyclin T1, it was clear that although IE2 was present at the viral transcriptosomes, the mutant cdk9 did not form aggregates in the infected cell nucleus and did not colocalize with the IE2 spots (Fig. 7, panels 16 to 18). Taken together, these data show that efficient cdk9 binding to cyclin T1 is required for cdk9 recruitment to the viral transcriptosomes.
FIG. 7.
Amino acid changes in the N terminus of cdk9HA result in differential localization to the viral transcriptosomes. The cell lines expressing constitutive E55A-, E57A, and R65A-, and Dox-induced E55A/E57A/R65A-cdk9HA virus were infected with HCMV Towne (MOI of 5) and seeded onto glass coverslips. At 8 h p.i., cells were washed with PBS, fixed with 2% FA, permeabilized, and costained with antibodies specific for IE2 and HA (to detect exogenous cdk9HA). Fluorescein isothiocyanate- and tetramethylrhodamine isothiocyanate-conjugated isotype-specific secondary antibodies were used. Nuclei were stained with Hoechst dye. The white arrows marked “n” indicate viral transcriptosomes with no colocalization between stained proteins, and the unmarked white arrows show examples of colocalization. All of the images represent confocal optical 0.2-μm sections at a magnification of ×1,000 under conditions of oil immersion.
DISCUSSION
In this study, we used inhibitor drug treatments, infection with recombinant HCMV expressing wt or mutant cdk9, and infection of cells that expressed exogenous mutant cdk9 to determine the cellular and viral factors that contribute to cdk9 recruitment. Data from the infection in the presence of CHX, which allows accumulation of viral IE transcripts as well as cellular RNA synthesis, show that there is an additional requirement for protein synthesis, at least initially, in the infection. The presence of the viral RNA in addition to the functional viral genome and input tegument proteins is still not sufficient for recruitment of cdk9 to the viral transcriptosomes.
The results of experiments with Act D raise the issue of why inhibition of viral transcription would affect cdk9 localization. One possibility is that active transcription at the viral transcriptosomes promotes cdk9 recruitment due to a high abundance of other components of the transcription machinery. For example, RNAP II, cyclin T1, DSIF, and Brd4 are factors that interact with cdk9 during productive transcription, and each of these proteins is highly concentrated at the viral transcriptosomes (27). Another possibility is that only newly synthesized cdk9 has the ability to be recruited to the viral transcriptosomes. Previously synthesized cdk9 that is already associated in inactive or active complexes may not be readily disrupted by the infection. There is an induction of cdk9 RNA during the infection, and all of the components of P-TEFb are upregulated (increased protein levels and activity), suggesting that there may be a virus-specific need for newly synthesized P-TEFb components (27, 49).
When Act D is added after IE2 and cdk9 have localized to the viral transcriptosomes (i.e., at 10 h p.i.), IE2, but not cdk9, is retained at those sites. This suggests that ongoing transcription is necessary not only for cdk9 recruitment to the viral transcriptosomes but also for maintaining its localization there. However, the requirement for cellular or viral transcription cannot be determined from this experiment. Just as differences in recruitment of factors to the viral transcriptosomes were previously observed in the presence of Roscovitine treatment, these results indicate that different mechanisms of recruitment are employed for IE2 and cdk9 (27).
It is still not clear what the nucleating factor(s) is for the establishment of the viral transcriptosomes. Since it is a virus-specific event, it seems that there must be some viral component that initiates the chain of events for the viral transcriptosome formation. In terms of cdk9 recruitment, we have considered likely viral candidates that may contribute to cdk9 localization to the viral transcriptosomes. We have previously shown that cdk9 recruitment is not an IE1-dependent process, since cdk9 localizes to the viral transcriptosomes during ΔIE1 HCMV infection (49). Another likely candidate is IE2 86, and the results of the IE2 86 ΔSX-EGFP infection in this study suggest that it may play an accessory role for recruiting cdk9. During infection with IE2 86 ΔSX-EGFP virus, significantly lower levels of IE2 protein are synthesized. Cdk9 recruitment was delayed compared to that observed with the wt virus and appeared to correspond to the accumulation of IE2 at the transcriptosomes. These results, coupled with those of our previous studies showing that accumulation of IE2 86 and cdk9 at these sites could be visualized at 2 h p.i. during the wt infection (49), suggest that a threshold level of IE2 may be required for cdk9 recruitment. We cannot exclude the possibility that the deleted region in this IE2 mutant is also involved in cdk9 recruitment independently of any effects on protein levels. However, since the level of cdk9 at the viral transcriptosomes in the HCMV IE2 86 ΔSX-EGFP infection did reach the level that was seen with the wt HCMV infection, albeit with some delay, this possibility seems less likely.
While there may be a viral or cellular factor that is universally required for initiating transcriptosome formation, it is likely that each component of the viral transcriptosome would also have individual requirements for recruitment to the sites. For cdk9, we focused on kinase activity, phosphorylation of the conserved T-loop, cyclin T1 binding, and Brd4 association with the P-TEFb complex. When cdk9 kinase activity is inhibited during infection with Flavopiridol and DRB, cdk9 protein still localizes to the viral transcriptosomes with IE2. Kinase activity was also examined through a the use of a D167N mutation. D167 is located at the ATP-binding pocket and is important for substrate recognition and kinase function. A point mutation, D167N, has been shown to be inactive with respect to kinase function (14, 30). As was seen in the presence of Flavopiridol and DRB, inhibition of cdk9 activity does not prevent its recruitment to the viral transcriptosomes.
We previously reported that infection in the presence of Roscovitine, another inhibitor of cdk9 activity, results in impaired localization of cdk9 (27). The difference observed with Roscovitine compared to other inhibitors of cdk9 may be accounted for by structural changes due to binding of the inhibitors to cdk9 or differential inhibition of other kinases by these drugs. Flavopiridol inhibits cdk9 activity by binding to the ATP-binding pocket in a noncompetitive and structurally tight fashion. Binding of Flavopiridol to cdk9 causes a conformational change in cdk9, burying Flavopiridol in the active site such that only 8% of the molecular surface remains exposed (6). In contrast, Roscovitine binds to cdk9 competitively with ATP. Structural studies of Roscovitine bound to cdk2 showed that the benzyl group of Roscovitine contacts a part of the protein surface that is in common to all cdks and that the purine group binds to the ATP-binding pocket of the cdk but in a different orientation than that seen with ATP (13). It is possible that cdk9 binding to the various inhibitors disrupts cdk9 interaction with other proteins or that differential effects of the inhibitors on other kinases influence recruitment of cdk9. This paper highlights that cdk9 recruitment to the viral transcriptosomes requires binding to cyclin T1. Roscovitine alone does not generally inhibit the interaction of cyclin T1 with cdk9 (unpublished data), but it still may specifically prevent cdk9 from forming a complex with cyclin T1 at the transcriptosome.
We studied the contributions of other properties of cdk9 that may affect its localization to the viral transcriptosomes through mutational analysis. There is increasing evidence that Brd4 can play a regulatory role during transcription through its interactions with P-TEFb via cyclin T1 binding. Brd4 interaction with cdk9-cyclin T1 counters the formation of the cdk9-inactive complex and enhances cdk9-cyclin T1 recruitment to transcription sites (52). Many viral infection studies have determined a key role for Brd4 in establishing viral gene transcription, and Brd4 has been found to concentrate at the viral transcriptosomes (7, 32, 53, 54). Although Brd4 interacts with P-TEFb via binding with cyclin T1, S175 of cdk9 has been shown to contribute to Brd4 binding to the complex. A mutation in S175 may alter the conformation of the active site due to its close proximity to the region, as well as affecting cdk9 kinase activity, since S175 has been shown to be an activating residue upon phosphorylation. Our data show, however, that Brd4 does not play a role in recruiting cdk9 to the viral transcriptosomes during the HCMV infection.
The results of the experiments performed with a series of HA-tagged cdk9 mutants with differential binding to cyclin T1 revealed that the localization of each cdk9HA mutant to the viral transcriptosomes correlated with the level of cyclin T1 in complex with the mutant. E57 is one of 15 identified contact points between cdk9 and cyclin T1 (6). The results of the IP experiments indicate that E57A can bind only to a low level of cyclin T1, and the IFA data show impairment of E57A localization compared to IE2 localization at the viral transcriptosomes. E55 does not contact cyclin T1, and greater levels of cyclin T1 are in complex with E55A than with E57A. According to the IFA results, E55A retains the ability to localize to the viral transcriptosomes. In similarity to the results seen with E57A, the R65A mutant binds to a very low level of cyclin T1, and localization of this mutant to the viral transcriptosomes was not detected. R65 is located at the surface of cdk9 and contributes a positive charge to that region. A likely scenario is that mutation of R65 to alanine alters the charged nature of the region, nonspecifically affecting surface interactions. Furthermore, the E55A/E57A/R65A triple mutant does not associate with cyclin T1 and is not recruited to the viral transcriptosomes. The role of cyclin T1 in cdk9 recruitment to the viral transcriptosomes may be similar to that of the cyclin T1-directed localization of cdk9 to the nuclear speckles (i.e., SC35 spliceosome domains) in uninfected cells (21).
Many other viral infections have been known to subvert the P-TEFb complex for viral gene expression. In human immunodeficiency virus type 1 infections, Tat protein interacts with cyclin T1, recruiting P-TEFb to viral RNA (8, 18, 22, 25, 31, 50, 56, 57). The herpes simplex virus type 1 ICP22 IE viral protein also binds to cdk9 and specifically alters RNAP II phosphorylation (15, 16). It has also been found that EBNA2 plays not only a role in initiation of viral transcription during Epstein-Barr virus infection but also a cdk9-dependent role in productive elongation of viral RNA synthesis (5, 38). In addition, cdk9 plays an integral role in p53 phosphorylation during Kaposi's sarcoma-associated herpesvirus infection via cdk9 interaction with a virus-encoded cyclin (11). P-TEFb activity is also modulated through Brd4 during human T-cell leukemia virus infection (12, 55).
This report focuses on the mechanism and requirements of cdk9 recruitment to the viral transcriptosomes during HCMV infection, and the results reported in this paper contribute to our understanding of one of the first key events that occurs during the infection. In summary, initiation of viral transcriptosome formation requires de novo protein synthesis. A threshold level of IE2 also appears to be required for cdk9 recruitment, and active transcription is necessary to maintain cdk9 localization. While cdk9 kinase activity, T-loop-activating residues, and Brd4 binding do not dictate cdk9 recruitment to the viral transcriptosomes, cdk9 binding to cyclin T1 appears to be essential.
Acknowledgments
We are grateful to William Britt for the pp65 antibody. We are also thankful to Inder Verma for the EF1α promoter-driven lentivector, to Xavier Graña for the pRc-CMV-cdk9HA plasmids, to Martin Messerle for the RpsL-neo cassette, to B. Plachter for the pcDNA-pp71 plasmid, and to Mel Simon for the pSLIK-neo and pEN-TRmiRc2 plasmids. We thank J. Brady for providing us with Flavopiridol. We also appreciate the use of the Deltavision microscope and SoftWorx software at the Cancer Center Digital Imaging Shared Resource at the University of California, San Diego.
This work was supported by NIH grants CA73490 and CA34729.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Ahn, J.-H., W.-J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 7310458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J.-H., E. R. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 184899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 714599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek, M.-C., P. M. Krosky, A. Pearson, and D. M. Coen. 2004. Phosphorylation of the RNA polymerase II carboxyl-terminal domain in human cytomegalovirus-infected cells and in vitro by the viral UL97 protein kinase. Virology 324184-193. [DOI] [PubMed] [Google Scholar]
- 5.Bark-Jones, S. J., H. M. Webb, and M. J. West. 2006. EBV EBNA 2 stimulates CDK9-dependent transcription and RNA polymerase II phosphorylation on serine 5. Oncogene 251775-1785. [DOI] [PubMed] [Google Scholar]
- 6.Baumli, S., G. Lolli, E. D. Lowe, S. Troiani, L. Rusconi, A. N. Bullock, J. E. Debreczeni, S. Knapp, and L. N. Johnson. 2008. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 271907-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 794806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 967791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 738320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, N. R., M. E. Noble, J. A. Endicott, E. F. Garman, S. Wakatsuki, E. Mitchell, B. Rasmussen, T. Hunt, and L. N. Johnson. 1995. The crystal structure of cyclin A. Structure 31235-1247. [DOI] [PubMed] [Google Scholar]
- 11.Chang, P. C., and M. Li. 2008. Kaposi's sarcoma-associated herpesvirus K-cyclin interacts with Cdk9 and stimulates Cdk9-mediated phosphorylation of p53 tumor suppressor. J. Virol. 82278-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, W. K., M. Zhou, M. K. Jang, K. Huang, S. J. Jeong, K. Ozato, and J. N. Brady. 2007. Modulation of the Brd4/P-TEFb interaction by the human T-lymphotropic virus type 1 tax protein. J. Virol. 8111179-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Azevedo, W. F., S. Leclerc, L. Meijer, L. Havlicek, M. Strnad, and S. H. Kim. 1997. Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. Eur. J. Biochem. 243518-526. [DOI] [PubMed] [Google Scholar]
- 14.de Falco, G., and A. Giordano. 1998. CDK9 (PITALRE): a multifunctional cdc2-related kinase. J. Cell Physiol. 177501-506. [DOI] [PubMed] [Google Scholar]
- 15.Durand, L. O., S. J. Advani, A. P. Poon, and B. Roizman. 2005. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J. Virol. 796757-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durand, L. O., and B. Roizman. 2008. Role of cdk9 in the optimization of expression of the genes regulated by ICP22 of herpes simplex virus 1. J. Virol. 8210591-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortunato, E. A., A. K. McElroy, V. Sanchez, and D. H. Spector. 2000. Exploitation of cellular signaling and regulatory pathways by human cytomegalovirus. Trends Microbiol. 8111-119. [DOI] [PubMed] [Google Scholar]
- 18.Gold, M. O., X. Yang, C. H. Herrmann, and A. P. Rice. 1998. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J. Virol. 724448-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 664452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, G., M. Jarosch, J. B. Wang, C. Berbes, and M. A. McVoy. 2003. Tn7-mediated introduction of DNA sequences into bacmid-cloned cytomegalovirus genomes for rapid recombinant virus construction. J. Virol. Methods 107185-194. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann, C. H., and M. A. Mancini. 2001. The CDK9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 1141491-1503. [DOI] [PubMed] [Google Scholar]
- 22.Isel, C., and J. Karn. 1999. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 290929-941. [DOI] [PubMed] [Google Scholar]
- 23.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 1385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. García-Martínez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J. Mol. Biol. 28841-56. [DOI] [PubMed] [Google Scholar]
- 26.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19523-534. [DOI] [PubMed] [Google Scholar]
- 27.Kapasi, A. J., and D. H. Spector. 2008. Inhibition of the cyclin-dependent kinases at the beginning of the human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Q., J. P. Price, S. A. Byers, D. Cheng, J. Peng, and D. H. Price. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 28028819-28826. [DOI] [PubMed] [Google Scholar]
- 29.Majello, B., and G. Napolitano. 2001. Control of RNA polymerase II activity by dedicated CTD kinases and phosphatases. Front. Biosci. 6D1358-D1368. [DOI] [PubMed] [Google Scholar]
- 30.Majello, B., G. Napolitano, A. Giordano, and L. Lania. 1999. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene 184598-4605. [DOI] [PubMed] [Google Scholar]
- 31.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 112633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 151401-1415. [DOI] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 35.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 737678-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 735137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Napolitano, G., P. Licciardo, R. Carbone, B. Majello, and L. Lania. 2002. CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J. Cell. Physiol. 192209-215. [DOI] [PubMed] [Google Scholar]
- 38.Palermo, R. D., H. M. Webb, A. Gunnell, and M. J. West. 2008. Regulation of transcription by the Epstein-Barr virus nuclear antigen EBNA 2. Biochem. Soc. Trans. 36625-628. [DOI] [PubMed] [Google Scholar]
- 39.Park, J. J., Y. E. Kim, H. T. Pham, E. T. Kim, Y. H. Chung, and J. H. Ahn. 2007. Functional interaction of the human cytomegalovirus IE2 protein with histone deacetylase 2 in infected human fibroblasts. J. Gen. Virol. 883214-3223. [DOI] [PubMed] [Google Scholar]
- 40.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell 1153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 723729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 762973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez, V., A. K. McElroy, J. Yen, S. Tamrakar, C. L. Clark, R. A. Schwartz, and D. H. Spector. 2004. Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J. Virol. 7811219-11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin, K. J., E. A. Wall, J. R. Zavzavadjian, L. A. Santat, J. Liu, J. I. Hwang, R. Rebres, T. Roach, W. Seaman, M. I. Simon, and I. D. Fraser. 2006. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. USA 10313759-13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shlapobersky, M., R. Sanders, C. Clark, and D. H. Spector. 2006. Repression of HMGA2 gene expression by human cytomegalovirus involves the IE2 86-kilodalton protein and is necessary for efficient viral replication and inhibition of cyclin A transcription. J. Virol. 809951-9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86 kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 686223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sourvinos, G., N. Tavalai, A. Berndt, D. A. Spandidos, and T. Stamminger. 2007. Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J. Virol. 8110123-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamrakar, S., A. J. Kapasi, and D. H. Spector. 2005. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J. Virol. 7915477-15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92451-462. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 791233-1245. [DOI] [PubMed] [Google Scholar]
- 52.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19535-545. [DOI] [PubMed] [Google Scholar]
- 53.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117349-360. [DOI] [PubMed] [Google Scholar]
- 54.You, J., V. Srinivasan, G. V. Denis, W. J. Harrington, Jr., M. E. Ballestas, K. M. Kaye, and P. M. Howley. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 808909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, M., H. Lu, H. Park, J. Wilson-Chiru, R. Linton, and J. N. Brady. 2006. Tax interacts with P-TEFb in a novel manner to stimulate human T-lymphotropic virus type 1 transcription. J. Virol. 804781-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, Q., D. Chen, E. Pierstorff, and K. Luo. 1998. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 173681-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 112622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]