Abstract
In 1979, a lineage of avian-like H1N1 influenza A viruses emerged in European swine populations independently from the classical swine H1N1 virus lineage that had circulated in pigs since the Spanish influenza pandemic of 1918. To determine whether these two distinct lineages of swine-adapted A/H1N1 viruses evolved from avian-like A/H1N1 ancestors in similar ways, as might be expected given their common host species and origin, we compared patterns of nucleotide and amino acid change in whole genome sequences of both groups. An analysis of nucleotide compositional bias across all eight genomic segments for the two swine lineages showed a clear lineage-specific bias, although a segment-specific effect was also apparent. As such, there appears to be only a relatively weak host-specific selection pressure. Strikingly, despite each lineage evolving in the same species of host for decades, amino acid analysis revealed little evidence of either parallel or convergent changes. These findings suggest that although adaptation due to evolutionary lineages can be distinguished, there are functional and structural constraints on all gene segments and that the evolutionary trajectory of each lineage of swine A/H1N1 virus has a strong historical contingency. Thus, in the context of emergence of an influenza A virus strain via a host switch event, it is difficult to predict what specific polygenic changes are needed for mammalian adaptation.
Swine influenza A viruses (IAVs) of the H1N1 subtype currently circulate as two distinct lineages within North American and European swine populations (2, 33). While the first clinical observations of swine IAV infection coincided with the 1918 human influenza pandemic (7, 25; reviewed in reference 59), the first North American swine IAV isolates were not obtained until 1930 (50). Termed classical swine A/H1N1 virus, this genetically and antigenically stable viral lineage presumably emerged by stable transfer of the human 1918 pandemic virus to swine (11, 59) and subsequently spread to swine in other parts of the world, including Europe, in 1976 (2, 32). Independently, a novel lineage of avian-like H1N1 swine IAV emerged in Europe in 1979 that essentially replaced classical swine IAV (2, 34, 45). To date, this second lineage of swine IAV is enzootic throughout swine-producing regions of Western Europe, where it cocirculates with swine IAVs of the H3N2 and H1N2 subtypes (28). All eight gene segments of the prototype H1N1 viruses of this lineage are thought to be derived from closely related Eurasian avian IAVs by a stable host switch without reassortment, and this lineage is phylogenetically and antigenically distinct from the classical swine H1N1 lineage (4, 10, 34, 49).
The processes by which avian IAVs stably switch hosts and acquire mutations that facilitate replication and efficient transmission in a new host species are fundamental to understanding the ecology of these viruses but are also of critical importance to public health and veterinary preparedness. IAVs from the genetically and antigenically divergent avian reservoir pool have been associated with stable host switch events to novel host species, including humans, swine, domestic poultry, and horses (1, 55, 61). The last three human influenza pandemic viruses all contained two or more novel genes that were very similar to those found in IAVs of wild birds, derived either by reassortment with circulating human strains in formation of the 1957 and 1968 pandemic viruses (23, 47) or possibly by whole-genome adaptation in the case of the 1918 pandemic virus (18, 36, 43, 60). Other novel influenza viruses derived by stable host switching from avian influenza viruses have also been isolated recently from pigs, including other independent introductions of A/H1N1 influenza viruses in China (19), A/H4N6 influenza viruses in Canada (22), and most recently, A/H2N3 influenza viruses in the United States (27). Similarly, a stable lineage of A/H3N8 influenza virus emerged in dogs in the United States following a host switch event without reassortment from the equine A/H3N8 lineage (9). The present concern that an avian influenza virus, especially the currently circulating lineages of highly pathogenic avian H5N1 influenza virus, could initiate a new pandemic if the virus stably adapts to humans is also a question of considerable biomedical importance (62). Together, these examples demonstrate that reassortment is not a prerequisite for IAV emergence in novel hosts.
Swine have been hypothesized to be the mixing vessel in which avian and human IAVs reassort, resulting in the emergence of novel human pandemic influenza virus strains (2, 46). However, direct or experimental data linking swine as intermediaries in the emergence of past pandemics are lacking. Swine are the only animals documented to be susceptible to infection with avian, swine, and human IAVs (2), and coinfections with both avian and human IAVs have been reported (3, 5, 20, 49, 64). This has been attributed to the fact that swine tracheal epithelium expresses both α2,3 (avian IAV preferred)- and α2,6 (mammalian IAV preferred)-N-acetylneuraminic acid-galactose-linked receptors (17), and it is believed that avian IAVs adapted to swine undergo a shift from α2-3 to α2-6 binding, a critical step required in the adaptation of an avian virus to a human host (52). A subset of amino acids that are invariant in all avian hemagglutinin (HA) subtypes but vary in mammalian-adapted HAs have been identified (29). It is possible that this set of mutations (or a subset thereof) play important roles in the adaptation of avian IAVs to swine.
Whether common genetic changes are associated with the adaptation to specific host species, such that they are predictive of future events, or if genetic changes are made up of unique constellations of mutations that occur independently in each host switch event is an important question. The process by which the 1918 pandemic A/H1N1 influenza virus emerged and adapted to both humans and swine is not yet fully elucidated, although the virus is avian-like in both its coding sequences (58, 60) and nucleotide composition (36). The European avian-like swine A/H1N1 viruses emerged independently of the 1918 pandemic virus from an avian-like source (34, 45, 49). We therefore sought to compare changes that might be associated with mammalian adaptation between these two swine H1N1 lineages.
To address whether the two swine H1N1 lineages were evolving in parallel, as might be expected given their common host species, we examined patterns of base composition variation to determine relative nucleotide usages. We examined in detail the amino acid sites that had previously been reported as important for mammalian adaptation to determine whether these mutations appeared as parallel genetic changes, and therefore were always required for avian H1N1 IAV to adapt to pigs, or whether there is more flexibility in the adaptive process.
MATERIALS AND METHODS
Viral culture, viral cDNA amplification, and sequencing.
The following 17 viral isolates of European avian-like swine H1N1 IAVs were selected for genomic sequencing from the International Reference Laboratory at the Veterinary Laboratories Agency, Weybridge, United Kingdom: A/swine/Belgium/1979 (H1N1), A/swine/Belgium/1983 (H1N1), A/swine/France (OMS)/1984 (H1N1), A/swine/France (OMS)/1985 (H1N1), A/swine/Belgium/1989 (H1N1), A/swine/Spain/1991 (H1N1), A/swine/France (OMS)/1992 (H1N1), A/swine/England/1992 (H1N1), A/swine/England/1993 (H1N1), A/swine/Denmark/1993 (H1N1), A/swine/England/1994 (H1N1), A/swine/France (OMS)/1995 (H1N1), A/swine/England/1995 (H1N1), A/swine/England/1996 (H1N1), A/swine/England/1997 (H1N1), A/swine/England/1998 (H1N1), and A/swine/Scotland/1999 (H1N1). Viruses were propagated by inoculation into the allantoic cavities of 9- to 11-day-old embryonated chicken eggs originating from a commercial specific-pathogen-free flock. Total RNA was extracted from infected allantoic fluid by use of a QIAamp viral RNA kit (Qiagen, Valencia, CA), and first-strand cDNA was reverse transcribed from viral RNA with the universal influenza virus primer (21). Subsequent PCR amplification of all IAV segments was performed using overlapping primer sets for each of the eight segments, using standard methods (14a, 34a).
Sequence analysis.
In addition to the 17 European swine IAV genomes sequenced for this study, 3 European swine avian-like H1N1 genomes, 38 classical swine H1N1 IAV genomes, other available swine H1N1 full-length gene sequences, and 81 human A/H1N1 virus genomes were downloaded from the Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) and/or GenBank. The genome sequences of the 1918 pandemic IAV and representative Eurasian avian influenza virus sequences were used to infer the ancestry of the two swine lineages. Whole genome sequences were not available for the Eurasian avian viral sequences analyzed. Therefore, the NCBI BLAST database was used to find highly similar avian sequences for each European swine influenza virus gene segment. Consensus sequences were derived from the Eurasian avian IAV sequences for each segment. Within these data, we compiled separate manually aligned gene segments (coding regions) using Se-Al (36a). Sequence alignments consisted of the following coding regions for each segment: 197 PB2 (2,277 nucleotides [nt]) sequences, 203 PB1 (2,271 nt) sequences, 198 PA (2,148 nt) sequences, 214 HA (1,218 nt) sequences, 234 NP (1,494 nt) sequences, 231 NA (1,410 nt) sequences, 193 M1/2 (1,002 nt) sequences, and 224 NS1/2 (831 nt) sequences. Sequence alignments are available upon request.
Phylogenetic analysis.
The best-fit GTR + I + Γ4 model of nucleotide substitution was determined using ModelTest 3.7 (35a), and resulting parameter estimates were imported into PAUP* (56) to create maximum likelihood trees through tree bisection-reconnection branch swapping (parameter values available upon request). Whole genome sequences and a consensus Eurasian avian sequence were concatenated (in the absence of reassortment [see below]) to infer the evolutionary relationship of swine H1N1 IAVs. Individual gene segment phylogenetic trees are available in the supplemental material.
To estimate the rates of evolutionary change and the time to the most recent common ancestor (TMRCA), we applied a Bayesian Markov chain Monte Carlo approach available in the BEAST package (13), employing a relaxed (uncorrelated log normal) molecular clock in all cases (12). For each data set, we utilized the Bayesian skyline coalescent prior (as demographic history was a nuisance parameter in our analysis) with a 10% burn-in, assuming a GTR + I + Γ4 model of nucleotide substitution. Uncertainty in parameter estimates is reflected in the 95% highest probability density (HPD) values, and all chains were run for sufficient length to ensure convergence, as assessed using the TRACER program (http://tree.bio.ed.ac.uk/software/tracer/). For the estimates of TMRCA, the most recent sequence used for a classical swine H1N1 virus was from 1991 (A/swine/Maryland/23239), and that for a European swine H1N1 virus was from 2004 (A/swine/Spain/53207/2004). Selective pressures on codon sites were estimated along the branches of the swine H1N1 phylogenetic trees for all eight genes, using Datamonkey (35; http://www.datamonkey.org/). The best-fit codon model was fitted to the data by using parameters obtained from the best-fit nucleotide substitution model. A 1-df likelihood ratio test was applied to the data to determine whether the instantaneous rates of synonymous (α) and nonsynonymous (β) substitutions differ and whether this difference is based on α > β (negative selection) or α < β (positive selection) and is significant.
Analysis of base composition.
With the exception of the Eurasian avian sequences, for which insufficient whole H1N1 genome sequences were available, genome sequences were used to calculate the base compositions of all lineages. A method similar to that of Shultes et al. (48) was used to calculate the frequencies of GU (G+U), GA (G+A), and GC (G+C) across all eight gene segments for the European swine, classical swine, human H1N1, Eurasian avian, and 1918 H1N1 IAV lineages. Unambiguous calculations of the base compositional space of these IAV genes were defined by the following three parameters: GU (frequency of G plus frequency of U), GA (frequency of G plus frequency of A), and GC (frequency of G plus frequency of C) for each gene segment. Base composition frequencies of complete and first and second codons were calculated using the PAUP* package (56). Base compositional data were then graphically plotted using the R, version 2.7.0, statistical program (2008). The third-position GC content for each gene segment of swine IAV was measured using the GCUA (General Codon Usage Analysis) package (30).
Amino acid analysis.
Amino acid differences among the lineages were recorded as changes at amino acid sites compared to the putative ancestral sequence. Given its location at the root of the human and classical swine H1N1 clades, we used the 1918 Brevig Mission sequence as the ancestral sequence to infer changes for classical swine and human H1N1 viruses. In the case of the European swine H1N1 viruses, we collected highly similar Eurasian avian sequences from 1977 to 1998 for each gene segment to infer amino acid changes in European swine H1N1 IAVs.
Nucleotide sequence accession numbers.
Sequences generated for this analysis have been deposited in GenBank (accession numbers CY037895 to CY038027).
RESULTS
Phylogenetic analysis of classical and European swine A/H1N1 virus sequences.
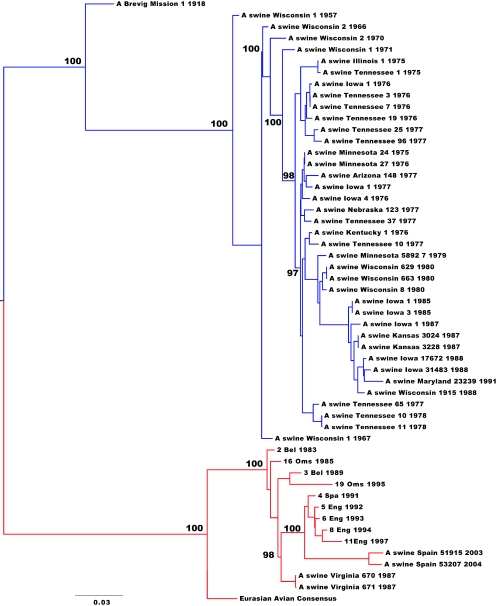
Maximum likelihood trees resulted in the same phylogenetic relationship across all eight gene segments between the two swine H1N1 IAV lineages (see the supplemental material), strongly suggesting that they are each monophyletic, with no major reassortment events. We therefore concatenated the major open reading frames of all eight gene segments and inferred an overall genomic maximum likelihood tree (Fig. 1). As expected, this revealed that the two swine lineages formed distinct clades, with the classical swine H1N1 IAV lineage derived from the 1918 pandemic influenza virus sequence and the European swine H1N1 IAV lineage derived from the Eurasian avian virus consensus sequence.
FIG. 1.
Maximum likelihood tree of concatenated genome sequences (54 whole genomes) of European and classical swine H1N1 IAVs. Horizontal branch lengths are drawn to scale (nucleotide substitutions per site). Bootstrap values (>75%) are shown next to the relevant nodes. The tree is midpoint rooted for clarity only. Classical swine H1N1 viruses are in blue, and European swine H1N1 viruses are in red.
Our estimates of rates of nucleotide substitution gave similar values for each gene segment in both swine lineages, ranging from 1.86 × 10−3 to 3.45 × 10−3 substitutions/site/year (Table 1) and broadly similar to those seen for other RNA viruses (14). At these substitution rates, the TMRCA of the European swine A/H1N1 lineage was 24 to 32 years before the most recent sample (95% HPD of 22 to 41 years), which indicates that the sampled isolates of European swine A/H1N1 viruses arose between the years 1963 and 1982 (Table 1). This is in strong concordance with previous studies that first detected this lineage of H1N1 IAV in pigs in Northern Europe in 1979 (34, 45). The TMRCA of classical swine viruses was estimated at 66 to 76 years before the most recent sample (95% HPD of 64 to 76 years), which is also historically consistent with the first isolate of H1N1 IAV in pigs in North America in 1930 (50).
TABLE 1.
Parameter estimates under the uncorrelated logistic Bayesian demographic model
| Gene | Lineage | Mean substitution rate (10−3) | HPD substitution rate (10−3) | Mean age (yr) | HPD agea (yr) | % Avian identity |
|---|---|---|---|---|---|---|
| PB2 | European | 2.56 | 1.95-3.26 | 26 | 23-30 | 98.7 |
| Classical | 3.15 | 2.71-3.58 | 63 | 61-65 | 96.4 | |
| PB1 | European | 2.66 | 2.20-3.10 | 25 | 22-28 | 98 |
| Classical | 2.89 | 2.41-3.35 | 62 | 60-68 | 95.9 | |
| PA | European | 3.08 | 2.66-3.51 | 24 | 23-25 | 97.9 |
| Classical | 2.45 | 2.02-2.87 | 64 | 61-69 | 95.1 | |
| HA | European | 3.45 | 2.73-4.14 | 27 | 24-30 | 90.6 |
| Classical | 3.33 | 2.87-3.83 | 61 | 61-63 | 86 | |
| NP | European | 1.99 | 1.63-2.35 | 27 | 24-30 | 96.8 |
| Classical | 2.82 | 2.28-3.37 | 62 | 61-66 | 96.2 | |
| NA | European | 2.01 | 1.49-2.50 | 32 | 25-41 | 92.1 |
| Classical | 2.90 | 2.34-3.46 | 64 | 61-69 | 87.4 | |
| MP | European | 2.49 | 1.79-3.20 | 25 | 23-27 | 97.6 |
| Classical | 1.86 | 1.30-2.39 | 65 | 61-77 | 99.2 | |
| NS | European | 2.80 | 2.10-3.55 | 27 | 24-31 | 93.8 |
| Classical | 2.77 | 2.13-3.38 | 63 | 61-66 | 89.1 |
The most recent sequence is from 1991 for classical swine H1N1 viruses and from 2004 for European swine H1N1 viruses.
Analysis of amino acid changes across lineages.
Amino acid site differences were used to determine how frequently amino acid changes occurred in parallel or convergently in the two swine lineages compared to the number that diverged across these lineages (see Table S1 in the supplemental material). Overall, 23.5% of the changes were parallel genetic changes between the two swine lineages (see Table S1 in the supplemental material). Similarly, there were only six (2.9%) convergent genetic changes (i.e., starting from a different ancestral amino acid site) noted across all gene segments. Thus, the largest class of changes (73.5%) were those that experienced divergent evolution across all genes, indicating that the swine A/H1N1 viral lineages experienced strikingly dissimilar evolutionary trajectories (see Table S1 in the supplemental material). Key amino acid changes in the internal gene segments that are unique to European swine H1N1 IAVs are listed in Table 2.
TABLE 2.
Key unique conserved changes in European avian-like swine influenza viruses
| Gene | Position | Most frequent amino acid
|
||||
|---|---|---|---|---|---|---|
| Avian | European avian-like swine | 1918 | Classical swine | Human | ||
| PB2 | 251 | R | K | R | R | R |
| 483 | M | T, A | M | M | M | |
| 649 | V | I | V | V, I | V | |
| 701 | D | N | D | D | D | |
| PB1 | 517 | I | V | I | I | I |
| 584 | R | H | R | R, H | R | |
| PA | 262 | K | R | K | K | K |
| 263 | T | E | T | T | T | |
| 712 | T | V, M | T | T | T | |
| NP | 284 | A | V, I | A | A | A |
| 384 | R | K | R | R | R | |
| M1 | 214 | Q | H | Q | Q | Q |
| NS1 | 25 | Q | R, W, L | Q | K, N, R, W | Q |
| 66 | E | K | E | E | E | |
| 227 | E | E, G | K | R, G | R, del | |
| NS2 | 26 | E | K | E | E | E |
| 49 | V | L | V | V | V | |
| 52 | M | T | M | M | M | |
| 70 | S | G | S | G | G | |
Internal gene changes.
A series of 32 changes consistently associated with human influenza viruses compared to avian IAVs were identified by Finkelstein et al. (15). Of the 10 changes identified in PB2 by Finkelstein et al., the European avian-like swine H1N1 IAV lineage shows only a single parallel change, at residue 271 (see Table S1 in the supplemental material), where the most recent isolates differ from the avian IAV with a T271I mutation (most human IAVs have a T271A mutation, but the 1918 virus has the avian 271T). In contrast, classical swine viruses maintain the human-associated changes at residues 64, 199, 475, 627, and 702 (this residue reverted back to the avian 702K after 1975). Of the 10 changes identified in PA, the European avian-like swine H1N1 IAV lineage shows only a single parallel change, S409N in some isolates, which is also observed in most classical swine isolates (see Table S1 in the supplemental material). In contrast, the classical swine isolates also maintain the D55N change seen in 1918 and subsequent human IAV strains. A total of nine changes in the NP protein have been proposed to be important in mammalian adaptation (15, 39). The European avian-like swine H1N1 IAV lineage contains none of these changes; however, a single strain, A/swine/England/1993 (H1N1), possesses the V33I change observed in both human and classical swine IAVs (see Table S1 in the supplemental material). Three changes have been proposed for M1 (15), but the European avian-like swine IAVs maintain the avian consensus at all three sites. In contrast, the classical swine IAV lineage shares the T121A change with human M1 (see Table S1 in the supplemental material). Four adaptive changes have been proposed for the M2 protein, in the extracellular domain, at residues 14, 16, 18, and 20 (40). Most of the European avian-like swine H1N1 strains share two of these changes, E16G and K18R, with human and classical swine H1N1 IAV lineages. Classical swine strains also share the G14E and S20N changes with human strains (see Table S1 in the supplemental material). It is also notable that a number of the European avian-like swine virus strains contain mutations associated with resistance to adamantane drugs (44), often with more than one resistance mutation at M2 residues 26, 27, 30, 31, and 34, within the ion channel domain. Three changes have been proposed for NS1 (15), and individual European avian-like swine virus strains bearing single changes at each of these three residues are observed (residues 81, 215, and 227), but they are not the same mutations seen in human strains and were not fixed in the swine lineage. Classical swine virus strains have the avian 227E residue, but most European avian-like swine virus strains possess an E227G change (see Table S1 in the supplemental material).
Changes in HA and NA.
Classical swine strains maintain the critical HA receptor binding domain mutation E190D (H3 numbering), as do the majority of European avian-like swine strains (A/swine/Netherlands/3/80 retains the avian consensus glutamic acid). Classical swine strains possess the avian glycine at 225, whereas this receptor binding domain residue is variable in European avian-like swine strains and includes the avian 225G, but also G225E and G225K (see Table S1 in the supplemental material). Interestingly, European avian-like swine strains also show variability in receptor binding residues 135, 137, and 138, unlike classical swine strains, which maintain the avian consensus at these sites. Thus, some European avian-like swine strains show V135I/A/S/T, A137I/V, and A138S changes. We also found evidence of positive selection at residue 145 (see Table S1 in the supplemental material).
European avian-like swine strains also show changes in or near mapped antigenic site regions in human H1, as previously reported (4). Most of these changes are in or near the mapped Ca and Sb antigenic regions (6, 37). These viruses also lose two potential N-linked glycosylation sites which are conserved in avian H1 sequences and the 1918 virus (38). In more recent strains, residues 104 to 106 (NGT) become NGA, and in most European avian-like swine strains, residues 304 to 306 (NSS) become NSN. However, these strains gain two potential glycosylation sites at residues 212 to 214, where ADA becomes NHT (in the antigenic Sb region), and at residues 291 to 293, where NCD becomes NCT in most strains.
The neuraminidase (NA) of the European avian-like swine strains maintains the 15 conserved amino acids making up the active site of the enzyme (8), and no mutations associated with NA inhibitor resistance are observed. The NA also maintains the full-length stalk and the seven potential N-linked glycosylation sites predicted for the 1918 influenza virus (41). Some European avian-like swine strains gain an additional potential glycosylation site at residues 386 to 388, where SFS becomes NFS or NYS.
Analysis of nucleotide compositional space of individual gene segments.
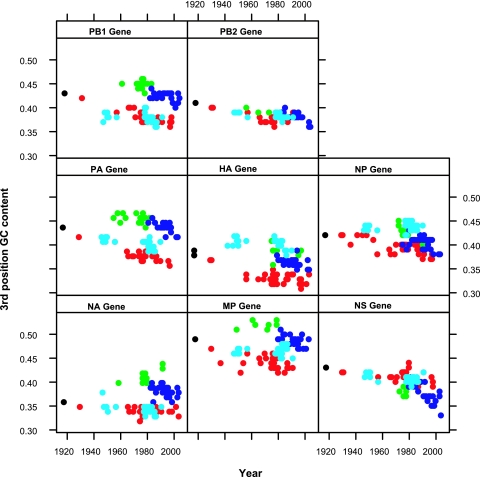
To investigate changes in base composition through time, an indicator of the evolutionary processes that shape genetic diversity in influenza virus, the percent GC content at the third position of each codon of each gene segment was plotted over time (Fig. 2). The directionality of third-position GC content change is measured as the percent change over time from the ancestral sequence.
FIG. 2.
Synonymous third-codon-position G+C contents over time for all eight genes across European and classical swine, Eurasian avian, 1918, and human H1N1 IAVs. Classical swine H1N1 viruses are in red, European swine H1N1 viruses are in blue, Eurasian avian virus sequences are in green, human H1N1 viruses are in light blue, and 1918 H1N1 virus is in black.
The GC content of each of the eight gene segments for the classical swine and European swine lineages tended to decrease over time from their ancestral sequences (1918 for classical swine viruses and Eurasian avian virus for the European swine viruses). Compared with the 1918 sequence, the changes in third-position base composition in the NA gene over time for classical swine and human H1N1 viruses are less marked than those for the other segments, and the trajectory of the HA and NP genes in the human H1N1 lineages shows an unexpected increase in GC content at the third position over time (Fig. 2). The GC contents for the PB2 gene are similar across lineages, suggesting functional and structural constraints on the gene segment.
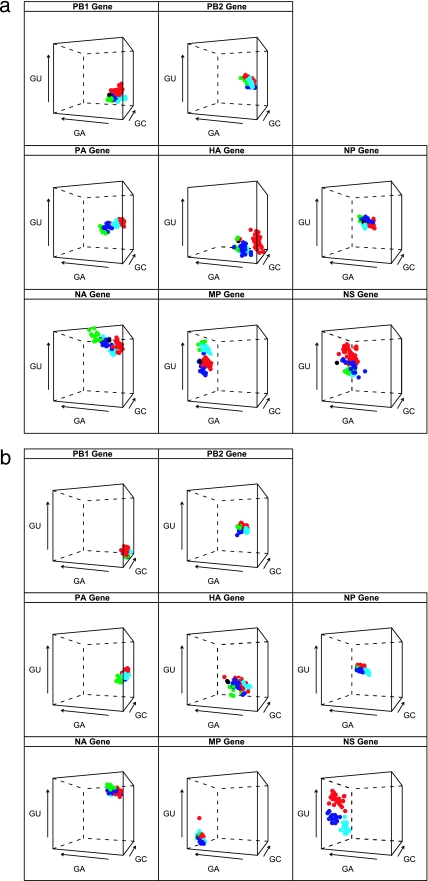
Some variation in nucleotide base compositional bias is expected for the eight gene segments, based on the different molecular functions of the gene products (which in turn affects amino acid usage). If base compositional bias for the eight gene segments is universal, such that all segments evolve in the same way irrespective of host, it is expected that no difference in the clustering of these genes in compositional space across the different A/H1N1 lineages over time would be observed. Nucleotide compositional analysis revealed that each gene segment has a unique clustering profile, revealing a powerful segment-specific bias (Fig. 3a). However, each A/H1N1 lineage within this overall bias subdivides the clustering profile in space, indicating that there is also a lineage-specific effect on gene composition. Furthermore, the 1918 sequence in general occupies contiguous compositional space with classical swine and human A/H1N1 lineages, as would be expected because classical swine and human A/H1N1 viruses are direct descendants of the 1918 virus. Similarly, the Eurasian avian and European swine A/H1N1 lineages occupy contiguous compositional space in this analysis. There is an overall lack of overlap in compositional space clustering between the two swine A/H1N1 lineages across most of the gene segments. The HA and MP genes show a greater spread in compositional space than do the other gene segments. Analysis of the first and second codon positions also revealed a considerable difference in nucleotide composition between the two swine H1N1 lineages (Fig. 3b). The compositional space is partitioned by a unique compositional profile with very little overlap. The Eurasian avian and European swine viruses showed more overlap for the polymerase genes. The NP and NS genes showed the most specific pattern for all lineages, with little or no overlap in compositional space. The HA gene segment profile showed the greatest spread, most likely indicative of antigenic drift. The base compositional results for M2 and NS2 (NEP) are shown in the supplemental material.
FIG. 3.
(a) Overall nucleotide compositions of eight gene segments by H1N1 IAV lineages. Axes correspond to the frequencies of G+U, G+A, and G+C for each gene segment. Classical swine H1N1 viruses are in red, European swine H1N1 viruses are in blue, Eurasian avian virus sequences are in green, human H1N1 viruses are in light blue, and 1918 H1N1 virus is in black. (b) Nucleotide compositions of eight gene segments at the first and second codon positions. Axes correspond to the frequencies of G+U, G+A, and G+C for each gene segment. Classical swine H1N1 viruses are in red, European swine H1N1 viruses are in blue, Eurasian avian virus sequences are in green, human H1N1 viruses are in light blue, and 1918 H1N1 virus is in black.
DISCUSSION
Our phylogenetic analysis supports the independent emergence of classical and European swine H1N1 IAVs, and the estimates for the TMRCA gave ranges of times of origin for both swine A/H1N1 lineages similar to those previously reported (2, 33, 50). Yet within this phylogenetic history, our analysis of whole genomes of swine H1N1 IAVs revealed that the lineages are experiencing largely divergent, rather than convergent or parallel, evolution; there were approximately three times as many mutations producing divergent evolution than those resulting in homoplasy.
The third-position base composition analysis revealed that each swine lineage is diverging from its putative ancestor by generally decreasing in GC content at the third codon position over time. The movement of human H1N1 viruses to a higher GC content for the HA gene implies that selection for antigenic differences may affect the trajectory of third-position GC content over time, although this will need to be explored in more detail. This trend is also reflected in the NP gene, which shows an overlap in both of the swine lineages, suggesting that both the HA and NP genes are highly host specific. Although synonymous changes at the third codon position can be attributed partially to neutral evolution (24, 31, 63), the similar patterns of decreasing GC content over time for all of the gene segments again argue for host specificity.
The nucleotide compositional analysis revealed several evolutionary patterns (Fig. 3a and b). First, each gene segment has a distinctive signature nucleotide compositional space profile. This trend strongly suggests that there are functional and structural constraints acting on each segment individually, which will clearly need to be explored further. However, within these segment-specific profiles, the compositional space is also partitioned by a lineage effect (Fig. 3). This strongly signifies that natural selection has played a major role in shaping nucleotide composition in IAV, reflective of the past history of each virus. Second, the wide distribution of points in the HA gene shows a strong host effect in compositional space, again likely driven by antigenic drift (such that selection for amino acid changes has a secondary effect on nucleotide composition). Third, the swine lineages share the same compositional space away from the human A/H1N1 and 1918 sequences, again supporting the idea that there is strong selection for host-specific antigenic change. The first- and second-codon-position compositional analysis revealed a more distinct pattern by clade. The classical swine and human H1N1 viruses show very little overlap, indicating host-specific compositional bias (Fig. 3b). The Eurasian avian and European swine clades overlap for the polymerase genes. This suggests that although European swine H1N1 viruses emerged almost 30 years ago, the polymerase genes are still very avian-like. Interestingly, the NP gene shows the most host-specific pattern, with no overlap among the groups. However, the lack of overlap between the two swine H1N1 lineages suggests that host specificity is contingent upon the history of the IAV. Similar to Rabadan et al. (36), we found that the 1918 virus is avian-like in the nucleotide composition of its gene segments.
The biased nucleotide composition at the first and second codon positions is reflective of nonsynonymous changes in amino acid usage. In the context of a single host switch event, the mutations identified in the 1918 influenza virus and subsequently maintained in human influenza viruses and in classical swine strains may represent a set of crucial functional changes from an ancestral avian IAV (15). However, the lack of parallel evolution in the independent emergence of the European avian-like swine strains suggests that the acquisition of a polygenic set of functional changes may be different between independent host switch events. The utility of identifying these mutations as proxies to define whether a future IAV is acquiring changes important in mammalian adaptation might be limited. For example, of the 10 amino acid changes identified in PB2 (15, 60), only more recent European avian-like swine strains share a single change from the avian consensus at one of these sites, at residue 271, with a T271I change. Crucially, they lack the PB2 E627K change (54), even in those strains isolated after 20 years of circulation in swine. Thus, this particular mutation may not be necessary for mammalian adaptation in general, or at least swine adaptation in particular. However, European avian-like swine strains do possess a D701N change from the avian consensus that may also play a role in mammalian adaptation (16), but they lack the K702R change associated with human PB2 genes and early classical swine H1N1 strains (60). Classical swine viruses after the mid-1970s reverted to the avian lysine at 702 but continued to possess the E627K change. The D701N change was observed as one of six changes after mouse adaptation of an avian H7N7 virus (16). None of the other five changes is observed in human, classical swine, or European avian-like swine lineages, and they may be specific to this mouse adaptation experiment. The D701N mutation has also been observed in a small minority of human H5N1 isolates but has been linked to increased pathogenicity in an experimental mouse H5N1 infection model (26). Recently, the structure of the C-terminal end of PB2 was resolved, and structural analysis suggests that this region of PB2 contains a nuclear localization signal and complexes with the importin α5 (57). The structure suggests that the changes observed at residues 701 and 702 may be important in the interaction with importin α5, suggesting a biological explanation for changes at these sites associated with host switch events.
In the H1 subtype, only a single amino acid change, E190D (using the H3 numbering), is required to alter receptor specificity from α2-3 to gain the ability to bind α2-6 receptors (53). The 1918 pandemic viruses possessed HAs with two receptor-binding variants—either with a single E190D change from the avian consensus or with two changes, E190D plus G225D (42). The form with two changes is highly specific for α2-6 binding (51, 53). Both the 1918 pandemic virus and its derivatives and the European avian-like swine virus HAs have the E190D mutation crucial for α2-6 binding (53). This indicates that H1 subtype HAs involved in switching from an avian host to a mammalian host may need to acquire this particular mutation for stable host adaptation. Other changes observed in the receptor binding domains of the European avian-like swine viruses (at residues 135, 137, 138, and 145) may also play a role in altering receptor specificity, but this has not been evaluated experimentally.
In summary, our study demonstrates that we should consider the role of historical contingency, reflected in a strong lineage-specific effect, in the emergence of IAVs from an avian reservoir into a new mammalian host and that mutations identified as important in prior host switch events may or may not be observed in future such events. The host switch events leading to the emergence of the European avian-like swine lineage from birds and the recent emergence of a canine H3N8 IAV lineage derived from equine H3N8 viruses (9) demonstrate that even in the absence of reassortment, stable host adaptation can occur in IAVs by acquisition of crucial mutations.
Acknowledgments
This work was supported in part by the intramural program of the NIAID and the NIH, by an Alfred P. Sloan Foundation graduate scholarship, and by the NIH/INRO fellowship program.
Footnotes
Published ahead of print on 18 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alexander, D. J., and I. H. Brown. 2000. Recent zoonoses caused by influenza A viruses. Rev. Sci. Tech. 19197-225. [DOI] [PubMed] [Google Scholar]
- 2.Brown, I. H. 2000. The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 7429-46. [DOI] [PubMed] [Google Scholar]
- 3.Brown, I. H., P. A. Harris, J. W. McCauley, and D. J. Alexander. 1998. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J. Gen. Virol. 792947-2955. [DOI] [PubMed] [Google Scholar]
- 4.Brown, I. H., S. Ludwig, C. W. Olsen, C. Hannoun, C. Scholtissek, V. S. Hinshaw, P. A. Harris, J. W. McCauley, I. Strong, and D. J. Alexander. 1997. Antigenic and genetic analyses of H1N1 influenza A viruses from European pigs. J. Gen. Virol. 78553-562. [DOI] [PubMed] [Google Scholar]
- 5.Castrucci, M. R., I. Donatelli, L. Sidoli, G. Barigazzi, Y. Kawaoka, and R. G. Webster. 1993. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 193503-506. [DOI] [PubMed] [Google Scholar]
- 6.Caton, A. J., G. G. Brownlee, J. W. Yewdell, and W. Gerhard. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31417-427. [DOI] [PubMed] [Google Scholar]
- 7.Chun, J. 1919-;-1920. Influenza including its infection among pigs. Nat. Med. J. 534-44. [Google Scholar]
- 8.Colman, P. M., J. N. Varghese, and W. G. Laver. 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 30341-44. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, P. C., E. J. Dubovi, W. L. Castleman, I. Stephenson, E. P. Gibbs, L. Chen, C. Smith, R. C. Hill, P. Ferro, J. Pompey, R. A. Bright, M. J. Medina, C. M. Johnson, C. W. Olsen, N. J. Cox, A. I. Klimov, J. M. Katz, and R. O. Donis. 2005. Transmission of equine influenza virus to dogs. Science 310482-485. [DOI] [PubMed] [Google Scholar]
- 10.Donatelli, I., L. Campitelli, M. R. Castrucci, A. Ruggieri, L. Sidoli, and J. S. Oxford. 1991. Detection of two antigenic subpopulations of A(H1N1) influenza viruses from pigs: antigenic drift or interspecies transmission? J. Med. Virol. 34248-257. [DOI] [PubMed] [Google Scholar]
- 11.Dorset, M., C. N. McBryde, and W. B. Niles. 1922. Remarks on hog flu. J. Am. Vet. Med. Assoc. 62162-171. [Google Scholar]
- 12.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9267-276. [DOI] [PubMed] [Google Scholar]
- 14a.Dugan, V. G., R. Chen, D. J. Spiro, N. Sengamalay, J. Zaborsky, E. Ghedin, J. Nolting, D. E. Swayne, J. A. Runstadler, G. M. Happ, D. A. Senne, R. Wang, R. D. Slemons, E. C. Holmes, and J. K. Taubenberger. 2008. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 4e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finkelstein, D. B., S. Mukatira, P. K. Mehta, J. C. Obenauer, X. Su, R. G. Webster, and C. W. Naeve. 2007. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 8110292-10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriel, G., B. Dauber, T. Wolff, O. Planz, H. D. Klenk, and J. Stech. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. USA 10218590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambaryan, A. S., A. I. Karasin, A. B. Tuzikov, A. A. Chinarev, G. V. Pazynina, N. V. Bovin, M. N. Matrosovich, C. W. Olsen, and A. I. Klimov. 2005. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res. 11415-22. [DOI] [PubMed] [Google Scholar]
- 18.Greenbaum, B. D., A. J. Levine, G. Bhanot, and R. Rabadan. 2008. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 4e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan, Y., K. F. Shortridge, S. Krauss, P. H. Li, Y. Kawaoka, and R. G. Webster. 1996. Emergence of avian H1N1 influenza viruses in pigs in China. J. Virol. 708041-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinshaw, V. S., R. G. Webster, B. C. Easterday, and W. J. Bean, Jr. 1981. Replication of avian influenza A viruses in mammals. Infect. Immun. 34354-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 1462275-2289. [DOI] [PubMed] [Google Scholar]
- 22.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 6871-85. [DOI] [PubMed] [Google Scholar]
- 23.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 634603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, MA.
- 25.Koen, J. 1919. A practical method for field diagnosis of swine diseases. Am. J. Vet. Med. 14468-470. [Google Scholar]
- 26.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 7912058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, W., A. L. Vincent, M. R. Gramer, C. B. Brockwell, K. M. Lager, B. H. Janke, P. C. Gauger, D. P. Patnayak, R. J. Webby, and J. A. Richt. 2007. Identification of H2N3 influenza A viruses from swine in the United States. Proc. Natl. Acad. Sci. USA 10420949-20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maldonado, J., K. Van Reeth, P. Riera, M. Sitja, N. Saubi, E. Espuna, and C. Artigas. 2006. Evidence of the concurrent circulation of H1N2, H1N1 and H3N2 influenza A viruses in densely populated pig areas in Spain. Vet. J. 172377-381. [DOI] [PubMed] [Google Scholar]
- 29.Matrosovich, M. N., A. S. Gambaryan, S. Teneberg, V. E. Piskarev, S. S. Yamnikova, D. K. Lvov, J. S. Robertson, and K. A. Karlsson. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233224-234. [DOI] [PubMed] [Google Scholar]
- 30.McInerney, J. O. 1998. GCUA: general codon usage analysis. Bioinformatics 14372-373. [DOI] [PubMed] [Google Scholar]
- 31.McVean, G. A. T., and B. Charlesworth. 1999. A population genetic model for the evolution of synonymous codon usage: patterns and predictions. Genet. Res. 74145-158. [Google Scholar]
- 32.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85199-210. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, C. W., I. H. Brown, B. C. Easterday, and K. Van Reeth. 2006. Swine influenza, p. 469-482. In B. E. Straw, J. J. Zimmerman, S. D'Allaire, and D. J. Taylor (ed.), Diseases of swine, 9th ed. Blackwell Publishing Professional, Ames, IA.
- 34.Pensaert, M., K. Ottis, J. Vandeputte, M. M. Kaplan, and P. A. Bachmann. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull. W. H. O. 5975-78. [PMC free article] [PubMed] [Google Scholar]
- 34a.Phipps, L. P., S. C. Essen, and I. H. Brown. 2004. Genetic subtyping of influenza A viruses using RT-PCR with a single set of primers based on conserved sequences within the HA2 coding region. J. Virol. Methods 122119-122. [DOI] [PubMed] [Google Scholar]
- 35.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 212531-2533. [DOI] [PubMed] [Google Scholar]
- 35a.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 36.Rabadan, R., A. J. Levine, and H. Robins. 2006. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. J. Virol. 8011887-11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Rambaut, A. 1996. Se-Al: Sequence alignment editor. http://tree.bioied.ac.uk/software/seal.
- 37.Raymond, F. L., A. J. Caton, N. J. Cox, A. P. Kendal, and G. G. Brownlee. 1983. Antigenicity and evolution amongst recent influenza viruses of H1N1 subtype. Nucleic Acids Res. 117191-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 961651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reid, A. H., T. G. Fanning, T. A. Janczewski, R. M. Lourens, and J. K. Taubenberger. 2004. Novel origin of the 1918 pandemic influenza virus nucleoprotein gene. J. Virol. 7812462-12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid, A. H., T. G. Fanning, T. A. Janczewski, S. McCall, and J. K. Taubenberger. 2002. Characterization of the 1918 “Spanish” influenza virus matrix gene segment. J. Virol. 7610717-10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. USA 976785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, A. H., T. A. Janczewski, R. M. Lourens, A. J. Elliot, R. S. Daniels, C. L. Berry, J. S. Oxford, and J. K. Taubenberger. 2003. 1918 influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerg. Infect. Dis. 91249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, A. H., J. K. Taubenberger, and T. G. Fanning. 2004. Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Nat. Rev. Microbiol. 2909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidtke, M., R. Zell, K. Bauer, A. Krumbholz, C. Schrader, J. Suess, and P. Wutzler. 2006. Amantadine resistance among porcine H1N1, H1N2, and H3N2 influenza A viruses isolated in Germany between 1981 and 2001. Intervirology 49286-293. [DOI] [PubMed] [Google Scholar]
- 45.Scholtissek, C., H. Burger, P. A. Bachmann, and C. Hannoun. 1983. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 129521-523. [DOI] [PubMed] [Google Scholar]
- 46.Scholtissek, C., H. Burger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147287-294. [DOI] [PubMed] [Google Scholar]
- 47.Scholtissek, C., V. von Hoyningen, and R. Rott. 1978. Genetic relatedness between the new 1977 epidemic strains (H1N1) of influenza and human influenza strains isolated between 1947 and 1957 (H1N1). Virology 89613-617. [DOI] [PubMed] [Google Scholar]
- 48.Schultes, E., P. T. Hraber, and T. H. LaBean. 1997. Global similarities in nucleotide base composition among disparate functional classes of single-stranded RNA imply adaptive evolutionary convergence. RNA 3792-806. [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz, U., W. M. Fitch, S. Ludwig, J. Mandler, and C. Scholtissek. 1991. Evolution of pig influenza viruses. Virology 18361-73. [DOI] [PubMed] [Google Scholar]
- 50.Shope, R. E. 1931. Swine influenza. I. Experimental transmission and pathology. J. Exp. Med. 54349-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srinivasan, A., K. Viswanathan, R. Raman, A. Chandrasekaran, S. Raguram, T. M. Tumpey, V. Sasisekharan, and R. Sasisekharan. 2008. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc. Natl. Acad. Sci. USA 1052800-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens, J., O. Blixt, L. Glaser, J. K. Taubenberger, P. Palese, J. C. Paulson, and I. A. Wilson. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 3551143-1155. [DOI] [PubMed] [Google Scholar]
- 53.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312404-410. [DOI] [PubMed] [Google Scholar]
- 54.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 671761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swayne, D. E. 2007. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 51242-249. [DOI] [PubMed] [Google Scholar]
- 56.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 57.Tarendeau, F., J. Boudet, D. Guilligay, P. J. Mas, C. M. Bougault, S. Boulo, F. Baudin, R. W. Ruigrok, N. Daigle, J. Ellenberg, S. Cusack, J. P. Simorre, and D. J. Hart. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14229-233. [DOI] [PubMed] [Google Scholar]
- 58.Taubenberger, J. K., and D. M. Morens. 2006. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 1215-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taubenberger, J. K., A. H. Reid, T. A. Janczewski, and T. G. Fanning. 2001. Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos. Trans. R. Soc. Lond. B 3561829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437889-893. [DOI] [PubMed] [Google Scholar]
- 61.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webster, R. G., and E. A. Govorkova. 2006. H5N1 influenza—continuing evolution and spread. N. Engl. J. Med. 3552174-2177. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Z., and R. Nielsen. 2008. Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25568-579. [DOI] [PubMed] [Google Scholar]
- 64.Zell, R., S. Motzke, A. Krumbholz, P. Wutzler, V. Herwig, and R. Durrwald. 2008. Novel reassortant of swine influenza H1N2 virus in Germany. J. Gen. Virol. 89271-276. [DOI] [PubMed] [Google Scholar]