Abstract
The internal hydrophobic sequence within the flaviviral capsid protein (protein C) plays an important role in the assembly of infectious virions. Here, this sequence was analyzed in a West Nile virus lineage I isolate (crow V76/1). An infectious cDNA clone was constructed and used to introduce deletions into the internal hydrophobic domain which comprises helix α2 and part of the loop intervening helices α2 and α3. In total, nine capsid deletion mutants (4 to 14 amino acids long) were constructed and tested for virus viability. Some of the short deletions did not significantly affect growth in cell culture, whereas larger deletions removing almost the entire hydrophobic region significantly impaired viral growth. Efficient growth of the majority of mutants could, however, be restored by the acquisition of second-site mutations. In most cases, these resuscitating mutations were point mutations within protein C changing individual amino acids into more hydrophobic residues, reminiscent of what had been observed previously for another flavivirus, tick-borne encephalitis virus. However, we also identified viable spontaneous pseudorevertants with more than one-third of the capsid protein removed, i.e., 36 or 37 of a total of 105 residues, including all of helix α3 and a hydrophilic segment connecting α3 and α4. These large deletions are predicted to induce formation of large, predominantly hydrophobic fusion helices which may substitute for the loss of the internal hydrophobic domain, underlining the unrivaled structural and functional flexibility of protein C.
The genus Flavivirus within the family Flaviviridae comprises important human pathogens such as Japanese encephalitis virus (JEV), the dengue viruses (DENV), yellow fever virus (YFV), tick-borne encephalitis virus (TBEV) and West Nile virus (WNV) (28). The ∼50-nm flavivirus virion is composed of two surface proteins, envelope (E) and membrane (M, derived from its precursor protein prM by furin-mediated cleavage), and the nucleocapsid consisting of the capsid protein (protein C) and the 11-kb positive-stranded RNA genome. In addition to the three structural proteins C, prM, and E, the genome encodes seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), which are necessary for replication of the RNA genome (28). Structural and nonstructural proteins are derived from a single polyprotein, which is co- and posttranslationally processed into mature proteins by viral and cellular proteases (6, 28).
The assembly of the virions is thought to occur at the membrane of the rough endoplasmic reticulum (ER) (28, 32). Protein C, which is the protein located at the very N terminus of the polyprotein, facilitates translocation of the subsequent protein prM into the lumen of the ER via an internal signal sequence located at its C terminus. Proteins prM and E remain attached to the host-derived membrane by spanning the lipid bilayer twice via their C-terminal anchor regions (38, 47, 48). Protein C is originally also anchored to the ER membrane via the C-terminal internal signal sequence. However, this signal sequence is cleaved off by the viral NS2B/3 protease, thereby producing the mature, cytoplasmic form of the protein (4, 30, 40). Multiple copies of protein C and one copy of the RNA genome form the nucleocapsid. In the virion, the nucleocapsid appears not to directly interact with the surrounding membrane and the embedded surface proteins prM and E (47) and furthermore lacks, in contrast to the icosahedrally arranged surface proteins, a well-ordered structure (25, 49). Instead, the nucleocapsid may nonspecifically interact with the ER membrane during budding by virtue of a hydrophobic, mostly helical sequence element which is present at a conserved position in all of the flavivirus protein C sequences (31, 35).
The recently solved three-dimensional (3D) structures of the DENV-2 and Kunjin virus C proteins (Kunjin virus is an Australian strain of WNV) (11, 31) support this notion. The nuclear magnetic resonance 3D structure of DENV-2 protein C indicates that the protein, composed of four α helices, forms a dimer in solution (31). The contact surfaces for dimerization are provided by helices α2 and α4. Helix α2 comprises most of the internal hydrophobic sequence within protein C. After dimerization, the interacting helices α2 form the bottom of a hydrophobic cleft. The highest density of positively charged residues is found on the opposite side of the dimer, on the surfaces of helices α4, which interact by forming a coiled coil. Accordingly, a model suggesting that the hydrophobic cleft presumably enables the nucleocapsid to attach to the membrane and that helices α4 play a potential role in interaction with the negatively charged RNA genome has been established (31).
The functional importance of the conserved internal hydrophobic domain in virus assembly and/or dimerization of protein C is supported by studies with a variety of flaviviruses. For instance, removal of major parts of this sequence element in TBEV resulted in an increased formation of capsidless subviral particles (19), the secretion of which is also observed in the course of natural infection or by expression of proteins prM and E only (1, 22-24, 36). Large deletions were tolerated only upon the acquisition of additional mutations increasing the hydrophobicity of the protein (21). These results are in good accordance with studies of YFV protein C (42) and WNV, in which case removal of the entire helix α2 produced a noninfectious phenotype (44). Furthermore, in DENV, removing large parts of the hydrophobic domain abolished both the ability to dimerize in vitro (46) and the ability to associate with the ER membrane (35). Taken together, these studies underlined the important roles of the conserved internal hydrophobic sequence in dimerization of protein C and virion assembly.
In the present study, we set out to systematically test the functional role of the hydrophobic sequence of WNV protein C for viral infectivity by introducing deletions ranging from 4 to 14 amino acids. Some of the smaller deletions were well tolerated, whereas growth with others was, consistent with previous findings with TBEV (21), dependent on the acquisition of second-site point mutations. Surprisingly, two well-replicating pseudorevertants were shown to have restored growth capability through spontaneously enlarged deletions. These mutants lacked more than one-third of the protein C sequence. The spontaneously deleted sequences included all of helix α3 and a hydrophilic loop connecting helices α3 and α4. Our data provide evidence that although removal of large parts or the entire internal hydrophobic domain usually causes severe defects in viral growth, truncating protein C even further can largely revert this impairment. A viable WNV mutant with capsid proteins less than two-thirds of the size of the natural protein was generated by reverse genetics, and its growth properties were analyzed in comparison to those of the wild-type virus. Secondary-structure predictions suggest that in these mutants, the formation of large, hydrophobic fusion helices might compensate for the loss of the conserved hydrophobic domain.
MATERIALS AND METHODS
Cells and virus.
Vero (ATCC CCL-81) cells were grown in Eagle′s minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS; PAA Laboratories), 1.5% glutamine (200 mM; Cambrex), 1% penicillin-streptomycin (10,000 U/ml penicillin and 10 mg/ml streptomycin; Sigma), and 15 mM HEPES, pH 7.4. Infections were performed in the presence of 2% instead of 10% FBS, and after infection, cells were maintained in medium lacking FBS. For virus stock production, FBS was replaced with 1% (wt/vol) bovine serum albumin (BSA). BHK-21 cells used for introduction of in vitro-transcribed RNA were handled in growth medium (EMEM supplemented with 5% FBS, 1% glutamine, 0.5% [10 mg/ml] neomycin, and 15 mM HEPES, pH 7.4) and maintenance medium (EMEM supplemented with 1% FBS, 1% glutamine, 0.5% neomycin, and 15 mM HEPES, pH 7.4) as described earlier (19, 33, 41).
The WNV strain used in this study was originally isolated from a dead crow collected during the summer of 1999 in New York City (crow V76/1). The virus was passaged three times in Vero cells and once in suckling mouse brain prior to the construction of the infectious cDNA clone.
Cloning procedures.
The two partial cDNA clones pWNV-K1 and pWNV-K4 were constructed as described in previous studies (7, 33, 45), with the exception that pBR322 (5) had been modified by replacing the tetracycline resistance gene with a multiple-cloning site (BspEI-SwaI-PacI-NotI-SwaI-AatII). For the introduction of deletions into the capsid protein within plasmid pWNV-K1, the Gene Tailor site-directed mutagenesis system (Invitrogen) was used. Detailed primer sequences for all constructs are available from the authors upon request.
All constructs were amplified in Escherichia coli strain DH5α cells and characterized by complete sequencing of both strands of the entire inserts.
In vitro RNA transcription and transfection.
In vitro transcription with T7 RNA polymerase (Ambion T7 Megascript transcription kit) and transfection of BHK-21 cells by electroporation were performed as described in previous studies (12, 19). In the case of transcription reactions required as standards in real-time PCR analysis, the pWNV-K1 template DNA was degraded by incubation with DNase I for 15 min at 37°C, and the RNA was purified and separated from unincorporated nucleotides by using an RNeasy Mini kit (Qiagen). RNA concentrations were estimated from band intensities or, for determination of the RNA standard concentration, measured spectrophotometrically.
Immunofluorescence staining.
Intracellular expression of WNV specific proteins was determined by indirect immunofluorescence staining of the envelope protein E. Accordingly, RNA-transfected BHK-21 cells were seeded into 24-well plates and supplied with growth medium (EMEM with supplements and 5% FBS), which was exchanged for maintenance medium (EMEM with supplements and 1% FBS) at 20 h posttransfection. After 24 or 48 h, cells were treated with 1:1 acetone-methanol for fixation and permeabilization. To specifically detect WNV protein E, a cross-reactive polyclonal antibody directed against JEV protein E was used (dilution, 1:50). Staining was performed with a secondary fluorescein isothiocyanate-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories) as suggested by the manufacturer.
Hemagglutination assay (HA).
For the detection of WNV viral and/or subviral particles in supernatants of infected cells, a rapid assay based on the agglutination of erythrocytes, which is induced by the interaction with viral particles, was applied (8, 13). Briefly, virus supernatants were diluted 1:1 in borate-buffered saline (120 mM sodium chloride, 50 mM sodium borate, pH 9.0) containing 0.4% BSA for particle stabilization. Subsequently, this mixture was further diluted to produce a geometrical dilution row. Fifty microliters of each of the diluted samples was mixed with the same amount of a 0.5% solution of goose erythrocytes in round-bottom 96-well plates and incubated for 3 h at room temperature. Virus-induced agglutination of erythrocytes was visible by the lack of sedimented erythrocytes; the examination of plates was performed by visual inspection.
Mutant stability.
To assay the genetic stability of transfected mutants, supernatants of transfected cells were diluted until the end point of infectivity was reached. The supernatant corresponding to the end point was then transferred onto fresh cells, and these passages were repeated at least twice. Subsequently, RNA was isolated and sequence analysis was performed by using the cDNA synthesis system of Roche Applied Science and standard PCR and sequencing protocols.
RNA replication and export.
Intracellular RNA replication was monitored by real-time PCR as described previously (20, 41) with minor modifications. Briefly, Vero cells grown in six-well plates were incubated with wild-type and mutant WNV stock preparations at a multiplicity of infection (MOI) of 1. After 1 h, the cell monolayer was washed and supplied with growth medium which contained 1% BSA and 15 mM HEPES instead of FBS. At selected time points, cells were detached by trypsin incubation and washed twice in phosphate-buffered saline (PBS) (pH 7.4) containing 1% BSA. Cytoplasmic RNA was purified from these cells (RNeasy Mini kit; Qiagen) and was subjected to real-time PCR (PE Applied Biosystems) quantification as described previously (20, 41). The primers (5′-TCAGCGATCTCTCCACCAAAG-3′ and 5′-GGGTCAGCACGTTTGTCATTG-3′) and probe (5′-Fam-TGCCCGACCATGGGAGAAGCT-Tamra-3′) targeted a region within the envelope gene of the WNV genomic RNA. RNA equivalents were finally determined from a standard curve based on an RNA preparation of known concentration which was serially diluted in cell lysates of negative control cells and purified according to the same protocol.
The RNA content in supernatants of transfected cells was measured as published recently (41). Accordingly, prior to quantification by real-time PCR, aliquots of supernatants were cleared by low-speed centrifugation and RNA was purified by using the QIAamp viral RNA Mini kit (Qiagen) as suggested by the manufacturer. RNA export was finally calculated by determining the percentage of total RNA (intracellular and extracellular) in the supernatant fraction.
Cytotoxicity assay.
Similar to the RNA replication and export experiments, Vero cells were seeded into six-well plates and infected with WNV stock preparations at an MOI of 1, but the growth medium did not contain BSA. Aliquots of supernatants were transferred into 96-well plates, and cytotoxicity was assessed by measuring the release of lactate dehydrogenase (LDH) using the CytoTox 96 nonradioactive cytotoxicity assay (Promega) according to the manufacturer′s instructions.
Plaque morphology and immunocytochemistry.
Vero cells were grown to 80% confluence in 12-well plates and incubated for 1 h with virus suspensions serially diluted in infection medium. The cells were subsequently overlaid with EMEM containing 5% FBS (PAA Laboratories), 1.5% glutamine (200 mM; Cambrex), 1% penicillin-streptomycin (10,000 U/ml penicillin and 10 mg/ml streptomycin; Sigma), 15 mM HEPES, and 0.25% agarose (Sigma). The plaque morphology was determined following an incubation period ranging from 6 to 9 days postinfection. Accordingly, cells were fixed and stained with a solution containing 4% formaldehyde and 0.1% crystal violet.
Focus-forming units (FFU) were determined by immunocytochemistry. After incubation for 6 days, the agarose overlay was removed and cells were fixed with 1:1 acetone-methanol. The cells were rehydrated with PBS (pH 7.4) containing 5% sheep serum for 30 min at room temperature. Subsequently, the cells were incubated for 1 h at 37°C with a WNV-specific polyclonal antiserum (gamma-WN/KIS/2) diluted 1:3,000 in PBS (pH 7.4) with 0.2% Tween and 3% sheep serum. Cells were washed twice with PBS (pH 7.4) containing 0.2% Tween and 3% sheep serum and once with TBS buffer (137 mM sodium chloride, 3 mM potassium chloride, 25 mM Tris, pH 8.0) containing 0.2% Tween and 3% sheep serum. The incubation with a 1:400 dilution of an anti-rabbit alkaline phosphatase-conjugated secondary antibody was performed in TBS buffer with 0.2% Tween and 3% sheep serum for 45 min at room temperature. Following two washes with the same buffer, WNV-specific foci were detected by incubating with Sigma Fast Red TR/naphthol AS-MX for 10 min.
Computer-assisted sequence analysis.
Secondary-structure predictions were performed using PsiPred (15). Hydrophobicity plots were generated according to the algorithm of Kyte and Doolittle (26) using PROTEAN (DNASTAR, Inc.) and a window size of 11.
Nucleotide sequence accession number.
The sequence of the WNV isolate (crow 76/1) was deposited under GenBank accession no. FJ151394.
RESULTS
Establishment of a two-component infectious cDNA clone for WNV isolate V76/1.
To generate a tool for WNV reverse genetics, the genome of a previously uncharacterized lineage I isolate (see Materials and Methods) (Table 1) was reverse transcribed, sequenced, and assembled into two plasmids from which, after in vitro ligation, full-length genomic RNA could be transcribed (Fig. 1A). To verify the functionality of the infectious cDNA clone, full-length RNAs were transcribed in several independent experiments and introduced into BHK-21 cells by electroporation. An apparent cytopathic effect (CPE) was observed in cells at day 2 posttransfection, indicating virus replication. The supernatants of transfected BHK-21 cells were harvested and used to inoculate Vero cells, which are more susceptible to infection by WNV than BHK-21 cells (unpublished observation). At day 2 postinoculation, the release of infectious virions into the supernatant was tested by plaque assays on fresh Vero cells, indicating that virus had grown to high titers of typically 5 × 108 PFU/ml. Thus, the growth properties of the recombinant virus were virtually indistinguishable from those of the parental wild-type virus, which was furthermore confirmed by repeated growth curve analyses (data not shown).
TABLE 1.
WNV V76/1 isolate-specific genomic sequence differences
| Nucleotide no.a | Nucleotide in:
|
Amino acid difference | Location | |
|---|---|---|---|---|
| NY99- flamingo382-99 (GenBank accession no. AF196835)b | WNV V76/1 (GenBank accession no. FJ151394) | |||
| 1118 | C | U | A → V | E |
| 1285 | C | U | Silent | E |
| 3138 | U | C | Silent | NS1 |
| 6735 | C | A | Silent | NS4A |
| 7015 | U | C | Silent | NS4B |
| 7491 | G | U | Silent | NS4B |
| 8811 | U | C | Silent | NS5 |
| 10851 | A | G | NAc | 3′ noncoding region |
Genome position numbers are the same for both isolates.
The genomic sequence of the isolate used for sequence comparison (GenBank accession no. AF196835) is published in reference 27.
NA, not applicable.
FIG. 1.
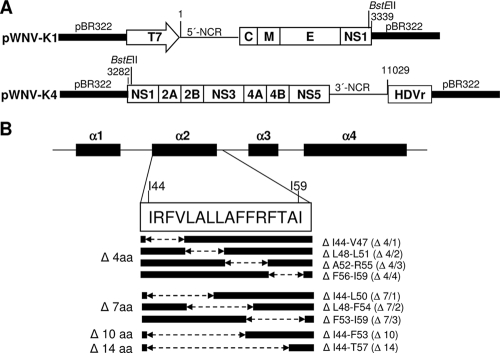
Capsid deletion mutants of WNV. (A) Schematic drawing of the two partial WNV cDNA clones (not to scale). The WNV genome was engineered as two partial cDNA clones into pBR322 using 5′ PacI and 3′ NotI restriction sites. pWNV-K1 contains the T7 promoter sequence (open arrow) and bp 1 to 3339 of the WNV genomic sequence. The second plasmid, pWNV-K4, contains the sequence corresponding to WNV bp 3282 to 11029 and the hepatitis δ virus ribozyme sequence (HDVr). To generate full-length DNA templates for in vitro transcription, the two clones are ligated in vitro subsequent to cleavage at the BstEII site at nucleotide position 3321/3326. In vitro transcription is driven by the T7 promoter, and the HDVr sequence ensures the production of an authentic 3′ end. (B) Schematic drawing of the positions and sizes of the engineered deletions. Deletions of 4, 7, 10, or 14 amino acids (aa) were introduced into helix α2 of WNV protein C as indicated by the broken arrows. The respective amino acid positions are indicated, as well as the nomenclature used for mutants throughout the study. NCR, noncoding region.
Deletions within helix α2 of WNV protein impair viral growth to various degrees.
To better characterize the functional importance of the hydrophobic helix α2 in WNV protein C, a set of nine deletions (Fig. 1B) was introduced into the infectious cDNA clone. Genomic RNAs were transcribed in vitro and used to transfect BHK-21 cells. Intracellular protein E expression was determined by immunofluorescence staining, using wild-type RNA and untransfected cells as positive and negative controls, respectively. At 24 h posttransfection, all of the samples presented a very similar picture, with approximately 10% of the cells being stained by immunofluorescence (data not shown). At 48 h posttransfection, the number of positive cells, however, had increased to 100% for deletion mutants Δ4/1, Δ4/3, and Δ4/4, thus being indistinguishable from wild-type-RNA-transfected cells (Fig. 2A). In contrast, cell culture spreading was reduced in all other mutants, with the most significant effect observed for mutants Δ7/2, Δ10, and Δ14 (Fig. 2A). Notably, deletions Δ4/2 and Δ7/2 exhibited a more distinct defect than the other deletions of the same length. Sequence inspection (Fig. 1B) indicates that both of these deletions are located within the most hydrophobic section of the helix (LALL-AFF) suggesting that the loss of hydrophobicity even more than the length of the deletion may cause the observed defect in cell culture spreading.
FIG. 2.
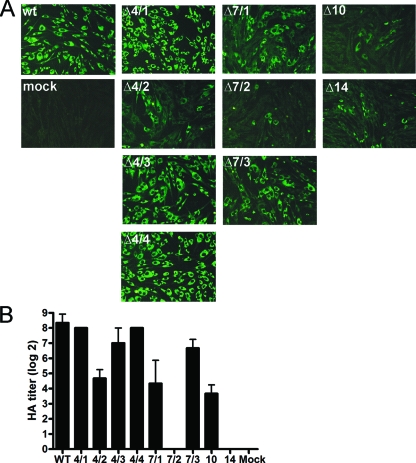
Infectious properties of WNV protein C deletion mutants. (A) Viral spread in cell culture. BHK-21 cells were transfected with wild-type (wt) or mutant in vitro-transcribed RNAs as indicated. As a control, mock-transfected cells were used. at 48 h posttransfection, intracellular protein E expression was visualized by immunofluorescence staining using a polyclonal antibody directed against JEV protein E which is cross-reactive to WNV protein E. As secondary antibody, an anti-rabbit fluorescein isothiocyanate conjugate was used. (B) Cell culture passage. Supernatants of transfected cells were used to inoculate fresh Vero cells. At 6 days postinoculation, the release of virus particles was assessed by subjecting aliquots of the supernatants to HA. Twofold serial dilutions of supernatants were tested, and the titers (indicated on the left) are expressed on a log2 scale (samples were measured in duplicate). Error bars indicate standard deviations.
To further evaluate the production of infectious virions and to assess the export of viral particles by the infected cells, we inoculated a monolayer of fresh Vero cells with supernatants of transfected cells. At day 6 postinoculation, virus particles in supernatants were quantified by HAs (Fig. 2B). Cells infected with mutants Δ4/1, Δ4/3, and Δ4/4 were capable of exporting viral particles as much as wild-type virus. In comparison, mutants Δ4/2, Δ7/1, Δ7/3, and Δ10 exhibited some degree of impairment, whereas mutants Δ7/2 and Δ14 were found to be incapable of producing infectious particles under these experimental conditions. Whereas these data mostly correlated well with the above-described immunofluorescence results, there are also discrepancies (such as with mutant Δ4/2, which produced more HA-reactive particles than one would have expected from the immunofluorescence data). This can be explained by the selection of pseudorevertants already in this first passage, a phenomenon which is analyzed in detail in a later section.
The supernatants tested in HA were also subjected to plaque and focus formation assays to quantify the infectious titers of the various mutants. However, only wild-type virus, but none of the mutants, formed visible plaques on Vero cells at 6 days postinfection. Infectious titers obtained by focus assay amounted to 1 × 106 FFU/ml for mutants Δ4/4 and Δ7/3, 5 × 105 FFU/ml for mutants Δ4/1 and Δ4/3, and 1 × 105 FFU/ml for mutants Δ4/2, Δ7/1, and Δ10, whereas no infectious particles were detected for mutants Δ7/2 and Δ14, in good agreement with the above-described HA data. Thus, the titers for all mutants were significantly lower than that obtained for wild-type virus (5 × 108 FFU/ml), suggesting that a large percentage of the particles produced by these mutants (as detected by HA) were not infectious or did not initiate the formation of visible foci.
Spontaneous mutations are selected during cell culture passages.
In order to test whether some of the WNV protein C mutants might revert to a better growth phenotype, end point dilution passages on Vero cells were performed and viral titers were monitored. Improved growth properties were indeed observed for mutants Δ4/2, Δ7/1, Δ7/2, Δ7/3, and Δ10 at different passage numbers, whereas several rounds of blind passaging of mutant Δ14 failed to produce a viable revertant. To investigate if these changes in phenotype were a direct result of additional alterations within the protein C sequence, viral RNA was isolated from supernatants of infected cells and subjected to reverse transcription-PCR and sequence analysis. Notably, sequencing of the protein C-coding region indeed verified the appearance of second-site mutations (Table 2). In most of the cases, and consistent with previous findings obtained with TBEV (21), point mutations that represented amino acid changes to more hydrophobic residues had evolved; however, most of these amino acid changes were, in contrast to findings with TBEV, located upstream of the original deletion (Fig. 3A). In total, five different point mutations were identified, with one of the point mutations, P22L, appearing more frequently than others and in the sequence context of two different engineered deletions.
TABLE 2.
Spontaneous mutations as determined by sequence analysis
| Mutantb | Mutation(s) at passagea:
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 5 | 8 | 11 | |
| Δ4/1 | None | None | NDc | ND | ND |
| Δ4/2 | P61L | P61L | ND | ND | ND |
| Δ4/3 | None | None | ND | ND | ND |
| Δ4/4 | None | None | ND | ND | ND |
| Δ7/1 | P22L | P22L | P22L/M34L | P22L/M34L | ND |
| Δ7/2 | NDc | NDc | P22L | K31M | K31M |
| Δ7/3 | None | Δ L51-E87 | R45L | R45L | ND |
| Δ10 | D39E/ΔG40-Q75 | D39E/ΔG40-Q75 | D39E/ΔG40-Q75 | D39E/ΔG40-Q75 | ND |
For the first two passages, concentrated samples were used. Subsequently, supernatants were diluted until the end point of infectivity was reached, and these samples were subjected to further end point passages.
The 4-amino-acid deletion mutants were passaged only twice.
ND, not done (sequence analysis was not performed).
FIG. 3.
Spontaneous mutations in the WNV capsid protein. (A) Second-site point mutations identified after passaging in Vero cells. The point mutations were identified at the indicated amino acid positions (marked on top) within a region corresponding to helix α1 (open box), helix α2 (gray box), and surrounding residues. The originally engineered deletions are shown by arrows, and the second-site point mutations are marked in bold and underlined. On the right, the corresponding nucleotide exchanges are listed. (B) Large deletions Δ36 and Δ37, identified after passaging of mutants Δ10 and Δ7/3, respectively. The helical parts of the capsid protein are indicated by boxes, and helix α2 is highlighted in gray. The positions of the large deletions are indicated by arrows. Furthermore, an artificial large deletion mutant (i.e., Δ48) was constructed, lacking all residues which had been spontaneously deleted in both Δ36 and Δ37. The precise nucleotide deletions are shown on the right. (C) Immunofluorescence analysis of large deletion mutants. The large deletions (i.e., Δ36, Δ37, and Δ48) were engineered into the infectious cDNA clone, and mutants were tested as described for Fig. 2A.
Furthermore, and much to our surprise, we also identified mutations in which the original deletions were enlarged to lengths of 36 and 37 residues, respectively (Fig. 3B). Passaging of mutants Δ10 and Δ7/3 resulted in the appearance of large deletions removing more than one-third of the entire amino acid sequence of protein C. Mutant Δ10 evolved into a deletion of residues G40 to Q75 (termed Δ36) and furthermore contains a conservative D-to-E exchange at the deletion border (position 39). This deletion removed all residues of helices α2 and α3 as well as flanking residues, thus producing a capsid protein lacking the entire internal hydrophobic sequence (31). The second large deletion mutant (termed Δ37) originated from mutant Δ7/3 and had residues L51 to E87 and thus part of helix α2, all of helix α3, and part of helix α4 removed. This mutant was predominant in the sequence pattern obtained after two cell culture passages (Table 2). However, apparently another pseudorevertant, which contained the original 7/3 deletion in combination with a single point mutation, R45L, arose in the same passaging experiment and outgrew the large deletion mutant during subsequent passages (Table 2, passage 5). A similar phenomenon was observed with mutant Δ7/2, in which case the original P22L mutation was replaced by a K31M mutation at later passages. In contrast, the same P22L mutation was also observed to arise in mutant Δ7/1 but there was complemented by a second amino acid change (M34L) at a later passage. These observations illustrate the competition of pseudorevertants with presumably variable evolutionary fitness during these cell culture passages.
Recombinant mutants Δ36 and Δ37 can be readily passaged in cell culture.
The unexpected tolerance of protein C toward deletions comprising 36 and 37 amino acids encouraged us to investigate these mutations in more detail. To ensure that the observed phenotypes were indeed a direct consequence of the identified alterations, the Δ36 and Δ37 deletions were engineered into the WNV wild-type backbone using the infectious cDNA clone. Immunofluorescence staining at 48 h posttransfection of in vitro-transcribed RNA (Fig. 3C) suggested that mutants Δ36 and Δ37 were indeed viable.
To further characterize the growth of these mutants, plaque and focus assays were performed on Vero cells. Even after incubation for 6 days, no plaques could be identified for both mutants, whereas wild-type plaques reached a size of between 8 and 15 mm (12.6 ± 2.4 mm) (Table 3). In contrast, both mutants were capable of forming foci on Vero cells, thus confirming their infectivity. These were, however, at least four times smaller than wild-type foci, indicating their reduced ability to spread in cell culture (Table 3). Nevertheless, both mutants achieved significantly higher titers (1 × 107 FFU/ml) than their respective parental mutants Δ10 (1 × 105 FFU/ml) and Δ7/3 (1 × 106 FFU/ml), thus confirming the notion that extension of the original deletions to 36 and 37 amino acids within protein C indeed caused improved cell culture growth properties.
TABLE 3.
Growth properties of mutants Δ36 and Δ37
| Virus | mm (mean ± SD)
|
Titer (FFU/ml) | |
|---|---|---|---|
| Plaque sizea | Focus sizeb | ||
| Wild type | 12.6 ± 2.4 | 19.8 ± 2.86 | 5 × 108 |
| Δ36 | 2.85 ± 0.63 | 1 × 107 | |
| Δ37 | 5.05 ± 1.07 | 1 × 107 | |
Mean plaque size was determined at day 6 postinfection. None of the capsid deletion mutants induced plaque formation, even when incubation was prolonged to 9 days postinfection.
Mean focus size was determined at day 6 postinfection.
Subsequently, we tested whether mutants Δ36 and Δ37 could be serially passaged in Vero cells and if that would provoke additional genetic alterations. A single passage using undiluted supernatant and three subsequent end point passages were performed, followed by sequence analysis of the entire genomes. No further sequence alterations were identified after the passages. These data indicated that the large deletion mutations present in mutants Δ36 and Δ37 are sufficient for providing efficient growth properties in cell culture and remain genetically stable.
A combined deletion mutant (Δ48) is severely impaired and genetically unstable.
The deletions present in mutants Δ36 and Δ37 affect overlapping but different regions of protein C. We wanted to investigate whether removal of the entire region extending from the 5′ border of the Δ36 deletion to the 3′ border of the Δ37 deletion would still yield a viable phenotype. To this end, mutant Δ48, including the D39E mutation and lacking residues G40 to E87 (Fig. 3B), was constructed and analyzed. Immunofluorescence analysis indicated a strongly impaired phenotype of mutant Δ48 (Fig. 3C), and no plaque or focus formation was observed with this mutant (not shown). However, a single blind passage was sufficient to rescue a viable phenotype in Vero cells. Sequence analysis after subsequent end point passages revealed a duplication of the residues flanking the 48-amino-acid deletion (i.e., DuM16-D39E+L88-A94), but the growth properties of this pseudorevertant remained restricted, achieving a titer of only 104 FFU/ml.
RNA export and specific infectivity of mutants Δ36 and Δ37 are moderately reduced compared to those of wild-type WNV.
To characterize in detail the capacity of mutants Δ36 and Δ37 to replicate, export, and infect, quantitative tests were performed in comparison to wild-type virus. As shown in Fig. 4A, intracellular RNA replication of both mutants was similar to that of the wild type at 24 and 48 h postinfection. At 72 and 96 h postinfection, intracellular RNA values of mutant Δ37 were still at wild-type levels, whereas those of mutant Δ36 decreased. This decrease was accompanied by strong CPE, causing a strong reduction of cell numbers at these time points. To confirm the visually observed CPE, cytotoxicity was quantitatively assessed by measuring the release of LDH into the supernatants of infected cells. As shown in Fig. 4E, LDH release from cells infected with mutant Δ36 was in the same range as for the wild type and mutant Δ37 until 48 h postinfection. In contrast, at the later time points, LDH levels in supernatants of Δ36-infected cells were significantly higher than others thus confirming its high cytotoxicity and suggesting that the decreased intracellular RNA values for Δ36 at 72 and 96 h postinfection were indeed a consequence of excessive cell deaths (compare Fig. 4A and E).
FIG. 4.
Characterization of mutants Δ36 and Δ37. Approximately 106 Vero cells were infected at an MOI of 1 with the indicated virus preparation. Wild-type virus and infection medium were used as the respective positive and negative controls. (A) RNA replication (intracellular RNA) was measured by real-time PCR at the indicated time points. (B) The RNA export kinetics (RNA in supernatant) of mutants Δ36 and Δ37 was monitored by real-time PCR. (C) The percentage of exported relative to total RNA (intra- plus extracellular RNA) was calculated for the 48-h time point. (D) Release of viral particles into the supernatant was assessed by HA. (E) Cytotoxicity was assessed by CytoTox 96 nonradioactive cytotoxicity assay (Promega) using supernatants of the same samples. The respective optical density at 490 nm (OD490) values, representing LDH release of disintegrating cells, are shown. (F) Specific infectivity of mutants Δ36 and Δ37 and wild-type virus. The specific infectivity was calculated by determining the ratio of RNA (real-time PCR) to infectious units (focus assay) in virus stock preparations. Mean values from two independent experiments with error bars indicating standard deviations are shown. wt, wild-type.
Quantification of RNA release into the supernatants revealed moderate differences between protein C deletion mutants and wild-type virus. Mutants Δ36 and, particularly, Δ37 released less viral RNA into the supernatant than the wild type at 24 h postinfection (Fig. 4B). Mutant Δ37 achieved wild-type levels at later time points; however, Δ36 remained approximately one order of magnitude below the wild-type control at all times. The decreasing values after 48 h (i.e., at 72 and 96 h) presumably reflect the loss of producing cells caused by the mutant's prominent cytotoxicity, thus calling into question the accuracy of the quantitative data at the later time points. To better compare the export efficiencies of mutant and wild-type RNAs, the percentage of total (extracellular and intracellular) RNA equivalents in the supernatant was calculated for the 48-h time point, at which time effects of cytotoxicity were still low and comparable among the samples. As illustrated in Fig. 4C, export efficiencies of mutants Δ36 and Δ37 were about two-thirds and half of the wild-type value, indicating that the mutated capsid proteins, although clearly less efficient in packaging of RNA and/or assembly of virions, were still able to facilitate the export of a significant percentage of the total RNA from infected cells. To further determine the export of viral particles, the same supernatants as used for the quantification of viral RNA were subjected to HA. As shown in Fig. 4D, the results of this analysis were in good agreement with the RNA data shown in Fig. 4B. For mutant Δ37, the release of viral particles was delayed but reached nearly the wild-type level at the latest time point. In contrast, the level for mutant Δ36 remained below that of the wild type by approximately 3 log2 dilutions (i.e., approximately 8-fold) at all times, similar to the approximately 10-fold difference observed in the RNA values.
To quantitatively compare the specific infectivities of mutant and wild-type viruses, virus preparations were subjected to quantitative PCR to determine the number of RNA equivalents (presumed to correlate to the number of virions) and to focus assays to quantify infectious units in these preparations. The ratio of RNA equivalents to FFU was then calculated, and results are plotted in Fig. 4F. Whereas this ratio was approximately 10 for wild-type virus (i.e., 1 out of 10 RNA equivalents/virions caused an infectious focus), it was between 10- and 100-fold higher in the case of the two deletion mutants, indicating reduced specific infectivity.
In conclusion, the quantitative comparisons indicated moderate but significant impairments of both viral export and entry caused by the deletion mutations.
Large deletions are predicted to cause complex rearrangements of the overall helical composition of protein C.
Notably, protein C deletion mutants Δ36 and Δ37 are capable of producing infectious virions, whereas mutants with deletions of fewer amino acids are much more severely impaired (i.e., Δ4/2, Δ7/1, Δ7/2, Δ7/3, and Δ10) or noninfectious in cell culture (i.e., Δ14). To obtain further insight into the structural consequences of deletions Δ36 and Δ37, the protein C sequences of these pseudorevertants were subjected to secondary-structure prediction analysis (PsiPred) (15) and compared to the wild-type sequence. In Fig. 5A, the secondary-structure prediction for the WNV V76/1 sequence is presented, with positions of helices being almost identical to those defined in the crystal structure (11). In addition, we determined the Kyte-Doolittle hydrophobicity profile (26). The spontaneous enlargements of the deletions apparently removed a hydrophilic region extending from α3 to the beginning of α4 (Fig. 5A, lower panel). This suggests that the spontaneous deletions compensate for the loss of the hydrophobic helical structure as represented by helix α2 by removing another, more hydrophilic region (Fig. 5A). Subsequently, the sequences of protein C deletion mutants (i.e., Δ36, and Δ37) were analyzed. As illustrated in the WNV crystal structure (11), the spontaneous deletion of 36 amino acids resulted in complete removal of helices α2 and α3 (Fig. 5B, lower panel). The secondary-structure prediction suggested the formation of a large single α helix (Fig. 5B, upper panel) by fusion of helices α1 and α4. Whether the mutant protein (i.e., a polypeptide of only 61 amino acids) is likely to adopt a stable conformation in solution remains elusive, and it will be interesting to investigate this using recombinant proteins. However, it is certainly conceivable that such a long fusion helix might form upon insertion of protein C into the ER membrane. Similarly, the formation of a fusion helix was predicted in the analysis of the second large deletion mutant (i.e., Δ37, illustrated in Fig. 5C). Notably, in the crystal structure, the remainders of helices α2 and α4 are oriented toward each other, thus suggesting, albeit not proving, that the formation of a large fusion helix might indeed be possible.
FIG. 5.
Structure predictions for sequences of the wild type (A), deletion mutation Δ36 (B), and deletion mutation Δ37 (C). (A) Secondary-structure prediction and hydrophobicity blot for the WNV V76/1sequence. Residues R23 to R98, which are also present in the Kunjin protein C crystal structure (11), are shown. The positions of the introduced deletions and the most hydrophilic part of the protein are shown by open boxes. The spontaneous deletions Δ36 and Δ37 are illustrated by black bars below. (B and C) Upper panels, secondary-structure predictions for mutant protein C sequences and corresponding hydrophobicity blots. Lower panels, deletions on the Kunjin protein C dimer (11). The four helices are shown, where present, with the same coloring in all three panels (i.e., α1 in green, α2 in blue, α3 in yellow, and α4 in red); the spontaneous large deletions (i.e., Δ36 and Δ37) are shown in gray. All secondary-structure predictions were performed using PsiPred (15), and the hydrophobicity blots were generated according to the algorithm of Kyte and Doolittle (26). The protein C 3D structure was adapted from the Protein Data Bank (accession no. 1SFK) using PyMOL software (10). wt, wild-type.
DISCUSSION
The internal hydrophobic domain of the flaviviral protein C is a functionally important region involved in protein dimerization and membrane interaction during assembly of the virion (19, 31, 34, 46). In earlier reports, it had been demonstrated that mutants lacking the entire internal hydrophobic sequence of protein C are either severely impaired or not viable at all (21, 42, 44). Although this was also observed with some of the WNV deletion mutants analyzed in this study, the pseudorevertant Δ36, lacking amino acids G40 to Q75 and thus the entire conserved hydrophobic domain, surprisingly demonstrates that efficient virion assembly and cell culture growth are in fact possible even in the complete absence of this region. Δ37, the second large, spontaneously emerged WNV capsid deletion mutant, lacks residues L51 to E87 and thus also large parts of this domain. Both mutants grew well in cell culture, indicating that highly truncated capsid proteins can be functional.
A remarkable flexibility of protein C toward deletions and sequence alterations has already been observed in earlier studies (19, 21, 42, 43, 50). Protein C is essential for binding and packaging of genomic RNA and contributes to particle assembly and stability (reviewed in reference 38). RNA binding has been assigned to the highly basic N- and C-terminal parts of the protein (17). Notably, as tested in a YFV trans-packaging system, one intact terminus is sufficient for the encapsidation of genomic RNA (42). Thus, nearly 40 residues of the N terminus of YFV protein C could be removed while the ability to package RNA was retained. Similarly, 27 residues of its C terminus, including the entire helix α4, were dispensable for packaging. Based on their observations with mutants containing deletions in both termini, Patkar et al. proposed a mechanism in which the N terminus is involved in initial binding of the genomic RNA, followed by binding of the C terminus (42). The capacity to bind and package viral RNA is largely preserved in both mutant Δ36 and mutant Δ37 proteins, as measured by their capacity to export RNA into the supernatants of infected cells. Another functional requirement of protein C is to mediate interaction with the ER membrane during budding (19, 35). It is believed that this interaction involves the hydrophobic cleft present on the surface of dimeric protein C (31). In the dimer of protein C, helices α2 from each monomer interact with each other, thus forming the bottom of this cleft. It is puzzling that mutants Δ36 and Δ37 still produced infectious particles even though the entire or large parts of helix α2 were missing. Secondary-structure predictions and hydrophobicity plots suggested the formation of new large, hydrophobic α helices in both of these mutants, whereas a hydrophilic stretch of residues was lost from both of these proteins. Taken together, these findings support the idea that the newly formed fusion helices might functionally substitute for the loss of helix α2. In addition, the protein C dimer serves as the basic building block in the assembly of the flaviviral nucleocapsid (18). In the 3D structures of flaviviral protein C (11, 31), the dimers resemble a three-layer structure with helices α1 on top, helices α2 in the middle, and helices α4 at the bottom. In contrast, helix α3 is not organized pairwise and seems to serve as a spacer. Mutant Δ36 lacks helices α2 and α3 and, as a consequence, the middle part of the three-layer structure is completely removed whereas the top layer of helices α1 and the bottom layer of helices α4 remain more or less unaffected (Fig. 5B). Similarly, mutant Δ37 lacks approximately half of helix α2, the entire helix α3, and approximately half of helix α4 (Fig. 5C). One might assume that the formation of a fusion helix (composed of the remainders of helices α2 and α4) results in the formation of two layers instead of three. Stacking of such two-layer structures might nevertheless enable multimer formation and nucleocapsid assembly, which has been shown as an intrinsic property of WNV C and DENV C proteins (11, 31). Therefore, it will be interesting to explore the oligomeric properties and the atomic structures of the mutant proteins in future experiments. Mutants Δ36 and Δ37, however, had a significantly reduced specific infectivity and formed only small foci and no plaques, indicating a significantly attenuated phenotype. This attenuation may be caused not only by an assembly defect but also by an impairment during entry and unpackaging and/or a reduced physico-chemical stability of the mutant particles. Indeed, a decreased thermal stability of particles containing C-terminal deletions [C(Δ77-96)] or deletions within the internal hydrophobic sequence [C(Δ43-48)] had recently been observed (42). The impairment of export and, potentially connected with this observation, an altered cytotoxicity (i.e., for mutant Δ36) are likely caused by a partial defect in particle assembly.
In this study, we also identified spontaneous mutations which resembled those described previously for TBEV (21), i.e., exchanges of individual amino acids to more hydrophobic residues and duplication mutations. In the TBEV study, the residue changes were, without exception, located downstream of the originally engineered deletion, whereas this was the case for only a single mutation in the WNV system. The P61L mutation in mutant Δ4/2 resembles a P57L mutation identified in TBEV mutant C(Δ28-46). Both of these mutations of P to L are located in the loop between the helix α2 containing the hydrophobic domain and the subsequent helix α3. All other WNV second-site point mutations, however, appeared upstream of the original deletion and affected residues preceding or located within helix α1. In addition, the TBEV study (21) identified two duplications as resuscitating mutations. Similarly, WNV mutant Δ48 was rescued by the emergence of a duplication mutation, DuM16-D39E+L88-A94, although growth of the resulting mutant was still highly restricted. Taken together, these findings suggest that similar mechanisms can work to compensate for deletion mutations in both TBEV and WNV and possibly flaviviruses in general.
Flaviviral RNA replication is dependent on the cyclization of the positive-stranded RNA genome, which is mediated via 5′ and 3′ cyclization sequences (reviewed in reference 34). In mosquito-borne flaviviruses, the 5′ cyclization sequence is located within the amino-terminal coding region of protein C (2, 3, 9, 14, 16, 20, 29, 39). The WNV 5′ cyclization sequence comprises nucleotides 137 to 144, encoding amino acids V14 to M16 of protein C. In good agreement with the functional importance of this region, none of the second-site mutations including also the large deletions affected this part of the sequence. Thus, intracellular RNA replication should not be impaired, and indeed, quantitative assessment showed no significant differences between mutants Δ36 and Δ37 and wild-type virus.
In conclusion, our data support a functional importance of the internal hydrophobic domain of the WNV protein C but demonstrate that this functionality can be substituted for in dramatically truncated forms of this protein. Deletions of more than one-third of the protein in the absence of additional mutations, which would increase hydrophobicity, can generate functional protein C. As suggested by secondary-structure predictions for WNV protein C deletion mutants Δ36 and Δ37, the loss of functional elements contained in the hydrophobic helix α2 was presumably compensated for by the formation of hydrophobic fusion helices and the extrusion of an intermittent hydrophilic loop region. The high immunogenicity of mutant flaviviruses containing deletions within protein C has been successfully demonstrated (19, 21, 37, 44). Taking into account the delayed growth kinetics together with the fact that high titers can be achieved with mutants Δ36 and Δ37 in cell culture, we propose that these large deletion mutants might be particularly useful vaccine candidates.
Acknowledgments
We thank Ernest Gould, Bob Shope, Bob Tesh, Eileen N. Oslund, and Franz X. Heinz for kindly providing the virus isolate WNV V76/1 and Paul Breit for photographic artwork. We are especially grateful to Franz X. Heinz for very helpful discussions.
This project was funded by the Wiener Wirtschafts Förderungs Fonds (“Co operate Vienna 2003”).
We declare a potential conflict of financial interests as employees of Intercell AG (P.S., W.S., A.V.G., and A.M.), a biotechnology company.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 695816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, D. E., M. F. Lodeiro, C. V. Filomatori, S. Fucito, J. A. Mondotte, and A. V. Gamarnik. 2006. Structural and functional analysis of dengue virus RNA. Novartis Found. Symp. 277120-132. (Discussion, 277:132-135, 251-253.) [PubMed] [Google Scholar]
- 3.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 796631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amberg, S. M., A. Nestorowicz, D. W. McCourt, and C. M. Rice. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 683794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 295-113. [PubMed] [Google Scholar]
- 6.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44649-688. [DOI] [PubMed] [Google Scholar]
- 7.Chowrira, B. M., P. A. Pavco, and J. A. McSwiggen. 1994. In vitro and in vivo comparison of hammerhead, hairpin, and hepatitis delta virus self-processing ribozyme cassettes. J. Biol. Chem. 26925856-25864. [PubMed] [Google Scholar]
- 8.Clarke, D. H., and J. Casals. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7561-573. [DOI] [PubMed] [Google Scholar]
- 9.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 772265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 11.Dokland, T., M. Walsh, J. M. Mackenzie, A. A. Khromykh, K. H. Ee, and S. Wang. 2004. West Nile virus core protein; tetramer structure and ribbon formation. Structure 121157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84183-191. [DOI] [PubMed] [Google Scholar]
- 13.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 16990-99. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 19833-41. [DOI] [PubMed] [Google Scholar]
- 15.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292195-202. [DOI] [PubMed] [Google Scholar]
- 16.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 756719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khromykh, A. A., and E. G. Westaway. 1996. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 141685-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiermayr, S., R. M. Kofler, C. W. Mandl, P. Messner, and F. X. Heinz. 2004. Isolation of capsid protein dimers from the tick-borne encephalitis flavivirus and in vitro assembly of capsid-like particles. J. Virol. 788078-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 763534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kofler, R. M., V. M. Hoenninger, C. Thurner, and C. W. Mandl. 2006. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 804099-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kofler, R. M., A. Leitner, G. O'Riordain, F. X. Heinz, and C. W. Mandl. 2003. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 77443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konishi, E., and A. Fujii. 2002. Dengue type 2 virus subviral extracellular particles produced by a stably transfected mammalian cell line and their evaluation for a subunit vaccine. Vaccine 201058-1067. [DOI] [PubMed] [Google Scholar]
- 23.Konishi, E., A. Fujii, and P. W. Mason. 2001. Generation and characterization of a mammalian cell line continuously expressing Japanese encephalitis virus subviral particles. J. Virol. 752204-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konishi, E., and P. W. Mason. 1993. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J. Virol. 671672-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157105-132. [DOI] [PubMed] [Google Scholar]
- 27.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 2862333-2337. [DOI] [PubMed] [Google Scholar]
- 28.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 29.Lo, M. K., M. Tilgner, K. A. Bernard, and P. Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 7710004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobigs, M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc. Natl. Acad. Sci. USA 906218-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, L., C. T. Jones, T. D. Groesch, R. J. Kuhn, and C. B. Post. 2004. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA 1013414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 7510787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandl, C. W., M. Ecker, H. Holzmann, C. Kunz, and F. X. Heinz. 1997. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 781049-1057. [DOI] [PubMed] [Google Scholar]
- 34.Markoff, L. 2003. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59177-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markoff, L., B. Falgout, and A. Chang. 1997. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233105-117. [DOI] [PubMed] [Google Scholar]
- 36.Mason, P. W., S. Pincus, M. J. Fournier, T. L. Mason, R. E. Shope, and E. Paoletti. 1991. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology 180294-305. [DOI] [PubMed] [Google Scholar]
- 37.Mason, P. W., A. V. Shustov, and I. Frolov. 2006. Production and characterization of vaccines based on flaviviruses defective in replication. Virology 351432-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 313-22. [DOI] [PubMed] [Google Scholar]
- 39.Nomaguchi, M., T. Teramoto, L. Yu, L. Markoff, and R. Padmanabhan. 2004. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 27912141-12151. [DOI] [PubMed] [Google Scholar]
- 40.Nowak, T., P. M. Farber, G. Wengler, and G. Wengler. 1989. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology 169365-376. [DOI] [PubMed] [Google Scholar]
- 41.Orlinger, K. K., V. M. Hoenninger, R. M. Kofler, and C. W. Mandl. 2006. Construction and mutagenesis of an artificial bicistronic tick-borne encephalitis virus genome reveals an essential function of the second transmembrane region of protein e in flavivirus assembly. J. Virol. 8012197-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patkar, C. G., C. T. Jones, Y. H. Chang, R. Warrier, and R. J. Kuhn. 2007. Functional requirements of the yellow fever virus capsid protein. J. Virol. 816471-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrauf, S., P. Schlick, T. Skern, and C. W. Mandl. 2008. Functional analysis of potential carboxy-terminal cleavage sites of tick-borne encephalitis virus capsid protein. J. Virol. 822218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seregin, A., R. Nistler, V. Borisevich, G. Yamshchikov, E. Chaporgina, C. W. Kwok, and V. Yamshchikov. 2006. Immunogenicity of West Nile virus infectious DNA and its noninfectious derivatives. Virology 356115-125. [DOI] [PubMed] [Google Scholar]
- 45.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 744394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, S. H., W. J. Syu, and S. T. Hu. 2004. Identification of the homotypic interaction domain of the core protein of dengue virus type 2. J. Gen. Virol. 852307-2314. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 7611645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., B. Kaufmann, P. R. Chipman, R. J. Kuhn, and M. G. Rossmann. 2007. Structure of immature West Nile virus. J. Virol. 816141-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, W., C. Qin, S. Chen, T. Jiang, M. Yu, X. Yu, and E. Qin. 2007. Attenuated dengue 2 viruses with deletions in capsid protein derived from an infectious full-length cDNA clone. Virus Res. 126226-232. [DOI] [PubMed] [Google Scholar]