Abstract
During lytic infection by herpes simplex virus type 1 (HSV-1), histones are present at relatively low levels on the viral genome. However, the mechanisms that account for such low levels—how histone deposition on the viral genome is blocked or how histones are removed from the genome—are not yet defined. In this study, we show that histone occupancy on the viral genome gradually increased with time when transcription of the viral immediate-early (IE) genes was inhibited either by deletion of the VP16 activation domain or by chemical inhibition of RNA polymerase II (RNAP II). Inhibition of IE protein synthesis by cycloheximide did not affect histone occupancy on most IE promoters and coding regions but did cause an increase at delayed-early and late gene promoters. IE gene transcription from HSV-1 genomes associated with high levels of histones was stimulated by superinfection with HSV-2 without altering histone occupancy or covalent histone modifications at IE gene promoters. Moreover, RNAP II and histones cooccupied the viral genome in this context, indicating that RNAP II does not preferentially associate with viral genomes that are devoid of histones. These results suggest that during lytic infection, VP16, RNAP II, and IE proteins may all contribute to the low levels of histones on the viral genome, and yet the dearth of histones is neither a prerequisite for nor a necessary result of VP16-dependent transcription of nucleosomal viral genomes.
During lytic infection of mammalian cells by herpes simplex virus type 1 (HSV-1), virion protein 16 (VP16) triggers the cascade of viral gene expression by stimulating the transcription of immediate-early (IE) genes (5). VP16 binds to the cis-regulatory sequences on viral IE gene promoters as part of a protein complex that also includes two host cell proteins, Oct-1 and HCF-1 (67). VP16, through its activation domain (AD), then interacts with various general transcription factors or coactivators and recruits the host RNA polymerase II (RNAP II) machinery (13, 23, 27, 42, 68). In turn, some of the IE proteins regulate expression of the delayed-early (DE) and late (L) genes, completing the viral gene expression cascade that results in production of infectious virions.
In eukaryotes, the packaging of DNA by histone proteins presents a significant impediment to the transcriptional machinery (41). This barrier can be overcome by activator-mediated recruitment of transcriptional coactivators that either covalently modify histones (29, 41) or remodel the position of nucleosomes along DNA (11, 56). Many covalent modifications of histones have been identified, including lysine acetylation and methylation (29, 41); the former is generally associated with active transcription, and the latter marks either active or inactive transcription depending on which lysine residue is methylated.
Unlike the small-DNA tumor viruses of the polyomavirus and papillomavirus families, the large (152-kbp) genome of HSV-1 is packaged in the virion nucleocapsid not with histones but with the polyamine spermine (7, 12, 50, 53). Long-standing evidence suggests that during lytic infection the HSV-1 genome is mainly nonnucleosomal (39, 40, 46), whereas during latency the viral genome is associated with evenly spaced nucleosomes (10). In support of these earlier findings, chromatin immunoprecipitation (ChIP) assays have indicated that histones, typically represented by histone H3, are present on the viral genome during lytic infection, albeit at relatively lower levels than on cellular genes (20, 21, 26, 35, 50). Furthermore, active-transcription marks, such as histone H3 acetylated on lysine 9 and lysine 14 (H3K9/K14ac) or trimethylated on lysine 4 (H3K4me3), are also associated with the viral genome during lytic infection (20, 21, 26, 35). In contrast, during latent or quiescent infections, histones associated with the viral genome carry modifications typical of heterochromatin, such as histone H3 trimethylated on lysine 9 (8, 66).
An exception to this pattern is the latency-associated transcript (LAT) gene, which during latency is associated with H3K9/K14ac, indicative of its transcriptionally active status (31, 32, 49). Moreover, upon reactivation from latency, lytic genes become associated with acetylated histone H3, and the LAT region becomes depleted of that mark (1, 49). Together, these observations suggest that regulation of chromatin might be an important mechanism for the switch between lytic and latent infections (28).
Although histone H3 occupancy is presumed to represent the existence of the nucleosome core particle on the viral genome, whether other core histones are present on the HSV-1 genome during lytic infection has not been established. This question becomes more compelling in light of the evidence that histone H2A-H2B dimers can be removed from nucleosome core particles, resulting in the presence of “hexasomes” at actively transcribed genes (19, 33, 58). In addition, the mechanisms regulating histone levels on the viral genome during lytic infection have not yet been defined. One key issue is whether histone deposition is inhibited or deposited histones are removed. Hints of potential mechanisms have arisen in several recent reports. For instance, an IE protein, ICP0, blocks the silencing of HSV-1 DNA by dissociating histone deacetylase 1 (HDAC1) from the REST/CoREST/HDAC repressor complex (14, 15). During lytic infection with a mutant virus that does not express ICP0, histone H3 occupancy on the viral genome increased and the fraction of acetylated histone H3 decreased, suggesting that the disruption of the REST/CoREST/HDAC complex by ICP0 might be relevant (6). Similarly, the tegument protein VP22 was suggested to block nucleosome assembly on the viral DNA (64), but no data yet indicate whether or how histone occupancy on the viral genome changes in the absence of VP22 during HSV-1 lytic infection.
Another factor that may mediate histone levels on the viral genome during lytic infection is VP16. We have shown previously that during lytic infection with a mutant virus that lacks the AD of VP16 (strain RP5), higher levels of histone H3 associate with RP5 genomes than with wild-type genomes (20). Moreover, during RP5 infections, transcriptional coactivators—such as p300 and CBP histone acetyltransferases or BRM and Brg-1 remodeling enzymes—are not efficiently recruited to IE gene promoters (20). This observation is consistent with prior knowledge about the VP16 AD in artificial experimental contexts, in which VP16 physically interacts with and recruits transcriptional coactivators—such as histone acetyltransferases p300/CBP (3, 17, 24, 34, 62, 65), PCAF (65), and GCN5 (22, 37, 59, 62, 63), as well as ATP-dependent chromatin remodeling enzymes (16, 43, 47, 48)—to potentiate transcription from nucleosomal templates. However, the hypothesis that the transcriptional coactivators recruited by VP16 are required for modifying and removing the histones from the viral genome, and thereby required for IE gene expression, is contradicted by our recent report that disruption of coactivator expression did not diminish IE gene expression during infection of cultured cells (36). Thus, the impact of histones and chromatin on viral gene expression remains incompletely defined.
In the present report, we show that during lytic infection of cultured cells, all four core histones associate with the viral genome. In the absence of VP16 transcriptional activation (i.e., during infection by the RP5 mutant virus), histone deposition throughout the viral genome increased gradually but dramatically to levels approaching that on cellular genes. Inhibition of RNAP II-mediated transcription by actinomycin D also increased the histone occupancy on the viral genome in a temporal pattern similar to that of RP5 infections. In contrast, inhibition of IE protein expression by cycloheximide had no significant effect on histone association with most of the actively transcribed IE gene promoters and coding regions but did increase histone occupancy on DE and L gene promoters. To address whether VP16 can stimulate the removal of histones that are already deposited on the viral genome, we asked whether providing wild-type VP16 in trans (by HSV-2 superinfection) results in histone depletion from RP5 IE gene promoters. Surprisingly, although HSV-2 superinfection stimulated IE gene expression from the RP5 genome, it did not lead to depletion of histones from IE genes. In addition, active-transcription marks, such as H3K9/K14ac or H3K4me3, did not increase on the RP5 IE gene promoters upon HSV-2 superinfection. Sequential ChIP (seq-ChIP) experiments indicated that RNAP II and histone H3 cooccupy RP5 genomes upon HSV-2 superinfection at a level similar to that of a constitutively expressed housekeeping gene, indicating that RNAP II does not preferentially associate with histone-free viral genomes.
Taken together, our results suggest that the low level of histone occupancy on the viral genome during lytic infection is the result of a complex process that involves VP16, active transcription by RNAP II, and IE proteins. However, histone removal or covalent modification of histones may not be necessary for the VP16-dependent transcription of IE genes from viral genomes heavily associated with histones.
MATERIALS AND METHODS
Cell lines and viruses.
HeLa (ATCC CCL-2) cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 110 mg/liter sodium pyruvate and 10% fetal bovine serum (Invitrogen). In some experiments, cycloheximide (Sigma) or actinomycin D (Sigma) was added to the cell culture medium prior to and during infection, as indicated in the figure legends. The RP5 strain of HSV-1, which lacks sequences encoding the AD of VP16, has been described previously (60). The RP5 and wild-type KOS strains of HSV-1 and the G strain of HSV-2 were prepared and titers were determined in Vero cells.
Gene expression assays and qRT-PCRs.
Total cellular RNA was isolated using Trizol reagent (Invitrogen). Total RNA was reverse transcribed by random primers using a reverse transcription system (Promega). The synthesized cDNA was used as the template for quantitative real-time PCR (qRT-PCR) analysis using SYBR green master mix (Roche) and an ABI 7500 RT-PCR system (Applied Biosystems). Gene expression was normalized first against 18S rRNA and then to proper controls by the 2−ΔΔCT method. For ChIP assays, data were analyzed using the standard-curve method as explained in the following section. Primer sequences for PCRs spanning the ICP27 promoter and the gC promoter are as follows: for the ICP27 promoter, the forward primer sequence is 5′-TGGTGTCTGATTGGTCCTTG, and the reverse primer sequence is 5′-CGGGTGGTGGATGTCCTTAT; for the gC promoter, the forward primer sequence is 5′-TCGGGCGATTGATATATTTTT, and the reverse primer sequence is 5′-TGTCCCCTTCCGGAATTTAT. Other primer pairs used in this study have been defined previously (20, 51).
ChIP and seq-ChIP.
ChIP was performed as described previously (20). In summary, confluent plates of HeLa cells were infected in the absence or presence of actinomycin D or cycloheximide, as indicated in the figure legends. Infections were stopped by the addition of formaldehyde to the cell culture plate at a final concentration of 1%. After cells were resuspended in a hypotonic buffer, nuclei were released by Dounce homogenization in order to minimize the background signals from cytoplasmic capsids or membrane-bound virions. Nuclei were collected by centrifugation and then disrupted by sonication using a Branson digital sonifier 450 to obtain 200- to 1,000-bp DNA fragments. Protein-DNA complexes were immunoprecipitated using 5 to 10 μg of antibodies against histone H2A (ab18255; Abcam), histone H2B (ab1790; Abcam), histone H3 (ab1791; Abcam), histone H4 (05-858; Upstate), RNAP II (8WG16; Covance), or VP16 (61). Protein-DNA complexes were collected by use of protein G-agarose beads (Invitrogen). After several washes, the protein-DNA complexes were eluted and cross-links were reversed overnight at 65°C in the presence of 200 mM NaCl and 10 μg RNase A. Samples were then precipitated with ethanol, digested with proteinase K (Roche) at 42°C for 2 h, and purified with Qiagen spin columns using the gel extraction protocol. The presence of viral and cellular DNA fragments in the immunoprecipitated material was analyzed by qRT-PCR as described above. A standard curve using serial threefold dilutions of input samples (1%, 0.3%, 0.1%, or 0.04%) was produced to quantitate the signals from immunoprecipitation samples. Background signals obtained from immunoprecipitation reactions performed in the absence of antibodies (no-antibody control) were subtracted from the signals obtained from immunoprecipitation samples. In some cases, data were further normalized against a cellular control gene (beta interferon [IFN-β] promoter or U3 snRNA promoter) by dividing the value described above for the viral DNA by that of the cellular DNA (we refer to this value as the input ratio) in order to account for the differences in immunoprecipitation efficiencies.
For seq-ChIP assays, after the first immunoprecipitation, protein-DNA complexes were eluted in 100 μl elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% sodium dodecyl sulfate) by incubation at 65°C for 15 min. The eluates were then subjected to the second immunoprecipitation as indicated above, whereby 30 to 45 μl of eluate was used as input in each immunoprecipitation reaction. The rest of the procedure is the same as the ChIP protocol described above.
RESULTS
Absence of the AD of VP16 causes an increase in histone occupancy on the HSV-1 genome during lytic infection.
We have reported previously that during lytic infection by wild-type HSV-1, the IE gene promoters are relatively free of histones, as represented by H3 (20). We have also shown that at IE times during infection by strain RP5 (lacking the VP16 AD), the HSV-1 genome more abundantly associates with histone H3 but lacks acetylated histone H3, an active-transcription mark (20). In order to get a more dynamic and quantitative picture of histone deposition on the viral genome in the presence or absence of the VP16 AD, we have compared levels of histone occupancy on the RP5 and KOS (wild-type) genomes at different stages of infection using ChIP assays coupled with quantitative PCR. Parallel plates of HeLa cells were infected with the KOS or RP5 virus at multiplicities of infection (MOIs) such that similar numbers of viral genomes entered cells at the beginning of infection, as shown by quantitative PCR assays of infected-cell nuclear extracts (see Fig. S1 in the supplemental material). At 2, 4, and 6 h postinfection (hpi), infections were stopped by formaldehyde cross-linking, and the occupancy of histones H2A, H2B, H3, and H4 and RNAP II on viral genes was analyzed by ChIP.
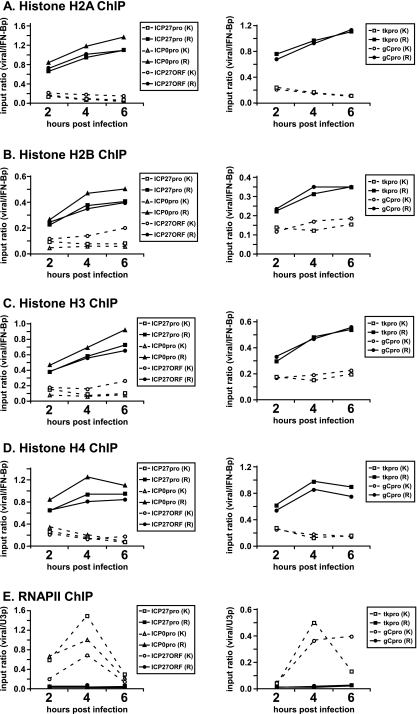
As indicated in Fig. 1A, occupancy of histone H2A on the RP5 genome at 2 hpi was higher than on the KOS genome at several viral gene fragments, such as IE promoters and open reading frames (ORFs) (Fig. 1A, left) as well as DE and L promoters (Fig. 1A, right). Histone H2A occupancy on the RP5 genome continued to increase at 4 and 6 hpi up to a point comparable to that of the cellular gene (i.e., an input ratio of approximately 1). Similar patterns were observed for histones H2B (Fig. 1B), H3 (Fig. 1C), and H4 (Fig. 1D). Occupancy by all core histones throughout the viral genome increased prominently but gradually in RP5 infections, whereas in KOS infections histone occupancy either stayed the same or decreased during the later stages of lytic infection. As expected, recruitment of RNAP II to IE promoters, and as a consequence to DE and L gene promoters, was severely impaired in RP5 infections (Fig. 1E). These results together suggest that the VP16 AD contributes to keeping the viral genome free of nucleosomes.
FIG. 1.
Core histone occupancy on the viral genome increases in RP5 lytic infections. HeLa cells were infected with the KOS (K) or RP5 (R) strain of HSV-1 at an MOI of 1 PFU/cell or 0.005 PFU/cell, respectively. ChIP was performed to detect the occupancy of histones H2A (A), H2B (B), H3 (C), and H4 (D) and RNAP II (E) on ICP27, ICP0, tk, and gC promoters (pro) and on the ICP27 ORF at 2, 4, and 6 hpi. The data were analyzed as explained in Materials and Methods. The graphs show the results of a representative experiment; similar results were observed for replicate experiments. IFN-Bp, IFN-β promoter; U3p, U3 snRNA promoter.
This effect could conceivably be mediated directly by the VP16 AD, i.e., by recruiting transcriptional coactivators that modify and remodel the chromatin structure. However, we have recently indicated that transcriptional coactivators recruited by VP16 do not contribute significantly to IE gene expression during lytic infection of human foreskin fibroblasts (36). Alternatively, VP16 may indirectly regulate chromatin, e.g., by stimulating the expression of IE genes that may in turn contribute to lower histone levels on the viral genome. This concept is supported by the observation that histone occupancy on DE (tk) and L (gC) gene promoters, where VP16 does not bind directly, also increased substantially in RP5 infections, with kinetics similar to that on IE gene promoters (Fig. 1). A further possibility is that active transcription by RNAP II itself may prevent histone deposition, a mechanism that has been suggested for several actively transcribed genes (4, 19, 33, 55). The last two possibilities are addressed in the next sections.
Inhibition of transcription increases histone occupancy on the viral genome during lytic infection.
In Saccharomyces cerevisiae, histones can be depleted from actively transcribed regions (19, 30, 33, 38, 58). Since the increase in histone occupancy on the viral genome in RP5 infections might be dependent on the inability to recruit the RNAP II machinery (Fig. 1E), we studied histone deposition on the viral genome during KOS infections under circumstances in which RNAP II-mediated transcription was blocked. To this end, HeLa cells were pretreated with actinomycin D and subsequently infected with KOS in the presence of actinomycin D. Infections of mock-treated cells were carried out in parallel. At 2, 4, and 6 hpi, infections were stopped by formaldehyde cross-linking and ChIP was performed to assay the presence of core histones, RNAP II, and VP16 on the viral genome.
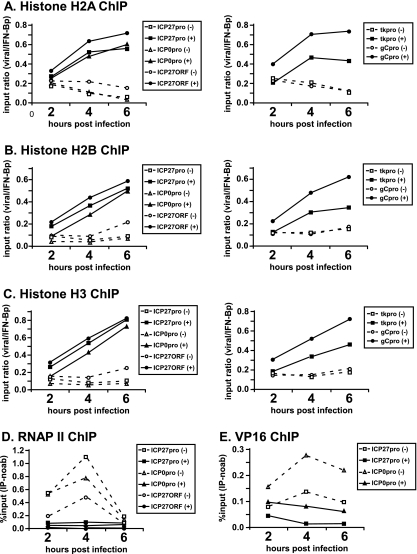
As indicated in Fig. 2A, the occupancy of histone H2A was not substantially higher on viral genes at 2 hpi in the presence of actinomycin D than in its absence. Similarly to results from RP5 infections, at later times in infection the H2A occupancy throughout the viral genome increased in the presence of actinomycin D and reached a level about half that of the cellular gene.
FIG. 2.
Inhibition of transcription by actinomycin D increases the core histone occupancy on the HSV-1 genome. HeLa cells were infected with HSV-1 strain KOS at an MOI of 1 PFU/cell in the presence (+) or absence (−) of actinomycin D (1 μg/ml). ChIP assays were performed to detect the occupancy of histone H2A (A), H2B (B), H3 (C), RNA polymerase II (D), and VP16 (E) on the ICP27, ICP1, tk, and gC promoters (pro) and on the ICP27 ORF at 2, 4, and 6 hpi. (A to C) Input ratios were calculated as follows. The percent input [% input (IP-noab)] values obtained for viral DNA fragments (after subtracting the values for the control samples without primary antibody) were normalized against the values for a cellular control gene fragment (IFN-β promoter [IFN-βp]). (D) Percent input values represent signals obtained from immunoprecipitated samples after subtracting background signals obtained in the absence of antibodies, expressed as a fraction of the signals obtained using input DNA. Graphs show the results of a representative experiment; similar results were observed in independent replicates.
Similarly, the occupancy of histones H2B (Fig. 2B), H3 (Fig. 2C), and H4 (data not shown) on the viral genome increased in the presence of actinomycin D, with kinetics similar to that observed for RP5 infections, although at 2 hpi, histone deposition on the viral genome was slightly less than that observed for RP5 infections. RNAP II ChIP served as a positive control, showing that actinomycin D effectively blocked the recruitment of RNAP II to the viral genome (Fig. 2D). These results suggest that transcription by RNAP II per se may contribute to the depletion of histones from the HSV-1 genome during lytic infection.
Interestingly, actinomycin D treatment also decreased the VP16 occupancy on IE gene promoters, particularly at 4 hpi (Fig. 2E). Although this could be due to the fact that actinomycin D is a DNA-intercalating drug and as such might prevent the binding of VP16 to DNA, it is also possible that the increase in histone occupancy on IE gene promoters diminishes the binding of VP16 to these regions.
IE proteins are not required for inhibition of histone deposition on IE genes during lytic infection.
During RP5 infections or in the presence of actinomycin D, histone occupancy increased not only at IE genes but also at DE and L gene promoters, where VP16 does not bind directly. Given that IE proteins play important roles in the transcriptional regulation of DE and L genes, we considered whether IE proteins also contribute to the depletion of histones from these genes. The hypothesis that histones are removed from DE and L genes by IE proteins leads to the prediction that inhibition of IE protein synthesis would increase the occupancy of histones on these promoters but would not affect IE genes, which are actively transcribed by VP16 and RNAP II in the absence of IE proteins. Therefore, we tested whether the inhibition of IE protein expression by cycloheximide affected histone deposition on the viral genome during lytic infection.
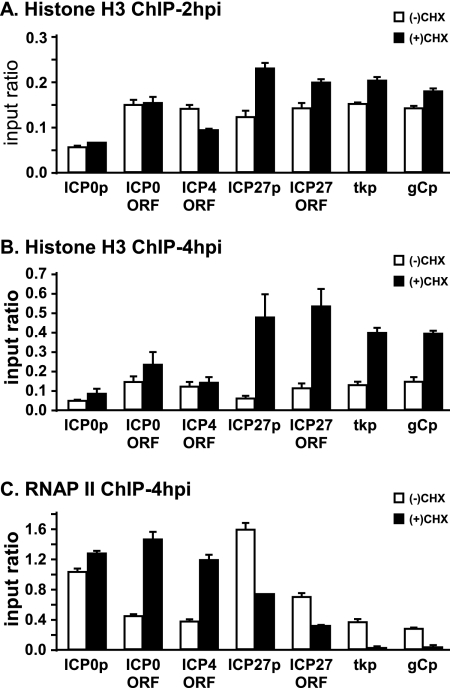
To this end, HeLa cells were infected with KOS in the presence of cycloheximide to block de novo viral protein synthesis. At 2 and 4 hpi, infections were stopped and ChIP assays were performed to analyze histone H3 and RNAP II occupancy on the HSV-1 genome. As expected, histone H3 occupancy on the ICP0 promoter and the ICP0 and ICP4 ORFs was minimally affected by inhibition of IE protein expression at 2 hpi (Fig. 3A). H3 occupancy increased modestly on the ICP27 promoter and the ICP27 ORF, as well as on the tk and gC promoters (Fig. 3A). Similarly, at 4 hpi histone deposition did not change substantially on the ICP0 promoter, the ICP0 ORF, or the ICP4 ORF (Fig. 3B) yet increased dramatically on the ICP27 promoter, the ICP27 ORF, and the tk and gC promoters (Fig. 3B). These results suggest that IE proteins are not involved in depleting the histones from IE promoters and ORFs, with the exception of ICP27.
FIG. 3.
Changes in histone occupancy on the HSV-1 genome upon inhibition of IE protein synthesis by cycloheximide. HeLa cells were pretreated with 60 μg/ml cycloheximide for 2 h and then were infected with HSV-1 KOS at an MOI of 1 PFU/cell in the presence of cycloheximide [(+)CHX]. (A) At 2 hpi, chromatin was cross-linked, and a ChIP assay probing histone H3 on the ICP0 promoter (ICP0p), the ICP27 promoter (ICP27p), the ICP0 ORF, the ICP4 ORF, the ICP27 ORF, the tk promoter (tkp), and the gC promoter (gCp) was performed as indicated before. (B and C) At 4 hpi, chromatin was cross-linked, and a ChIP assay probing histone H3 (B) and RNAP II (C) on the indicated viral gene fragments was performed as indicated in the legend to Fig. 2. The data shown represent the averages from two independent experiments; the error bars indicate the ranges between the averages from these experiments.
Interestingly, the increase in histone H3 occupancy on the ICP27 promoter, the ICP27 ORF, and the tk and gC promoters corresponded to a significant decrease in RNAP II occupancy (Fig. 3C). As expected, RNAP II occupancy on other IE genes either did not change or increased in the presence of cycloheximide (Fig. 3C). These results together suggest that during early times in lytic infection, VP16-mediated transcription keeps the IE genes relatively free of histones, and at later stages IE proteins are involved in the removal of histones from other regions of the viral genome.
Histones are neither modified nor removed from the RP5 genome upon superinfection with HSV-2.
We designed an experiment to test whether VP16 can induce the removal of histones that are already deposited on the viral genome. The VP16 proteins of HSV-1 and HSV-2 share 93% sequence similarity (9). Moreover, HSV-2 superinfection can reactivate quiescent HSV-1 infections in vitro (8), and the VP16 gene from HSV-2 can function effectively when recombined into the HSV-1 genome (52). These observations indicate that the HSV-2 VP16 protein is capable of activating transcription from the HSV-1 genome. Given that RP5 genomes associate with higher levels of histones at later stages of infection (Fig. 1), we asked whether VP16 provided in trans by HSV-2 superinfection can overcome the nucleosomal barrier and induce IE gene expression from the quiescent and histone-laden RP5 genome.
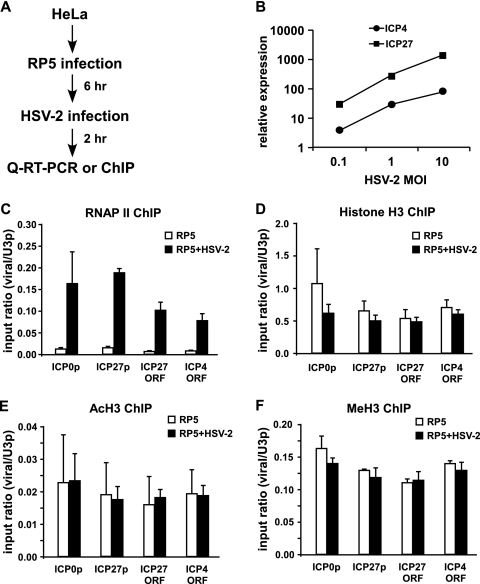
We first tested whether VP16 from HSV-2 superinfection could stimulate IE gene expression from RP5 genomes. HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell. At 6 hpi, when the RP5 viral genomes are highly associated with histones (Fig. 1), the cells were superinfected with HSV-2 at a range of MOIs. RP5 IE gene expression was analyzed after 2 h of HSV-2 superinfection (Fig. 4A). As expected, transcription of the RP5 IE genes ICP4 and ICP27 was induced significantly upon HSV-2 superinfection in a dose-responsive manner with respect to the HSV-2 MOI (Fig. 4B). RP5 IE gene expression was also stimulated when the HSV-2 superinfections were performed in the presence of cycloheximide (see Fig. S2 in the supplemental material), indicating that the induction of IE gene expression is largely attributable to VP16 but does not depend on de novo synthesis of HSV-2 IE proteins such as ICP0. These results together indicate that VP16 of HSV-2 can efficiently stimulate IE gene expression from viral genomes that are predominantly nucleosomal.
FIG. 4.
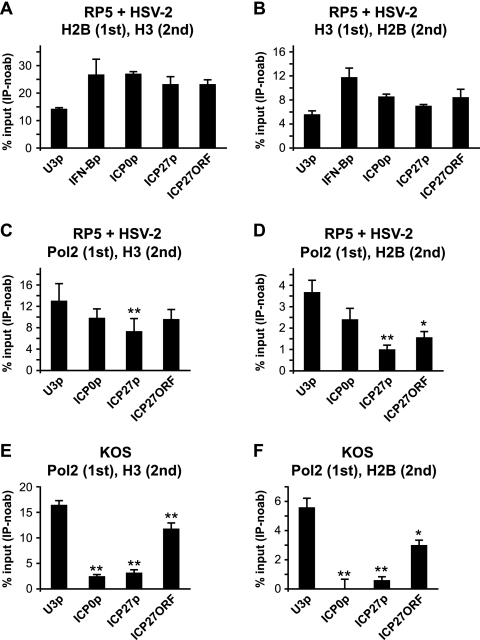
HSV-2 superinfection does not cause major changes in histone occupancy or covalent histone modifications on the RP5 genome. (A) Summary of the assays performed. (B) HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell and, at 6 hpi, were superinfected with HSV-2 at an MOI of 0.1, 1, or 10 PFU/cell. At 2 h after HSV-2 superinfection, levels of ICP4 and ICP27 expression were analyzed by qRT-PCR. (C to F) HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell and, at 6 hpi, were superinfected with HSV-2 at an MOI of 10 PFU/cell. At 2 h after HSV-2 superinfection, ChIP was performed, assaying the presence of RNAP II (C), histone H3 (D), H3K9/K14ac (E), and H3K4me3 (F) on the ICP0 promoter (ICP0p), the ICP27 promoter (ICP27p), the ICP4 ORF, and the ICP27 ORF. The data shown represent the averages from three independent experiments, where percent input values from viral genes are normalized against those from the U3 snRNA promoter (U3p). Error bars represent the standard deviations for the averages from these experiments.
We next tested whether VP16 can induce changes in the chromatin structure of RP5 genomes upon superinfection with HSV-2. We hypothesized that if VP16 is involved in the modification and removal of histones, then active-transcription marks, such as H3K9/K14ac or H3K4me3, would increase and the histone H3 occupancy on the RP5 genomes would decrease upon HSV-2 superinfection.
To address these questions, we performed ChIP assays with cells infected with RP5 and superinfected with HSV-2. HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell. At 6 hpi, these cells were superinfected with HSV-2 at 10 PFU/cell, which robustly induced RP5 IE gene expression (Fig. 4B). Infections were stopped by formaldehyde cross-linking after 2 h of HSV-2 superinfection, and the occupancy of RNAP II, histone H3, H3K9/K14ac, and H3K4me3 on RP5 IE promoters was analyzed by ChIP.
As expected, RNAP II occupancy increased significantly at IE gene promoters and ORFs upon superinfection with HSV-2 (Fig. 4C). Even so, RNAP II occupancy on the IE promoters was still 10 to 20% of that observed on the cellular control U3 snRNA promoter, indicating that the relative rate of transcription was lower on IE genes than on the U3 snRNA gene. This result may suggest that only some, but not all, RP5 genomes are transcriptionally active upon HSV-2 superinfection. Interestingly, this increase in RNAP II occupancy did not correlate with a significant decrease (P > 0.05 by Student's t test) in histone H3 occupancy on IE promoters and ORFs (Fig. 4D), suggesting that neither the VP16 AD nor RNAP II is sufficient to deplete histones from the viral genome. No change in the occupancy of H3K9/K14ac (Fig. 4E) or H3K4me3 (Fig. 4F) on RP5 IE genes was observed after superinfection with HSV-2, indicating that VP16 does not induce major changes in covalent histone modifications on the viral genome. These results collectively suggest that VP16 and RNAP II are not sufficient to alter the chromatin structure on IE promoters. Perhaps more importantly, neither histone removal nor covalent modification of histones seems to be a prerequisite for VP16-mediated transcription from nucleosomal viral genomes.
Histones and RNAP II partially cooccupy the RP5 genome upon HSV-2 superinfection.
Although the results shown in Fig. 1 indicated a significant increase in histone occupancy on the RP5 genome at 6 hpi, we cannot be certain that all RP5 viral genomes are occupied by histones (i.e., the input ratio may be somewhat less than 1). In addition, it is likely that not all RP5 genomes were reactivated in response to HSV-2 superinfection, as indicated by the lower occupancy of RNAP II on IE genes than on the U3 snRNA promoter (Fig. 4C). These observations raise the possibility that VP16 and RNAP II may preferentially associate with viral genomes that remain nonnucleosomal. This could explain why no changes in histone occupancy or covalent histone modifications were evident on the RP5 genomes after HSV-2 superinfection (Fig. 4). Therefore, we performed seq-ChIP assays to probe this issue.
Since histones H2B and H3 are expected to cooccupy a given cellular gene at high levels, as control assays for histone-RNAP II seq-ChIPs we first addressed whether histones H2B and H3 cooccupy cellular and viral genes. HeLa cells were infected with RP5 and superinfected with HSV-2 as in previous experiments. The cross-linked chromatin from infected cells was subjected first to immunoprecipitation using a histone H2B antibody, followed by a second immunoprecipitation with a histone H3 antibody (Fig. 5A). As expected, histones H2B and H3 cooccupied both the IFN-β and the U3 snRNA promoter (Fig. 5A). We noted that the level of histone cooccupancy on the U3 snRNA promoter was less than that on the IFN-β promoter, which might indicate active transcription on the former promoter. Histones H2B and H3 also cooccupied the viral IE genes at levels similar to those on cellular promoters (Fig. 5A). Similar results were observed when the first immunoprecipitation was performed using H3 antibodies, followed by a second immunoprecipitation by H2B antibodies (Fig. 5B). These results collectively indicate the feasibility of seq-ChIP assays in this experimental context. Interestingly, the association of H3 and H2B at viral genes was comparable to that at the cellular genes, indicating coassociation of these histones on viral DNA. This observation is consistent with the idea that entire nucleosomes form on the viral genome.
FIG. 5.
Histone and RNAP II cooccupancy on RP5 IE genes upon HSV-2 superinfection. HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell and, at 6 hpi, were superinfected with HSV-2 at an MOI of 10 PFU/cell. At 2 h after HSV-2 superinfection, seq-ChIP was performed, assaying the cooccupancy of histone H3, histone H2B, and RNAP II on the U3 snRNA promoter (U3p), the IFN-β promoter (IFN-Bp), the ICP0 promoter (ICP0p), the ICP27 promoter (ICP27p), and the ICP27 ORF. (A) Histone H2B (first immunoprecipitation) and H3 (second immunoprecipitation) cooccupancy. (B) Histone H3 (first immunoprecipitation) and H2B (second immunoprecipitation) cooccupancy. (C) RNAP II (first immunoprecipitation) and H3 (second immunoprecipitation) cooccupancy. (D) RNAP II (first immunoprecipitation) and H2B (second immunoprecipitation) cooccupancy. (E) HeLa cells were infected with KOS at an MOI of 1 PFU/cell, and seq-ChIP was performed at 2 hpi, as described above. RNAP II (first immunoprecipitation) and H3 (second immunoprecipitation) cooccupancy. (F) seq-ChIP was performed with HeLa cells infected as described for panel E. RNAP II (first immunoprecipitation) and H2B (second immunoprecipitation) cooccupancy. The data shown in panels A, C, and D represent the averages from three independent experiments, with error bars representing the standard deviations between the averages from these experiments. The data shown in panel B represent the averages from two independent experiments, with error bars representing the ranges between the averages from these experiments. The data shown in panels E and F represent three independent immunoprecipitations done in parallel, where the error bars represent the standard deviations. Samples with mean values that vary significantly from those for the U3 snRNA promoters are indicated by asterisks (**, P ≤ 0.01; *, 0.01 ≤ P ≤ 0.05; paired Student's t test). Percent input [% input (IP-noab)] values represent signals obtained from immunoprecipitated samples after subtracting background signals obtained in the absence of antibodies, expressed as a fraction of the signals obtained using input DNA.
Cooccupancies of histone H3 and RNAP II (Fig. 5C) and of histone H2B and RNAP II (Fig. 5D) were analyzed similarly by seq-ChIP with RP5-infected and HSV-2-superinfected cells. The cross-linked chromatin was subjected first to immunoprecipitation with RNAP II antibodies, followed by a second immunoprecipitation with histone H3 or histone H2B antibodies using chromatin material obtained from the first immunoprecipitation. We used the U3 snRNA promoter as a positive control. As expected, RNAP II and histone H3 (Fig. 5C) or RNAP II and histone H2B (Fig. 5D) cooccupied the U3 snRNA promoter. Interestingly, RNAP II and histone H3 cooccupied the RP5 ICP0 promoter and the ICP27 ORF at levels similar to that of the U3 promoter, yet the cooccupancy was significantly lower on the ICP27 promoter (Fig. 5C). RNAP II and histone H2B also cooccupied IE genes (Fig. 5D), yet the degree of cooccupancy was significantly lower than on the U3 snRNA promoter for some IE genes, such as the ICP27 promoter and the ICP27 ORF. These results suggest that although RNAP II does not preferentially associate with “histone-free” RP5 genomes upon HSV-2 superinfection, VP16- and RNAP II-mediated transcription may lead to the disruption of the nucleosome structure. One possible mechanism is the eviction of a histone H2A-H2B dimer from the nucleosome core particle, as observed with other systems (19, 33).
In order to gain more confidence in these results, we analyzed RNAP II and histone cooccupancy on genes that associate with either RNAP II or histones, but not both. The promoter region of the transcriptionally inactive IFN-β gene associates with histones but not RNAP II. As expected, neither of the histones cooccupied the IFN-β promoter with RNAP II, as indicated by the absence of IFN-β-specific amplification in PCR samples parallel to those presented in Fig. 5C and D (data not shown). As a second negative control, we analyzed the cooccupancy of RNAP II and histones on genes that are associated with RNAP II but low levels of histones. To this end, we repeated the seq-ChIP assays with KOS-infected cells and asked whether histones and RNAP II cooccupy the viral IE genes. Given that histone levels are low on the KOS genomes (Fig. 1 and 2), we expected that the viral genomes that are associated with RNAP II would be low in histones also. As expected, histone H3 was underrepresented on all viral IE promoters that are associated with RNAP II (Fig. 5E). Interestingly, histone H3 was depleted from the ICP27 ORF to a lesser extent than from IE promoters. Similarly, the cooccupancy of histone H2B and RNAP II on IE promoters and the ICP27 ORF was significantly lower than on the U3 snRNA promoter (Fig. 5F). These results together reinforce our conclusions from seq-ChIP assays performed with RP5-infected and HSV-2-superinfected cells.
DISCUSSION
An increasing amount of evidence indicates the important role of chromatin regulation during the lytic and latent stages of HSV-1 infection. Recent studies have shown that histone H3 associates with the HSV-1 genome at lower levels than with cellular genes during lytic infection (20, 21, 26, 35, 50). Active-transcription marks, such as H3K9/K14ac and H3K4me3, are also present on the viral genome during lytic infection (20, 21, 26). In contrast, during latency the viral genome is chromatinized to a larger extent, and the histones associated with the viral genome carry inactive-chromatin marks, with the exception of the actively transcribed LAT gene (10, 32, 66). In addition, during reactivation from latency and quiescence, acetylated histones associate with IE gene promoters (8, 49). These results all indicate that chromatin is dynamically regulated during different stages of HSV-1 infection and that the transcriptional status of the viral genome correlates with the type of covalently modified histones associated with it. On the other hand, the mechanism by which the viral genome stays predominantly nonnucleosomal during lytic infection has not been studied in detail.
In this study, we addressed potential mechanisms to account for the dearth of histones on the viral genome during lytic infection. We have previously analyzed the role of transcriptional coactivators that are recruited by VP16 in this process and shown that disrupting the expression of a number of coactivators did not reduce IE gene expression during lytic infection, suggesting that these coactivators are not likely involved in modulating histone occupancy on IE genes (36). We thus turned our attention to other possible mechanisms and analyzed whether the VP16 AD, transcription by RNAP II per se, or IE proteins affect histone levels on the viral genome.
Our results show that the VP16 AD contributes to low histone levels on the HSV-1 genome, as the absence of the VP16 AD resulted in a dramatic increase in the occupancy of all core histones on the RP5 genome (Fig. 1). An indirect mechanism whereby the VP16 AD might mediate the absence of histones from the viral genome is simply by recruitment of host RNAP II machinery, as it has been shown that histones are underrepresented on actively transcribed genes (4, 19, 44, 45, 55). In support of this notion, histone occupancy dramatically increased throughout the viral genome in the presence of the RNAP II inhibitor actinomycin D (Fig. 2). This result indicates that transcription per se reduces histone occupancy on viral genes. This result might be further interpreted to mean that the VP16 AD and whatever coactivators it recruits are not sufficient to maintain low histone levels on the viral genome without active transcription by RNAP II. This interpretation is complicated by the observation that VP16 occupancy on IE promoters at later times postinfection was also reduced by actinomycin D. The loss of VP16 binding might be a consequence of increased histone occupancy on these promoters, blocking access by the VP16-induced protein complex. An alternative possibility is that actinomycin D directly prevents the binding of VP16 and its partners to IE promoters, which may in turn lead to an increase in histone occupancy.
IE proteins themselves may also affect histone levels on the viral genome, as indicated by the increase in histone occupancy on DE and L gene promoters in the absence of IE protein expression (Fig. 3). As expected, inhibition of IE protein expression did not affect histone occupancy on most IE genes; the exception was the ICP27 gene, both the promoter and the ORF. We currently do not have a clear explanation of why the ICP27 gene is regulated differently than other IE genes. An important point to note is that RNAP II occupancy on the ICP27 gene was reduced substantially, much like on the DE and L promoters in the presence of cycloheximide, in contrast to the enhanced RNAP II recruitment to other IE genes (Fig. 3C). Therefore, it is likely that de novo IE protein synthesis is necessary for the maximal transcription of ICP27 but not of the other IE genes.
One mechanism by which IE proteins might mediate histone depletion is the recruitment of RNAP II and the induction of active transcription. Alternatively, IE proteins might be directly involved in regulating histone deposition on the viral genome. For instance, ICP0 was shown to disintegrate the REST/CoREST/HDAC repressor complex (14, 15) and thus was suggested to prevent the formation of inactive chromatin on the HSV-1 genome. Cliffe and Knipe (6) have recently shown that this function of ICP0 may be relevant during lytic infection, since in the absence of ICP0, histone H3 occupancy increased and the fraction of acetylated histones on the viral genome decreased significantly. Whether the REST/CoREST/HDAC complex is the only target of ICP0 and whether the changes in chromatin on the viral genome in the absence of ICP0 are directly mediated by REST/CoREST/HDAC still need to be determined.
Although our results indicate that VP16 and RNAP II contribute to the dearth of histones on IE gene promoters and ORFs during lytic infection, reduced histone levels are not required for IE gene transcription. Histones were neither modified on nor depleted from RP5 genomes upon superinfection with HSV-2 (Fig. 4D), in spite of the induction of RP5 IE gene expression by HSV-2 (Fig. 4B). These results suggest that the presence of histones may not interfere with the ability of VP16 and RNAP II to initiate transcription from the viral genome.
A potential complication in these assays is the possibility that not all viral genomes are activated by HSV-2 superinfection, as indicated by the low levels of RNAP II occupancy on IE genes (Fig. 4C). This might explain why no change in histone occupancy or modifications was observed on the RP5 genome after HSV-2 superinfection. We think that similar problems might be faced when studying reactivation from latency in vivo, where only a fraction of latent viral genomes might get reactivated.
Given the possibilities that not all RP5 genomes become transcriptionally active upon HSV-2 infection and that some RP5 genomes may escape histone deposition, we hypothesized that the RP5 genomes that are activated by HSV-2 superinfection might actually be the ones that escape nucleosome deposition. However, seq-ChIP assays showed that RNAP II has no clear preference for histone-free RP5 genomes (Fig. 5C and D). We conclude that the removal of histones from viral templates is not a prerequisite for transcriptional activation by VP16. Interestingly, histones H3 and H2B were significantly underrepresented in some RNAP II-associated IE genes (Fig. 5C and D), suggesting that nucleosomes might be partially disrupted at actively transcribed IE genes. A similar phenomenon was suggested for transcriptionally active eukaryotic genes (19, 30, 33, 38, 58).
Another important question addressed in this study is the nature of chromatin on the viral genome during lytic infection. We suggest that the presence of all core histones on the viral genome during KOS and RP5 infections is indicative of the formation of canonical nucleosomes on the viral genome. This conclusion is supported by our findings from seq-ChIP assays, which indicated that histones H2B and H3 cooccupied the RP5 genomes in HSV-2-superinfected cells (Fig. 5A and B).
A crucial question is whether the low histone occupancy on the HSV-1 genome during lytic infection matters for transcription. A related issue is whether the dearth of histones on the viral genome during lytic infection is due to the prevention of histone deposition or the effective removal of histones that are deposited. In contrast to a recent report (6), we have noted that even at very early times in infection (30 min to 1 h), the number of histones on the viral genome is much lower than on cellular genes (data not shown), suggesting that histones may not be deposited at all on most viral genomes during lytic infection.
One possible model is that efficient transcription, which starts soon after the viral genome is released into the nucleus, might block histone deposition on the HSV-1 genome. If transcription does not start efficiently, as in the case of RP5 infections or in the presence of actinomycin D, then the histones are deposited on the viral genomes rather slowly, as indicated by the time course assays with results shown in Fig. 1 and 2. Therefore, we propose that histones are prevented from being deposited on the viral genome, rather than being deposited first and then actively removed. If this is the case, then the small number of histones on the viral genome may not matter for viral transcription during lytic infection. In this scenario, the small number of histones might represent randomly positioned histones on the viral genome. A second alternative is that during wild-type lytic infection, a small fraction of viral genomes are silenced by the deposition of nucleosomes. In either case, histones may not matter for the outcome of lytic infection, as a large fraction of actively transcribed viral genomes stay nonnucleosomal.
Given that histones are not present in the HSV-1 nucleocapsid (7, 50, 53) and are underrepresented on the HSV-1 genome during lytic infection, it is tempting to speculate that histone chaperones and assembly factors may be involved in regulating the viral chromatin structure. Some of the candidate chaperones include Spt6 and FACT, which associate with RNAP II and enable its elongation on nucleosomal templates (2, 25). Interestingly, the histone chaperone HIRA, which is involved in replication-independent histone deposition (18, 57), was shown to be present in promyelocytic leukemia bodies in senescent cells (69) and as such is likely to be involved in chromatin assembly on the viral genome. A recent study suggested that disruption of HIRA expression reduces both the association of histone H3.3 with the HSV-1 genome and the viral gene expression at later stages of infection, concomitant with a reduction in viral replication (54). On the other hand, it is not clear whether HIRA contributes to viral gene expression at early times in infection.
Nucleosome assembly on the viral genome at early times may be blocked by VP22, a tegument protein that interacts with TAF-I, a protein homologous to histone chaperone NAP-1 (64). However, our results (Fig. 1 and 2) suggest that VP22 delivered by the incoming virion is not sufficient for histone depletion, because the levels of VP22 are presumably unaffected by the absence of the VP16 AD or the presence of actinomycin D. Given these findings, it will be important to address whether histone chaperones are modulated in a way that leads to low levels of histones on the HSV-1 genome during different stages of infection.
Overall, we show that during lytic infection of HSV-1, histones are found at low levels on the viral genome; the cause of such levels may involve active transcription by RNAP II, as well as VP16 and IE proteins. Future studies should address the details of the underlying molecular mechanism in terms of histone deposition on or histone removal from the HSV-1 genome. In addition, our results suggest that IE proteins contribute to keeping other regions of the viral genome free of nucleosomes at later stages of lytic infection. Therefore, it will be crucial to define the cellular factors involved in depositing the histones on the viral genome. Lastly, it will be interesting to see whether and how chromatin assembly factors are regulated during establishment of and reactivation from latency of HSV-1.
Supplementary Material
Acknowledgments
This research was supported by the Van Andel Research Institute and by NIH grant AI064634. S.B.K. was supported by a predoctoral fellowship from the American Heart Association.
We thank Martha Roemer for initiating the ChIP analyses of core histones. We also thank David Nadziejka for assistance in preparing the manuscript and members of the Triezenberg lab for their comments on this work.
Footnotes
Published ahead of print on 25 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amelio, A. L., N. V. Giordani, N. J. Kubat, J. E. O'Neil, and D. C. Bloom. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 802063-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 3011090-1093. [DOI] [PubMed] [Google Scholar]
- 3.Black, J. C., J. E. Choi, S. R. Lombardo, and M. Carey. 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23809-818. [DOI] [PubMed] [Google Scholar]
- 4.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 111587-1598. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. [DOI] [PubMed] [Google Scholar]
- 6.Cliffe, A. R., and D. M. Knipe. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 8212030-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, H. M., V. Connor, Z. S. Cheng, F. Grey, C. M. Preston, and S. Efstathiou. 2008. Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression. J. Gen. Virol. 8968-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cress, A., and S. J. Triezenberg. 1991. Nucleotide and deduced amino acid sequences of the gene encoding virion protein 16 of herpes simplex virus type 2. Gene 103235-238. [DOI] [PubMed] [Google Scholar]
- 10.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14165-173. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 682818-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodrich, J. A., T. Hoey, C. J. Thut, A. Admon, and R. Tjian. 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75519-530. [DOI] [PubMed] [Google Scholar]
- 14.Gu, H., Y. Liang, G. Mandel, and B. Roizman. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. USA 1027571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, H., and B. Roizman. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. USA 10417134-17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez, J. L., M. Chandy, M. J. Carrozza, and J. L. Workman. 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 26730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, S., M. Brand, G. Mittler, J. Yanagisawa, S. Kato, M. Meisterernst, and L. Tora. 2002. TATA-binding protein-free TAF-containing complex (TFTC) and p300 are both required for efficient transcriptional activation. J. Biol. Chem. 27732875-32882. [DOI] [PubMed] [Google Scholar]
- 18.Haushalter, K. A., and J. T. Kadonaga. 2003. Chromatin assembly by DNA-translocating motors. Nat. Rev. Mol. Cell Biol. 4613-620. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff, S. 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 915-26. [DOI] [PubMed] [Google Scholar]
- 20.Herrera, F. J., and S. J. Triezenberg. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 789689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, J., J. R. Kent, B. Placek, K. A. Whelan, C. M. Hollow, P. Y. Zeng, N. W. Fraser, and S. L. Berger. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 805740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda, K., D. J. Steger, A. Eberharter, and J. L. Workman. 1999. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 19855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingles, C. J., M. Shales, W. D. Cress, S. J. Triezenberg, and J. Greenblatt. 1991. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351588-590. [DOI] [PubMed] [Google Scholar]
- 24.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 141899-1907. [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 3011096-1099. [DOI] [PubMed] [Google Scholar]
- 26.Kent, J. R., P. Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 7810178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemm, R. D., J. A. Goodrich, S. Zhou, and R. Tjian. 1995. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc. Natl. Acad. Sci. USA 925788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knipe, D. M., and A. Cliffe. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6211-221. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 30.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 234243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubat, N. J., A. L. Amelio, N. V. Giordani, and D. C. Bloom. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 7812508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 781139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulaeva, O. I., D. A. Gaykalova, and V. M. Studitsky. 2007. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat. Res. 618116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6551-561. [DOI] [PubMed] [Google Scholar]
- 35.Kutluay, S. B., J. Doroghazi, M. E. Roemer, and S. J. Triezenberg. 2008. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology 373239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutluay, S. B., S. L. DeVos, J. E. Klomp, and S. J. Triezenberg. 2009. Transcriptional coactivators are not required for herpes simplex virus type 1 immediate-early gene expression in vitro. J. Virol. 833436-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 151946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 39.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 5145-59. [DOI] [PubMed] [Google Scholar]
- 40.Lentine, A. F., and S. L. Bachenheimer. 1990. Intracellular organization of herpes simplex virus type 1 DNA assayed by staphylococcal nuclease sensitivity. Virus Res. 16275-292. [DOI] [PubMed] [Google Scholar]
- 41.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 42.Lin, Y. S., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1991. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353569-571. [DOI] [PubMed] [Google Scholar]
- 43.Memedula, S., and A. S. Belmont. 2003. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol. 13241-246. [DOI] [PubMed] [Google Scholar]
- 44.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 371090-1097. [DOI] [PubMed] [Google Scholar]
- 45.Morillon, A. 2006. Is histone loss a common feature of DNA metabolism regulation? Biochem. Cell Biol. 84450-462. [DOI] [PubMed] [Google Scholar]
- 46.Muggeridge, M. I., and N. W. Fraser. 1986. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J. Virol. 59764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 221615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4649-655. [DOI] [PubMed] [Google Scholar]
- 49.Neumann, D. M., P. S. Bhattacharjee, N. V. Giordani, D. C. Bloom, and J. M. Hill. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J. Virol. 8113248-13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh, J., and N. W. Fraser. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 823530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ottosen, S., F. J. Herrera, J. R. Doroghazi, A. Hull, S. Mittal, W. S. Lane, and S. J. Triezenberg. 2006. Phosphorylation of the VP16 transcriptional activator protein during herpes simplex virus infection and mutational analysis of putative phosphorylation sites. Virology 345468-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pichyangkura, R. 1995. The effect of deletion mutations of the activation domain of VP16 upon herpes simplex virus type 1 lytic infection. Ph.D. dissertation. Michigan State University, East Lansing.
- 53.Pignatti, P. F., and E. Cassai. 1980. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J. Virol. 36816-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Placek, B. J., J. Huang, J. R. Kent, J. Dorsey, L. Rice, N. W. Fraser, and S. L. Berger. 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 831416-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 111599-1607. [DOI] [PubMed] [Google Scholar]
- 56.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 57.Sarma, K., and D. Reinberg. 2005. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 6139-149. [DOI] [PubMed] [Google Scholar]
- 58.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2410111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stafford, G. A., and R. H. Morse. 2001. GCN5 dependence of chromatin remodeling and transcriptional activation by the GAL4 and VP16 activation domains in budding yeast. Mol. Cell. Biol. 214568-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tal-Singer, R., R. Pichyangkura, E. Chung, T. M. Lasner, B. P. Randazzo, J. Q. Trojanowski, N. W. Fraser, and S. J. Triezenberg. 1999. The transcriptional activation domain of VP16 is required for efficient infection and establishment of latency by HSV-1 in the murine peripheral and central nervous systems. Virology 25920-33. [DOI] [PubMed] [Google Scholar]
- 61.Triezenberg, S. J., R. C. Kingsbury, and S. L. McKnight. 1988. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 2718-729. [DOI] [PubMed] [Google Scholar]
- 62.Uhlmann, T., S. Boeing, M. Lehmbacher, and M. Meisterernst. 2007. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J. Biol. Chem. 2822163-2173. [DOI] [PubMed] [Google Scholar]
- 63.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394498-502. [DOI] [PubMed] [Google Scholar]
- 64.van Leeuwen, H., M. Okuwaki, R. Hong, D. Chakravarti, K. Nagata, and P. O'Hare. 2003. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J. Gen. Virol. 842501-2510. [DOI] [PubMed] [Google Scholar]
- 65.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Q. Y., C. Zhou, K. E. Johnson, R. C. Colgrove, D. M. Coen, and D. M. Knipe. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 10216055-16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28294-304. [DOI] [PubMed] [Google Scholar]
- 68.Xiao, H., A. Pearson, B. Coulombe, R. Truant, S. Zhang, J. L. Regier, S. J. Triezenberg, D. Reinberg, O. Flores, C. J. Ingles, et al. 1994. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol. Cell. Biol. 147013-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye, X., B. Zerlanko, R. Zhang, N. Somaiah, M. Lipinski, P. Salomoni, and P. D. Adams. 2007. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 272452-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.