Abstract
Viruses utilize host factors in many steps of their life cycles. Yet, little is known about host factors that contribute to the life cycle of hepatitis B virus (HBV), which replicates its genome by reverse transcription. To identify host factors that contribute to viral reverse transcription, we sought to identify cellular proteins that interact with HBV polymerase (Pol) by using affinity purification coupled with mass spectrometry. One of the HBV Pol-interacting host factors identified was DDX3 DEAD-box RNA helicase, which unwinds RNA in an ATPase-dependent manner. Recently, it was shown that DDX3 is essential for both human immunodeficiency virus and hepatitis C virus infection. In contrast, we found that the ectopic expression of DDX3 led to significantly reduced viral DNA synthesis. The DDX3-mediated inhibition of viral DNA synthesis did not affect RNA encapsidation, a step prior to reverse transcription, and indicated that DDX3 inhibits HBV reverse transcription. Mutational analysis revealed that mutant DDX3 with an inactive ATPase motif, but not that with an inactive RNA helicase motif, failed to inhibit viral DNA synthesis. Our interpretation is that DDX3 inhibits viral DNA synthesis at a step following ATP hydrolysis but prior to RNA unwinding. Finally, OptiPrep density gradient analysis revealed that DDX3 was incorporated into nucleocapsids, suggesting that DDX3 inhibits viral reverse transcription following nucleocapsid assembly. Thus, DDX3 represents a novel host restriction factor that limits HBV infection.
Viruses rely on host factors to complete their life cycles. These factors facilitate many steps of the viral life cycle, including entry, uncoating, genome replication, viral assembly, and virus release (3, 13). Recently, some host factors that contribute to the life cycles of some clinically important human viruses were identified by full-genome small interfering RNA knockdown experiments (7, 14, 31). For instance, nearly 300 host factors that contribute to human immunodeficiency virus (HIV) infection were identified (7). Yet, little is known about host factors that contribute to the genome replication of hepatitis B virus (HBV).
HBV, the prototypical member of the hepadnavirus family, is a major cause of liver disease worldwide (34). HBV-mediated disease manifestations range from acute and chronic hepatitis to liver cirrhosis and hepatocellular carcinoma (HCC). Although HBV contains a DNA genome, the replication of the genome occurs by reverse transcription of the pregenomic RNA (pgRNA) template. HBV polymerase (Pol), or reverse transcriptase, acts as an RNA binding protein by specifically recognizing an RNA stem-loop structure called the 5′ ɛ encapsidation signal (5′ ɛ), and this interaction is required for pgRNA encapsidation (5, 15, 17). Viral reverse transcription occurs entirely within nucleocapsids following encapsidation. HBV reverse transcription has two steps for DNA synthesis: (i) minus-strand DNA synthesis and (ii) plus-strand DNA synthesis. During the first step, the pgRNA is converted into the minus-strand DNA. Then, the minus-strand DNA serves as the template for plus-strand DNA synthesis, producing a form of circular double-stranded DNA (relaxed circular [RC] DNA). In addition to the RC DNA, double-stranded linear (DL) DNA is synthesized from in situ priming during the plus-strand DNA synthesis (34).
The members of the DEAD-box family are involved in all aspects of RNA metabolism, including pre-mRNA splicing, mRNA translation, and RNA export from the nucleus (20, 21, 32). In particular, DEAD-box RNA helicases, including DDX3, are RNA helicases that unwind double-stranded RNA in an energy-dependent manner. Both HIV and hepatitis C virus (HCV) have been shown to utilize DDX3 as a cofactor for genome replication. Specifically, DDX3 was shown to be critical for the Rev/Rev-responsive element export of unspliced HIV genomic RNA from the nucleus (39). In addition, the interaction between DDX3 and the HCV core protein was shown to be required for HCV genome replication (4, 29). Despite the common use of DDX3 by HIV and HCV, these viruses employ distinct mechanisms to subvert DDX3 for their own RNA metabolism needs.
Extensive genetic analysis has provided many mechanistic details of hepadnaviral reverse transcription (1, 2, 24, 25, 27, 35, 36). However, little is known about host factors that contribute to viral genome replication. We utilized an affinity pull-down analysis coupled with mass spectrometry to search for host factors that bind to HBV Pol. We found that DDX3 specifically interacted with HBV Pol; however, unlike HIV and HCV replication, which is enhanced by DDX3, HBV reverse transcription was inhibited by DDX3. Thus, DDX3 is a newly identified host restriction factor for HBV replication.
MATERIALS AND METHODS
Cell culture and transfection.
HepG2, HeLa, and HEK293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco-BRL) and 10 μg of gentamicin per ml at 37°C in 5% CO2 and were passaged every third day. Cells were transfected using polyethylenimine (25 kDa; Sigma-Aldrich) as described previously (33). The amounts of plasmid DNA with which cells were transfected (12 μg per 60-mm plate and 30 μg per 100-mm plate) were kept constant by the inclusion of DNA vector pcDNA3. Transfection efficiencies of over 50% were routinely obtained by using the polyethylenimine transfection protocol.
Plasmids.
All DNA constructs were generated by overlap extension PCR protocols as described previously (23). The details of the molecular cloning of any plasmid construct will be provided upon request. The HBV overlength 1.3-mer replicon construct (i.e., 1.3 U of the HBV ayw subtype genome) was made as described previously (10). The HBV Pol null construct was made by the introduction of a frameshift mutation at the unique EcoRI site of the HBV genome (nucleotide 3182) by EcoRI digestion followed by filling in with the Klenow fragment. The HBV ΔL42 Pol null construct encodes a capsid assembly-defective core protein, which is generated by the deletion of leucine 42 of the core protein and is expressed by the HBV Pol null construct (33). The HBV C null, Pol null construct that was used to express the pgRNA was made by mutating the 34th and 35th codons of the C (core) gene in the HBV Pol null construct to stop codons. To generate a Pol expression construct (pCEP4-Pol), the Pol open reading frame was inserted into pCEP4, an Epstein-Barr virus-derived episomal vector (Invitrogen). Subsequently, three copies of a sequence encoding the Flag epitope were inserted in frame at the end corresponding to the N terminus, as described previously (33). pHA-Luc and pFlag-Luc constructs, which express a firefly luciferase having either three copies of a hemagglutinin (HA) tag or three copies of a Flag tag at its N terminus, were made into the pcDNA3 plasmid (Invitrogen). All DDX3 constructs (HA-DDX3, the antisense DDX3 [AS-DDX3] construct, and the DDX3-Mut1 and DDX3-Mut2 constructs, which have been characterized previously [39]) were gifts from Kuan-Teh Jeang.
Western blot analysis.
Western blot analysis was performed as described previously (33). For the detection of the Flag-tagged Pol protein, mouse anti-Flag M2 antibody (Sigma; 1:5,000) was used. Rabbit anti-HBV core antibody (Dako) was used to detect HBV core protein. Mouse anti-DDX3 antibody was purchased from Abcam.
Stable cell lines and immunoaffinity purification.
To generate a cell line that stably expresses HBV Pol, HEK293 cells were transfected with the pCEP4-Pol construct and transformants were selected with hygromycin (400 μg/ml). Following selection, cells were maintained in medium containing 50 μg/ml hygromycin. Immunoaffinity purification was performed as described previously (26). Cytosolic extracts were prepared as described previously (12). Briefly, cell pellets harvested from 10 100-mm plates were lysed with 4 ml of NP-40 lysis buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail). Cell extracts were then incubated with anti-Flag M2-agarose (Sigma) at 4 °C for 3 to 6 h. After extensive washes with the NP-40 lysis buffer, proteins were eluted with 100 μg of 3× Flag peptides (Sigma) per ml in the NP-40 lysis buffer containing 100 mM NaCl. Immunopurified protein complexes were resolved on sodium dodecyl sulfate (SDS)-4 to 12% polyacrylamide gel electrophoresis (PAGE) gels (Invitrogen). After staining of the gels with a silver-staining kit (GE Healthcare), protein bands were excised and subjected to mass spectrometry.
Protein identification by mass spectrometry.
Nano-liquid chromatography (nano-LC)-tandem mass spectrometry (MS-MS) analysis was performed on an Agilent 1100 series nano-LC linear trap quadrupole mass spectrometer (Thermo Electron, San Jose, CA) as described previously (22). The SEQUEST system (Thermo Electron, San Jose, CA) was used to identify peptide sequences present in the protein sequence database as described previously (22).
Coimmunoprecipitation.
Immunoprecipitation was performed essentially as described previously (30). After transient transfection, the medium was removed and the cells were rinsed twice in cold phosphate-buffered saline, incubated for 30 min at 4°C in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, and 1% NP-40), and collected by scraping. Cell debris was removed by centrifugation at 10,000 × g for 10 min at 4°C. Extracts were precleared with protein G-agarose beads for 1 h at 4°C. The primary antibody was added for 1 h at 4°C, and immunoglobulin complexes were collected on protein G-agarose beads for 1 h at 4°C. The beads were washed five times with 1 ml of lysis buffer each time. Protein complexes were generally recovered by boiling in Laemmli sample buffer and were analyzed by SDS-PAGE.
Southern blot analysis.
Southern blotting was performed as described before (35).
Extraction of viral RNA.
Cells were harvested for RNA extraction 4 days posttransfection as described previously (17), with some modifications. To reduce the variability among plates during RNA extraction, both capsid and total cytoplasmic fractions were isolated from a single plate. HepG2 cells were washed twice with cold HBS-EGTA buffer (2 mM HEPES [pH 7.5], 100 mM NaCl, 0.5 mM EGTA) and lysed in lysis buffer (50 mM Tris-Cl [pH 8.0], 1 mM EDTA, 1% NP-40, and 100 mM NaCl) containing 5 mM vanadyl ribonucleoside complexes (Fluka). Then nuclei were removed by centrifugation. Half of the lysate was used to isolate total cytoplasmic RNA, and the other half of the lysate was used to isolate the capsid RNA. Capsids were collected by immunoprecipitation using anticore antibody (Dako). Briefly, cell lysate was mixed with anticore antibody for 1 h at 4 °C. Protein G-Sepharose was added, and the mixture was incubated for an additional 2 h at 4°C. Samples were washed with lysis buffer, and immunoprecipitates were collected by centrifugation. Total and capsid-associated RNAs were extracted with Trizol (Gibco-BRL).
RPA.
RNA was extracted as described previously (17) and evaluated by RNase protection analysis (RPA) according to the instructions of the RNase manufacturer (Ambion). The riboprobe was derived from the core region, as described previously (33). Briefly, each sample of RNA was hybridized with the equivalent of 105 cpm of a probe labeled with [α-32P]UTP (3,000 Ci/mmol; Amersham) for 16 h at 42°C. RNase digestion was carried out with a mixture of RNase A and RNase T1 for 30 min at 37°C. The digested products were separated on a 5% acrylamide-8 M urea gel. Radiographic phosphorimages were quantified using a bioimaging analyzer (BAS-2500; Fujifilm).
Density gradient analysis.
Density gradient analysis was performed with an OptiPrep (60% [wt/vol] iodixanol; Axis-Shield) velocity gradient, essentially as described previously (18). OptiPrep density gradients ranging from 10 to 50% were prepared in lysis buffer as five steps of 10% increments. An aliquot of 200 μl of cell lysate was carefully loaded onto the top of the gradient and centrifuged for 45 min at 55,000 rpm in a TLS 55 swing-out rotor (Beckman Instruments) at 20°C. Fourteen 80-μl fractions from the top were collected, and the protein content of each fraction was examined by immunoblot analysis. When indicated, cell lysates were treated with RNase A at a concentration of 100 μg/ml for 1 h at 37°C or proteinase K at a concentration of 60 μg/ml for 1 h at 37°C or both.
RESULTS
Experimental strategy.
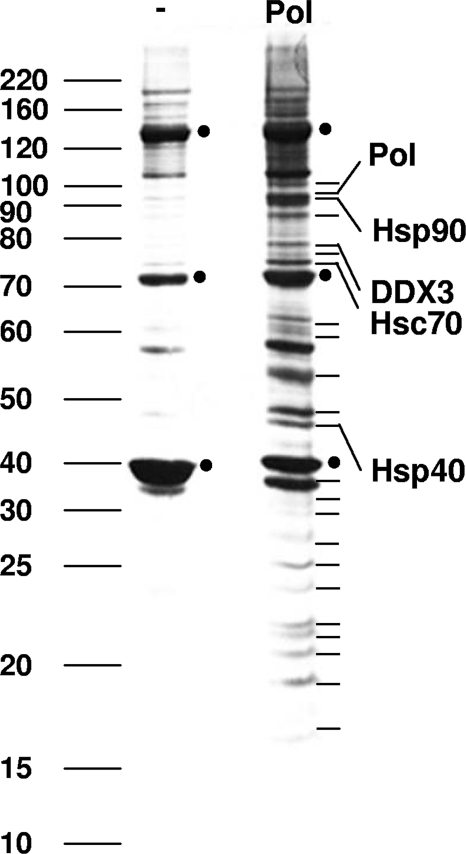
To search for host factors that bind to HBV Pol, we utilized affinity purification of HBV Pol followed by mass spectrometry to identify copurified host factors. A cell line that stably expresses HBV Pol was generated, as detailed in Materials and Methods, using an episomal Epstein-Barr virus vector, which has been employed previously to express a modest level of a bait protein (26). For affinity purification, HBV Pol was N-terminally tagged with three copies of the Flag epitope (Flag-tagged HBV Pol is hereinafter referred to as Flag-HBV Pol or HBV Pol). The N-terminal Flag tag did not interfere with HBV Pol function because it complemented the HBV Pol null mutation and induced viral genome replication to a level comparable to that occurring with untagged HBV Pol (data not shown). Flag-specific immunoaffinity isolation from the HBV Pol-expressing HEK293 cells was performed, and copurified HBV Pol-associated proteins were analyzed by SDS-PAGE and detected by silver staining. More than a dozen cellular proteins were copurified with HBV Pol but not isolated from parental HEK293 cells (Fig. 1). Protein bands that were specifically copurified by HBV Pol isolation were excised and identified by LC-MS-MS mass spectrometry. The p90 band was identified as HBV Pol. In addition, heat shock protein 90 (Hsp90), heat shock constitutive protein 70 (Hsc70), and Hsp40 were identified as HBV Pol binding proteins. These chaperones are well-characterized HBV Pol binding proteins, indicating that our pull-down analysis isolated cellular HBV Pol binding proteins effectively (6, 16). Some of the HBV Pol binding factors, such as the DDX3 DEAD-box RNA helicase (Fig. 1), appeared to be novel. Since DDX3 is important in HIV and HCV infections, we focused on determining the effect of DDX3 on HBV replication. While this report focuses on DDX3, several other cellular proteins were identified and will be discussed in a future publication (H. Wang and W.-S. Ryu, unpublished results).
FIG. 1.
Identification of DDX3 as an HBV Pol binding protein. Lysates of HEK293 cells stably expressing Flag-tagged HBV Pol were purified over anti-Flag M2-agarose beads, and the bound proteins were eluted by using 3× Flag peptides (lane Pol). Cell lysates from untransfected HEK293 cells were prepared in parallel (lane −). Eluted proteins were resolved by SDS-10% PAGE and visualized by silver staining. The proteins indicated by lines to the right were excised and subjected to LC-MS-MS. The bands identified, including those for HBV Pol, Hsp90, Hsc70, Hsp40, and DDX3, are labeled. Molecular weight markers are indicated to the left. Three immunoglobulin G bands are denoted by dots.
HBV Pol binds to DDX3 independently of the pgRNA.
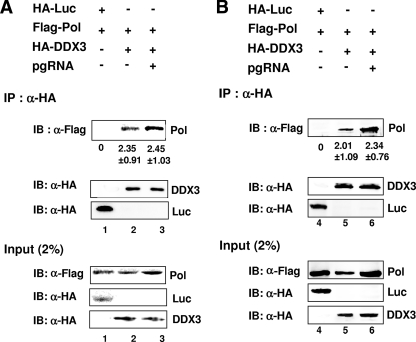
To confirm the interaction between HBV Pol and DDX3, we performed a coimmunoprecipitation assay (Fig. 2). To facilitate detection, DDX3 was tagged with the HA epitope at its N terminus. Cells were transfected with the constructs indicated in Fig. 2A, and coimmunoprecipitation was performed using an anti-HA antibody, followed by immunoblot analysis using an anti-Flag antibody. The immunoprecipitation of DDX3 with an anti-HA antibody coprecipitated HBV Pol (Fig. 2A, lane 2 of IP panel), indicating that HBV Pol interacts with DDX3. HBV Pol was not detected when the negative-control HA-tagged luciferase (HA-Luc) was immunoprecipitated, indicating that the interaction between HBV Pol and HA-DDX3 was specific (Fig. 2A, lane 1 of IP panel). HA-tagged DDX3 and luciferase were precipitated equally by the anti-HA antibody (Fig. 2A, IP panel). DDX3 and HBV Pol are both RNA binding proteins, and the pgRNA may mediate this interaction; therefore, we next examined the influence of the pgRNA on the interaction between HBV Pol and DDX3. The presence of pgRNA enhanced the interaction between HBV Pol and HA-DDX3 only modestly, but this result is probably because the expression of HBV Pol was slightly higher in cells containing the pgRNA (Fig. 2A, lane 3 of input panel). Thus, the HBV Pol-DDX3 interaction did not require the pgRNA. These data are supported by the stable interaction between DDX3 and HBV Pol in the initial mass spectrometry screen, in which pgRNA was not expressed (Fig. 1). The results of reciprocal immunoprecipitation assays using anti-Flag antibodies confirmed these findings (data not shown). Since DDX3 is an RNA binding protein, it is possible that the HBV Pol-DDX3 interaction is mediated by cellular RNAs. To address this issue, coimmunoprecipitation was performed following RNase and DNase treatment. The HBV Pol-DDX3 interaction was detected following RNase and DNase treatment (Fig. 2B), suggesting that the Pol-DDX3 interaction was not mediated by DNA or RNA.
FIG. 2.
Coimmunoprecipitation of DDX3 and HBV Pol. (A) HEK293 cells were transfected with 10 μg of each plasmid as indicated. The pgRNA carrying the 5′ ɛ stem-loop structure was provided by transfecting cells with the HBV C null, Pol null construct that could express the pgRNA but not the viral proteins. Anti-HA antibody (α-HA) was used to immunoprecipitate proteins from the cell lysates. The immunoprecipitated proteins (IP) were blotted with anti-HA antibody to detect HA-tagged proteins and anti-Flag antibody to detect coimmunoprecipitated HBV Pol. The fraction of immunoprecipitated HBV Pol out of the total cell lysate (as a percentage ± standard deviation) is denoted below each lane. Cell lysates (input, 2%) were analyzed in parallel by immunoblotting (IB). (B) An additional analysis was the same as that described in the legend to panel A, except that cell lysates were pretreated with RNase A (100 μg/ml) and DNase I (100 μg/ml) for 1 h at 37°C prior to immunoprecipitation. +, present; −, absent.
DDX3 inhibits HBV genome replication.
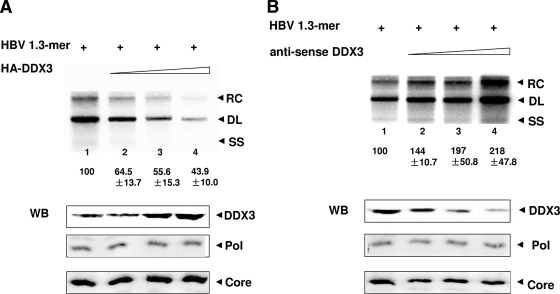
Having confirmed the interaction between HBV Pol and DDX3, we next examined the impact of DDX3 on HBV genome replication. Increasing amounts of the HA-DDX3 expression vector and an overlength 1.3-mer HBV construct that is capable of initiating viral DNA synthesis were used to transfect cells of the HepG2 line, a human hepatoma cell line (Fig. 3A). Three days following transfection, the HBV replication intermediates within capsids were extracted and analyzed by Southern blotting. Unexpectedly, the analysis indicated that the overexpression of DDX3 significantly diminished viral DNA synthesis in a dose-dependent manner (Fig. 3A), indicating that DDX3 inhibits viral DNA synthesis. Immunoblot analysis performed in parallel confirmed that both HBV Pol and core protein levels remained unchanged and that the DDX3 protein level increased accordingly (Fig. 3A).
FIG. 3.
DDX3 inhibits HBV genome replication. (A) Southern blot analysis. HepG2 cells were cotransfected with the HBV 1.3-mer construct and increasing amounts of the HA-DDX3 construct. (Top panel) Viral DNA strands within capsids were extracted and analyzed by Southern blotting. Viral replication intermediates, the RC, DL, and single-stranded (SS) DNA forms, are labeled. The value for lane 1 was set to 100, and values shown below the other lanes are relative to this standard. Data represent the means ± standard deviations of results from four independent transfections. (Bottom panel) DDX3, HBV Pol, and core protein levels were measured by Western blot analysis (WB) using anti-DDX3, anti-Flag, and anticore antibodies, respectively. (B) Southern blot analysis was performed similarly to that described in the legend to panel A, except that the AS-DDX3 construct was used in place of the HA-DDX3 construct for transfection. Western blot analysis was done as described in the legend to panel A. +, present.
Based upon the results described above, we predicted that viral DNA synthesis would increase if the expression of endogenous DDX3 was reduced. Thus, we examined the impact of endogenous DDX3 knockdown on HBV DNA synthesis. The AS-DDX3 (antisense) construct was used to decrease endogenous DDX3 levels, as described previously (39). HepG2 cells were cotransfected with increasing amounts of the AS-DDX3 construct and the HBV 1.3-mer construct (Fig. 3B). Three days following transfection, the viral replication intermediates within capsids were extracted and analyzed by Southern blotting. Consistent with our prediction, viral DNA levels increased upon the expression of the AS-DDX3 construct, in a dose-dependent manner (Fig. 3B). These data indicate that DDX3 inhibits HBV DNA synthesis. Immunoblot analysis performed in parallel confirmed that the HBV Pol and core protein levels remained unchanged but that the DDX3 protein level was diminished by the expression of the AS-DDX3 construct (Fig. 3B), as reported previously (39). Since the HBV Pol-DDX3 interaction was initially identified in HEK293 cells (Fig. 2), we examined the impact of DDX3 overexpression or knockdown on HBV genome replication in HEK293 cells. The effects of DDX3 on HBV replication in HEK293 cells were essentially identical to the results obtained with HepG2 cells (data not shown). Overall, these data indicate that DDX3 inhibits HBV DNA synthesis in both hepatoma cells and nonhepatoma cells.
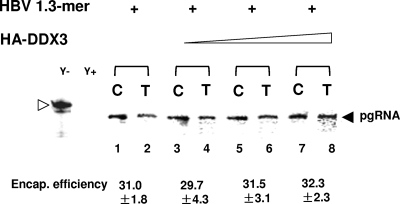
DDX3 does not affect pgRNA encapsidation.
We next wanted to determine at which step in HBV DNA synthesis DDX3 manifests its inhibitory effect. Since (i) DDX3 can unwind RNA secondary structure and (ii) DDX3 bound HBV Pol, which specifically recognizes the 5′ ɛ stem-loop structure of the pgRNA (see Fig. 9), it was possible that DDX3 inhibited HBV DNA synthesis by blocking encapsidation through the alteration of the structure of the 5′ ɛ stem-loop. Thus, we examined pgRNA encapsidation, the step preceding viral DNA synthesis or reverse transcription. To determine the effect of DDX3 overexpression on encapsidation, HepG2 cells were transfected with increasing amounts of the HA-DDX3 expression plasmid and the 1.3-mer construct, as indicated in Fig. 4. Three days following transfection, the cells were harvested and the cytoplasm was divided into two fractions: (i) total cytoplasm and (ii) capsid. RNAs from the fractions were extracted and analyzed by RPA (Fig. 4). The RPA data indicated that the encapsidation efficiency remained unchanged, even when DDX3 was overexpressed (Fig. 4). These data indicated that DDX3 did not inhibit encapsidation. Instead, DDX3 appeared to exert its inhibitory effect on HBV genome replication downstream of encapsidation, most likely by inhibiting reverse transcription.
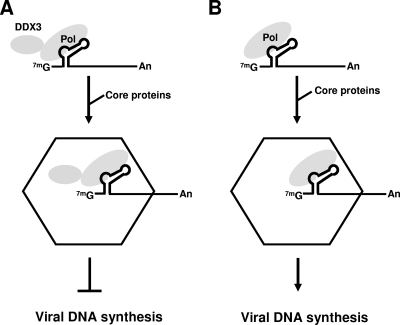
FIG. 9.
Schematic model illustrating the inhibitory role of DDX3 in HBV reverse transcription. The structure of the pgRNA is shown on top, with an emphasis on the 5′ ɛ stem-loop structure. It is most likely that the RNP complex (pgRNA-HBV Pol), rather than free HBV Pol, interacts with DDX3, although the data presented in Fig. 2 demonstrated that HBV Pol could interact with DDX3 in the absence of the pgRNA. Following the incorporation of DDX3 into the nucleocapsid (hexagon), DDX3 inhibits viral reverse transcription, perhaps disrupting a secondary structure of the pgRNA that is necessary for viral genome replication (1, 28, 36). Nucleocapsids assembled with DDX3 represent the replication-incompetent particles (A), whereas nucleocapsids assembled without DDX3 represent the replication-competent particles that can execute viral reverse transcription (B).
FIG. 4.
DDX3 does not inhibit HBV pgRNA encapsidation. HepG2 cells were cotransfected with the HBV 1.3-mer construct and increasing amounts of the HA-DDX3 construct. Approximately three times more cell equivalents of the core particles (C) than of total cytoplasmic RNA (T) were loaded. Extracted RNAs were analyzed by RPA, as detailed in Materials and Methods. The riboprobes for the detection of the pgRNA and the protected fragment of the pgRNA are labeled by open and closed arrowheads, respectively. Yeast RNA served as a negative control and was analyzed without (Y−) and with (Y+) RNase treatment. The encapsidation (encap.) efficiency (expressed as a percentage) was estimated by comparing the amount of the pgRNA detected in the C fraction to that detected in the T fraction. RPA results representative of those from at least four experiments are shown. Data represent the means ± standard deviations of results from four independent transfections. +, present.
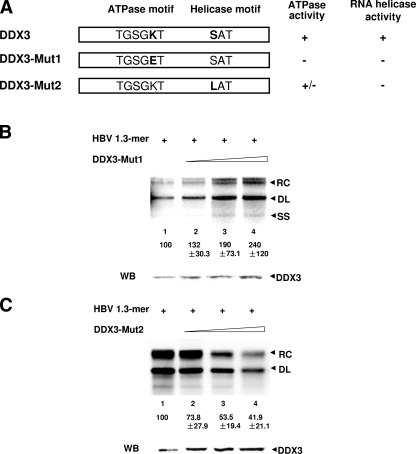
DDX3 ATPase activity is required for the inhibition of HBV genome replication.
Since DDX3 is an RNA helicase, we next sought to determine if RNA helicase activity is required for the inhibition of HBV DNA synthesis. To test this issue, we utilized two DDX3 mutants, DDX3-Mut1 and DDX3-Mut2, that have been characterized previously (39). DDX3-Mut1 has an inactive ATPase motif, while DDX3-Mut2 has a mutation of the SAT motif that unlinks ATPase activity from the RNA helicase activity (Fig. 5A). Since DDX3-Mut1 has no ATPase activity, it also lacks RNA helicase activity (39). In contrast, DDX3-Mut2 retains its ATPase activity but lacks RNA helicase activity (39). HBV synthesis of the 1.3-mer construct was examined following the ectopic expression of these two mutants. Unexpectedly, Southern blot analysis showed that HBV DNA synthesis was enhanced upon the expression of DDX3-Mut1 (Fig. 5B). The upregulation of HBV DNA by the expression of DDX3-Mut1 suggested that DDX3-Mut1 may act in a dominant-negative manner, antagonizing endogenous DDX3 in the suppression of viral DNA synthesis (see Discussion). On the other hand, HBV DNA synthesis was reduced upon the expression of DDX3-Mut2 (Fig. 5C). Thus, DDX3-Mut2 retained HBV replication-inhibiting activity similar to that of wild-type (WT) DDX3. Since DDX3-Mut2 retained ATPase activity, this function may be responsible for the inhibitory activity on viral DNA synthesis (see Discussion).
FIG. 5.
DDX3 inhibits viral genome replication in a manner that requires ATPase activity. (A) Schematic representation of the two DDX3 mutants. The lysine (K) in the ATPase motif was changed into glutamic acid (E) in DDX3-Mut1, while in DDX3-Mut2, the serine (S) in the helicase motif was changed into leucine (L). DDX3-Mut1 loses both ATPase and helicase activity, while DDX3-Mut2 lacks RNA helicase activity but maintains ATPase activity, as shown by the chart to the right. +, present; −, absent. (B) Southern blot analysis was performed as described in the legend to Fig. 3A but with increasing amounts of the DDX3-Mut1 construct. The value for lane 1 was set to 100, and values shown below the other lanes are relative to this standard. Data represent the means ± standard deviations of results from four independent transfections. The DDX3 level was determined by Western blot analysis (WB) using an anti-DDX3 antibody (bottom panel). SS, single-stranded DNA. (C) Southern blot analysis was performed as described in the legend to Fig. 3A but with increasing amounts of the DDX3-Mut2 construct. The details are the same as those given in the legend to panel B.
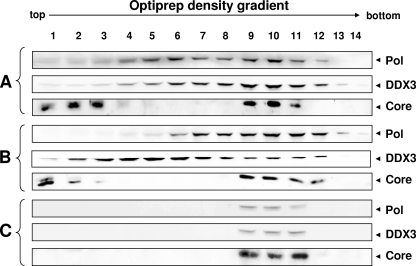
DDX3 is incorporated into the nucleocapsid.
The DDX3-mediated inhibition of reverse transcription led us to speculate that DDX3 may exert its inhibitory activity inside of the nucleocapsid. To determine if DDX3 is incorporated into capsid particles, we carried out density gradient analyses of nucleocapsids. We employed the 1.3-mer HBV Pol null construct, which can induce viral genome replication only when complemented by HBV Pol expression, as described previously (33). HEK293 cells were transfected with the HBV Pol null, HBV Pol, and HA-DDX3 expression constructs. Cell lysates were subjected to OptiPrep density gradient centrifugation, and DDX3, HBV Pol, and the core protein in each fraction were detected by immunoblot analysis with the respective antibodies (Fig. 6A). The core protein was detected in fractions 9 to 11, indicating that these three fractions contained HBV nucleocapsids. In contrast, HBV Pol and DDX3 were detected primarily in fractions 5 to 12. Because DDX3 is involved in RNA metabolism, we speculated that the DDX3 detected may represent DDX3 associated with various cellular RNP complexes. To remove these RNP complexes, cell lysates were treated with RNase A prior to density gradient analysis (Fig. 6B). Following RNase A treatment, DDX3 was abundantly detected in upper fractions, with a possible peak in fraction 5, reflecting the dissociation of DDX3 from the RNP complexes. HBV Pol and core proteins, similar to those in the analysis described above, were detected in fractions 5 to 12 and fractions 9 to 11, respectively (Fig. 6B). To determine whether DDX3 enzymes detected in fractions 9 to 11 are incorporated into nucleocapsid particles, cell lysates were treated with proteinase K and RNase A. Remarkably, the data revealed that DDX3 and HBV Pol were detected only in the fractions containing HBV nucleocapsids (Fig. 6C). Weak detection of both HBV Pol and DDX3, in comparison to that of the core protein, may reflect the fact that only a few molecules of these two proteins are incorporated into nucleocapsid particles, as opposed to 180 or 240 molecules of the core proteins, depending on the triangulation number of the capsids (37). Further, the data suggested that the vast majority of HBV Pol proteins remained unencapsidated, since they were sensitive to proteinase K treatment (Fig. 6C). Parallel findings for unencapsidated Pol proteins in duck HBV, as well as HBV, were reported previously (9, 38). Finally, a subset of DDX3 detected in the fractions containing nucleocapsids was resistant to proteinase K treatment, indicating that DDX3 was incorporated into the nucleocapsid particles.
FIG. 6.
DDX3 is incorporated into nucleocapsids. An OptiPrep density gradient analysis was performed as detailed in Materials and Methods. HEK293 cells were cotransfected with the 1.3-mer HBV Pol null construct and the HBV Pol and HA-DDX3 expression constructs. Three days following transfection, the cells were lysed with NP-40 lysis buffer and the lysates were subjected to a 10 to 50% OptiPrep density gradient. Results obtained after no treatment (A), after RNase A treatment (B), and after RNase A and proteinase K treatments (C) are shown. The HBV Pol, DDX3, and core proteins were detected by Western blot analysis with anti-Flag, anti-HA, and anticore antibodies, respectively.
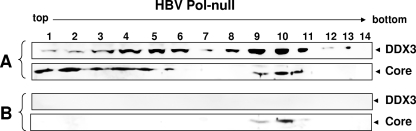
DDX3 is incorporated into nucleocapsids in an HBV Pol-dependent manner.
It was possible that DDX3 was incorporated into nucleocapsids independently of HBV Pol, because HBV core protein can assemble into capsids without HBV Pol (8). To assess this possibility, we carried out density gradient analyses of nucleocapsids following transfection with the HBV Pol null construct and the HA-DDX3 plasmid but without the HBV Pol expression plasmid (Fig. 7). Capsids were detected in fractions 9 to 11 (Fig. 7A), while DDX3 was detected in fractions 3 to 11, with two possible peaks in fractions 5 and 10 (Fig. 7A). However, DDX3 became undetectable following proteinase K treatment (Fig. 7B), indicating that DDX3 was not incorporated into nucleocapsids. Together, these data support the likelihood of the HBV Pol-mediated incorporation of DDX3 into nucleocapsids.
FIG. 7.
The incorporation of DDX3 into nucleocapsids depends on HBV Pol. Cells were cotransfected with the 1.3-mer HBV Pol null construct and HA-DDX3 expression constructs. Note that the HBV Pol expression plasmid was not included for transfection. OptiPrep density gradient analysis was done as described in the legend to Fig. 6. Results obtained after no treatment (A) and after RNase A and proteinase K treatments (B) are shown. The DDX3 and core proteins were detected by Western blot analysis with anti-HA and anticore antibodies, respectively.
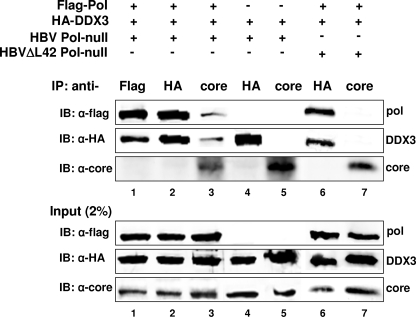
Encapsidated DDX3 is detected following immunoprecipitation with anticore antibody.
Since DDX3 was relatively abundant in the fractions containing capsids (fractions 9 to 11) prior to proteinase K treatment (Fig. 6 and 7), it is possible that the trace amount of DDX3 detected following proteinase K treatment may represent residual DDX3 molecules remaining due to incomplete digestion. However, if DDX3 is incorporated into capsids, then DDX3 should be detectable following the immunoprecipitation of capsid particles by using anticore antibodies in a method similar to that for the detection of duck HBV Pol in capsids (38). HEK293 cells were transfected with the HBV Pol null, HBV Pol, and HA-DDX3 expression constructs. Immunoprecipitation was carried out with the antibodies indicated above the lanes in Fig. 8, and HBV Pol, DDX3, and core protein in each fraction were detected by immunoblotting with the respective antibodies. Immunoprecipitation with anti-Flag or anti-HA antibody showed that the HBV Pol-DDX3 interaction occurs in the context of the replicon system, as anticipated (Fig. 8, upper panel, lanes 1 and 2). In contrast, the lack of detection of the core protein suggested that Pol-core or DDX3-core interactions did not measurably occur (Fig. 8, lanes 1 and 2). However, both HBV Pol and DDX3 were modestly detected by immunoprecipitation with the anticore antibody in a manner that depended on HBV Pol expression (Fig. 8, upper panel, lane 3 versus lane 5). This result is consistent with the detection of HBV Pol and DDX3 molecules incorporated into nucleocapsids. Further, the modest detection of both Pol and DDX3 after immunoprecipitation with anticore antibody was consistent with the incorporation of one or a few of these molecules into nucleocapsids. Finally, Pol and DDX3 were not detectable when the capsid assembly-defective ΔL42 mutant, which has a deletion of leucine 42 from the core protein, was expressed from the HBV ΔL42 Pol null construct (Fig. 8, upper panel, lane 7) (19). These results further supported the notion that DDX3 detected after the immunoprecipitation of the core protein (Fig. 8, upper panel, lane 3) was contained within capsid particles. Overall, the data presented here indicate that DDX3 is incorporated into capsid particles.
FIG. 8.
Evidence that DDX3 is incorporated into nucleocapsid particles. Cells were transfected as indicated above each lane, and cell lysates were immunoprecipitated with the antibodies indicated above each lane. The immunoprecipitated proteins (IP) were detected by immunoblotting (IB) using the antibodies denoted to the left. A fraction of total lysate (input, 2%) was analyzed in parallel by immunoblotting. α-Flag, α-HA, and α-core, anti-Flag, anti-HA, and anticore antibodies, respectively; +, present; −, absent.
DISCUSSION
This study identified HBV Pol binding proteins by using affinity purification coupled with mass spectrometry. The work presented here focused on one identified host factor, the DDX3 DEAD-box RNA helicase. We provided evidence that DDX3 inhibited HBV reverse transcription. Further, we showed that DDX3 is incorporated into nucleocapsids in an HBV Pol-dependent manner. Based upon this model, HBV Pol in nucleocapsids assembled without DDX3 would be capable of viral reverse transcription, while Pol in nucleocapsids assembled with DDX3 would not (Fig. 9). To our knowledge, DDX3 is the first host factor that inhibits HBV reverse transcription following incorporation into nucleocapsids.
These data indicate that DDX3 inhibits viral reverse transcription. First, we showed that the ectopic expression of DDX3 diminished HBV DNA synthesis (Fig. 3A). This finding was further strengthened by the observation that the knockdown of endogenous DDX3 enhanced HBV DNA synthesis (Fig. 3B). Given that DDX3 possesses ATPase-dependent RNA helicase activity, it is conceivable that a secondary structure in the RNA template, such as the 5′ ɛ stem-loop structure, may be the target for DDX3's inhibitory action (39). However, the observation that DDX3 does not block pgRNA encapsidation (Fig. 4), a process that requires the integrity of the stem-loop structure, indicated that the 5′ ɛ stem-loop structure is not the target of the inhibitory activity of DDX3. Instead, the data suggested that DDX3 exerts its inhibitory effect on viral reverse transcription. Southern blot analysis indicated that RC and DL DNA levels were reduced to similar extents (Fig. 3A), suggesting a defect in minus-strand DNA synthesis, which is the first step of reverse transcription. We speculated that a secondary structure arising during the minus-strand DNA synthesis, such as one involving base pairing between the 5′ ɛ sequence and the 3′ Φ sequence, might be the target for DDX3 (1, 28, 36). To determine the actual target of DDX3, further investigation is needed to uncover the exact step of viral reverse transcription in which DDX3 interferes.
Importantly, we demonstrated that DDX3 is incorporated into nucleocapsid particles (Fig. 6). A density gradient analysis, together with RNase and proteinase K protection analyses, revealed that DDX3 is packaged into nucleocapsids in an HBV Pol-dependent manner (Fig. 6 and Fig. 7). This finding is further strengthened by the detection of DDX3 from immunoprecipitated capsids in a manner dependent on HBV Pol expression and the functional assembly of capsid particles (Fig. 8). Thus, we speculate that DDX3 inhibits HBV reverse transcription following incorporation into nucleocapsids.
Finally, genetic analysis of DDX3 provided further insights into the mechanism by which DDX3 interferes with HBV reverse transcription. Similar to that of WT DDX3, the expression of DDX3-Mut2 inhibited viral DNA synthesis (Fig. 5C). Since both WT DDX3 and DDX3-Mut2 retain ATPase activity, the simplest interpretation would be that the ATPase activity exerts the inhibitory effect on HBV DNA synthesis (39). In contrast, the expression of DDX3-Mut1 enhanced viral DNA synthesis (Fig. 5B). Perhaps the incorporation of DDX3-Mut1 into nucleocapsid particles (data not shown) precludes the incorporation of endogenous DDX3, resulting in enhanced DNA synthesis. Thus, the incorporation of the noninhibitory DDX3-Mut1 into nucleocapsids acts in a dominant-negative manner against the inhibitory activity of endogenous DDX3. Previously, DDX3 was shown to exhibit ATPase-dependent RNA helicase activity such that ATP hydrolysis, by ATPase activity, precedes RNA helicase activity (32). Thus, it is possible that the inhibition of HBV reverse transcription occurs at a step following ATP hydrolysis but prior to helicase activity. We favor this possibility.
DDX3 is a novel cellular host factor that restricts HBV genome replication. Recently, DDX3 was found to be deregulated in hepatitis virus-associated HCC, a finding that defines DDX3 as a potential tumor suppressor gene (11). It is of interest to determine whether the deregulation of DDX3 is linked to the advancement of HBV-associated HCC. In addition, the extent to which DDX3 may contribute to the tissue tropism of HBV infection remains to be determined.
Acknowledgments
We thank K. T. Jeang (NIH) for providing DDX3 constructs and Byung-Yoon Ahn (Korea University) for critical reading of the manuscript.
This work was supported by a Korea Research Foundation grant (KRF-2008-C00271).
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Abraham, T. M., and D. D. Loeb. 2006. Base pairing between the 5′ half of ɛ and a cis-acting sequence, Φ, makes a contribution to the synthesis of minus-strand DNA for human hepatitis B virus. J. Virol. 804380-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham, T. M., and D. D. Loeb. 2007. The topology of hepatitis B virus pregenomic RNA promotes its replication. J. Virol. 8111577-11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariumi, Y., M. Kuroki, K. Abe, H. Dansako, M. Ikeda, T. Wakita, and N. Kato. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 8113922-13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., M. Junker-Niepmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 645324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck, J., and M. Nassal. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 27836128-36138. [DOI] [PubMed] [Google Scholar]
- 7.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319921-926. [DOI] [PubMed] [Google Scholar]
- 8.Bruss, V. 2007. Hepatitis B virus morphogenesis. World J. Gastroenterol. 1365-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, F., and J. E. Tavis. 2004. Detection and characterization of cytoplasmic hepatitis B virus reverse transcriptase. J. Gen. Virol. 853353-3360. [DOI] [PubMed] [Google Scholar]
- 10.Cha, M.-Y., D.-K. Ryu, H.-S. Jung, H.-E. Chang, and W.-S. Ryu. 2009. Stimulation of hepatitis B virus genome replication by HBx is linked to both nuclear and cytoplasmic HBx expression. J. Gen. Virol. 90978-986. [DOI] [PubMed] [Google Scholar]
- 11.Chang, P. C., C. W. Chi, G. Y. Chau, F. Y. Li, Y. H. Tsai, J. C. Wu, and Y. H. Wu Lee. 2006. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene 251991-2003. [DOI] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff, S. P. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5253-263. [DOI] [PubMed] [Google Scholar]
- 14.Hao, L., A. Sakurai, T. Watanabe, E. Sorensen, C. A. Nidom, M. A. Newton, P. Ahlquist, and Y. Kawaoka. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344552-555. [DOI] [PubMed] [Google Scholar]
- 16.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 931060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong, J. K., G. S. Yoon, and W. S. Ryu. 2000. Evidence that the 5′-end cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J. Virol. 745502-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kock, J., M. Kann, G. Putz, H. E. Blum, and F. Von Weizsacker. 2003. Central role of a serine phosphorylation site within duck hepatitis B virus core protein for capsid trafficking and genome release. J. Biol. Chem. 27828123-28129. [DOI] [PubMed] [Google Scholar]
- 19.Koschel, M., D. Oed, T. Gerelsaikhan, R. Thomssen, and V. Bruss. 2000. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J. Virol. 741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, M. C., Y. H. Lee, and W. Y. Tarn. 2008. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell 193847-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C. S., A. P. Dias, M. Jedrychowski, A. H. Patel, J. L. Hsu, and R. Reed. 2008. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 364708-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, H. J., M. J. Kang, E. Y. Lee, S. Y. Cho, H. Kim, and Y. K. Paik. 2008. Application of a peptide-based PF2D platform for quantitative proteomics in disease biomarker discovery. Proteomics 83371-3381. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J., H.-J. Lee, M.-K. Shin, and W.-S. Ryu. 2004. Versatile PCR-mediated insertion or deletion mutagenesis. BioTechniques 36398-400. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J., M.-K. Shin, H.-J. Lee, G. Yoon, and W.-S. Ryu. 2004. Three novel cis-acting elements required for efficient plus-strand DNA synthesis of the hepatitis B virus genome. J. Virol. 787455-7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, N., R. Tian, and D. D. Loeb. 2003. Base pairing among three cis-acting sequences contributes to template switching during hepadnavirus reverse transcription. Proc. Natl. Acad. Sci. USA 1001984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min, K. W., J. W. Hwang, J. S. Lee, Y. Park, T. A. Tamura, and J. B. Yoon. 2003. TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 27815905-15910. [DOI] [PubMed] [Google Scholar]
- 27.Mueller-Hill, K., and D. D. Loeb. 2002. cis-Acting sequences 5E, M, and 3E interact to contribute to primer translocation and circularization during reverse transcription of avian hepadnavirus DNA. J. Virol. 764260-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oropeza, C. E., and A. McLachlan. 2007. Complementarity between epsilon and phi sequences in pregenomic RNA influences hepatitis B virus replication efficiency. Virology 359371-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owsianka, A. M., and A. H. Patel. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257330-340. [DOI] [PubMed] [Google Scholar]
- 30.Pyronnet, S., H. Imataka, A. C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randall, G., M. Panis, J. D. Cooper, T. L. Tellinghuisen, K. E. Sukhodolets, S. Pfeffer, M. Landthaler, P. Landgraf, S. Kan, B. D. Lindenbach, M. Chien, D. B. Weir, J. J. Russo, J. Ju, M. J. Brownstein, R. Sheridan, C. Sander, M. Zavolan, T. Tuschl, and C. M. Rice. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 10412884-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocak, S., and P. Linder. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5232-241. [DOI] [PubMed] [Google Scholar]
- 33.Ryu, D. K., S. Kim, and W. S. Ryu. 2008. Hepatitis B virus polymerase suppresses translation of pregenomic RNA via a mechanism involving its interaction with 5′ stem-loop structure. Virology 373112-123. [DOI] [PubMed] [Google Scholar]
- 34.Seeger, C., F. Zoulim, and W. Mason. 2007. Hepadnaviruses, p. 2977-3029. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 35.Shin, M.-K., J. Lee, and W.-S. Ryu. 2004. A novel cis-acting element facilitates minus-strand DNA synthesis during reverse transcription of the hepatitis B virus genome. J. Virol. 786252-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin, M.-K., J.-H. Kim, D.-K. Ryu, and W.-S. Ryu. 2008. Circularization of an RNA template via long-range base pairing is critical for hepadnaviral reverse transcription. Virology 371362-373. [DOI] [PubMed] [Google Scholar]
- 37.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3771-780. [DOI] [PubMed] [Google Scholar]
- 38.Yao, E., Y. Gong, N. Chen, and J. E. Tavis. 2000. The majority of duck hepatitis B virus reverse transcriptase in cells is nonencapsidated and is bound to a cytoplasmic structure. J. Virol. 748648-8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yedavalli, V. S., C. Neuveut, Y. H. Chi, L. Kleiman, and K. T. Jeang. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119381-392. [DOI] [PubMed] [Google Scholar]