Abstract
The structures of canine parvovirus (CPV) and feline parvovirus (FPV) complexed with antibody fragments from eight different neutralizing monoclonal antibodies were determined by cryo-electron microscopy (cryoEM) reconstruction to resolutions varying from 8.5 to 18 Å. The crystal structure of one of the Fab molecules and the sequence of the variable domain for each of the Fab molecules have been determined. The structures of Fab fragments not determined crystallographically were predicted by homology modeling according to the amino acid sequence. Fitting of the Fab and virus structures into the cryoEM densities identified the footprints of each antibody on the viral surface. As anticipated from earlier analyses, the Fab binding sites are directed to two epitopes, A and B. The A site is on an exposed part of the surface near an icosahedral threefold axis, whereas the B site is about equidistant from the surrounding five-, three-, and twofold axes. One antibody directed to the A site binds CPV but not FPV. Two of the antibodies directed to the B site neutralize the virus as Fab fragments. The differences in antibody properties have been linked to the amino acids within the antibody footprints, the position of the binding site relative to the icosahedral symmetry elements, and the orientation of the Fab structure relative to the surface of the virus. Most of the exposed surface area was antigenic, although each of the antibodies had a common area of overlap that coincided with the positions of the previously mapped escape mutations.
During a virus infection, host antibodies are raised against viral proteins that are recognized as foreign, although most of these antibodies do not neutralize the virus. The small fraction of antibodies that are neutralizing bind specifically to exposed structures on the surface of the viral capsid and can interfere with viral functions such as attachment, entry, or subsequent processing of the viral proteins critical to infectivity (20). Among the many ways to neutralize viruses, a frequently encountered mechanism is cross-linking of virus capsids by multivalent antibodies, resulting in aggregation and perhaps precipitation (8, 55). Alternatively, antibodies can bind bivalently across the icosahedral twofold axes of symmetry to prevent uncoating (12, 21, 48). Antibodies may also neutralize infectivity by interfering with receptor attachment to the viral surface, sterically blocking viral attachment to cells or occluding the receptor binding site (22, 27, 53). Antibodies have also been shown to induce a conformational change or rearrangement of viral capsid proteins, causing the receptor binding site to become inaccessible (33).
Epitopes recognized by host antibodies on viral proteins may overlap with other functional sites, such as those recognized by receptors. Receptor binding sites may be found in cavities that are not readily accessible to antibodies (5, 50) or sequestered within structures that are not exposed until after binding of another receptor (31). In other cases, the receptor binding sites appear to be exposed on the surface of the virus and to overlap substantially with antibody binding sites that have been defined (16, 23).
Parvoviruses have small, 260-Å-diameter, icosahedral, nonenveloped capsids that package a single-stranded DNA genome of about 5 kb. Each of the 60 subunits consists of the same eight-stranded antiparallel β-barrel motif found in numerous viral capsid structures (6, 34, 51). In canine parvovirus (CPV) (58) and feline panleukopenia virus (FPV) (2), large insertions between strands of the β-barrel form most of the capsid surface and create small, protruding “spikes” around the icosahedral threefold axes, which are involved in host recognition and antigenicity (11, 19, 25, 32, 38, 58). Whereas both viruses can utilize the feline transferrin receptor (TfR) for attachment and infection (25), CPV gained the ability to bind canine TfR and to infect canine cells and dogs (26). Residues 93 and 323 within the major capsid protein, located in the vicinity of the threefold spikes, allow the CPV capsids to bind canine TfR (25).
CPV and FPV are conserved in sequence, with little variation in most of the major viral capsid protein (24). However, a number of antigenic variants have arisen during the evolution of CPV in dogs. The antigenic sites of CPV and FPV have been characterized and mapped to the virus surface by use of monoclonal antibodies (MAbs) (54), peptide analysis of polyclonal sera, and cryo-electron microscopy (cryoEM) (61) and by comparison of naturally occurring antigenic variants (10, 28, 29, 44, 57). Competition assays and escape mutations divided the MAbs into two groups that mapped to sites designated sites A and B (40). Site A was near the top of the threefold spike, and site B was about equally distant from the surrounding five-, three-, and twofold axes (54) (Fig. 1). The role of the antigenic selection of viruses is not clear, as maternal antibodies initially protect animals against virus infection (42). Antibodies that develop after infection are protective against reinfection for many years, and there is strong cross-protection between antigenically variant viruses (1). Variations in both the A and B regions that affect antibody interactions also change the specific binding of the capsids to canine and feline TfR and consequently alter the virus host ranges (25, 37). Therefore, it is possible that selection is driven more by altered receptor binding than by the inherent antigenic properties of the capsid.
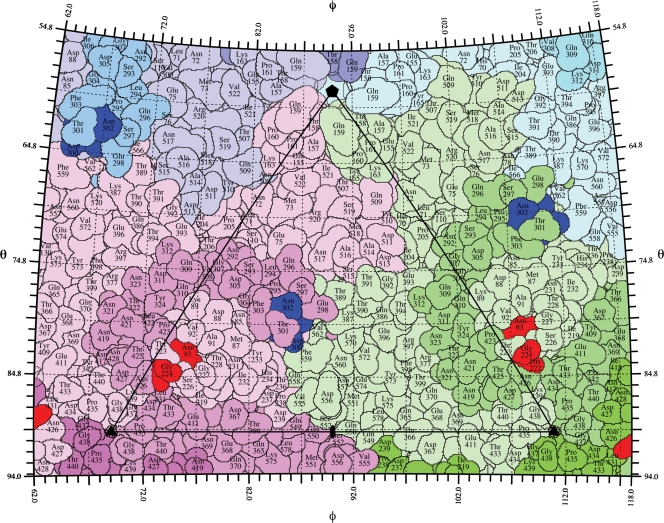
FIG. 1.
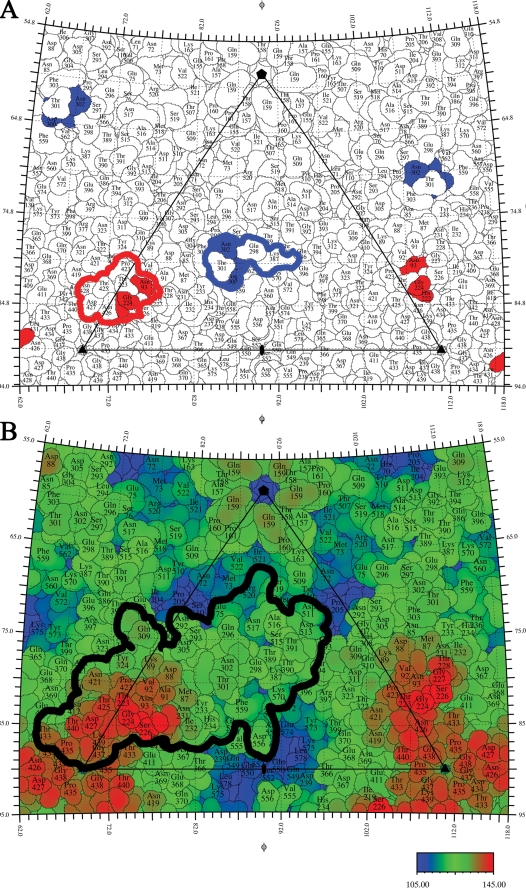
The viral surface is shown as a stereographic projection where the polar angles φ and θ represent the latitude and longitude of a point on the viral surface, respectively (63). The virus surface is represented as a quilt of amino acids (52), and the icosahedral asymmetric unit of the virus is indicated by the triangular boundary. To visualize the different copies of the capsid protein in the road map that comprise the asymmetric unit, VP2 molecules are displayed in tints of magenta, green, and blue. Escape mutations from neutralizing antibodies are colored red for site A and dark blue for site B.
Eight MAbs and the fragment antibody binding (Fab) portion generated from each were previously characterized by biochemical analyses (35). Three of the antibodies were from rats immunized with FPV, and five were from mice immunized with CPV. The antibodies were from animals that had been immunized repeatedly with purified viral capsids in adjuvants and therefore represented the high-affinity B-cell responses. All eight antibodies were found to neutralize virus infectivity when tested as immunoglobulins G (IgGs) (35). All of the Fabs bound to virus with similar affinities and competed with the cellular receptor (35). Three of the eight Fabs (14, B, and 6) attached to the A site, although Fab 14 bound to and neutralized only CPV, not FPV. Five of the eight antibodies (8, 15, 16, E, and F) attached to the B site. From the latter group, Fabs E and F neutralized virus at a low ratio of only 25 Fabs per capsid and inhibited binding of TfR to the virus (35). Only these two antibodies were found to generate Fab that neutralized virus. Escape mutations have never been isolated for Fab F.
Here we present cryoEM studies of the eight Fab molecules complexed with CPV or FPV capsids. We also present the crystal structure of the CPV-specific Fab 14. Although each antibody bound differently, all three Fabs directed to the A site had footprints that overlapped a common region that included the previously identified residues that altered MAb binding to that epitope. Similarly, the five Fabs directed to the B site had a common overlapped region coincident with the B epitope amino acids. The structure of Fab 14 was docked into the appropriate cryoEM density and demonstrated that the change in residue 93 on the viral surface allows the Fab to bind to CPV and not to FPV. The structure of the virus complexed with Fabs E and F suggests an alternative mechanism of neutralization from that for the other six antibodies.
MATERIALS AND METHODS
Preparation of viruses and Fabs.
Capsids of CPV and FPV were prepared from infected feline cells. Full (DNA-containing) and empty capsids were purified by sucrose gradient centrifugation as previously described (39). Eight different hybridomas produced antibodies (three rat IgGs and five mouse IgGs) against CPV or FPV capsids (Table 1; see Fig. 4) (40, 41). Hybridomas were grown in 500-ml volumes in gas-permeable bags (Nexell, Irving, CA) containing Dulbecco's minimal essential medium with 5% fetal calf serum. The IgGs were purified using protein G (GE Healthcare, Piscataway, NJ). The Fabs were generated by papain digestion, the Fc portion was separated with protein A (GE Healthcare), and the monomeric Fabs were purified by chromatography in a Sephadex G100 column in phosphate-buffered saline. Protein concentrations were determined by measuring the A280 and by bicinchoninic acid assay.
TABLE 1.
MAbs used to generate Fabs for structural study and cryoEM data and reconstruction statistics
| Virus-Fab complex | Source of antibody | Antigenic sitea | Specificity of Fab | No. of micrographs | Range of defocus level | No. of selected particles | No. of particles used in map | Resolutionb (Å) | Resolutionc (Å) |
|---|---|---|---|---|---|---|---|---|---|
| CPV-14d | Mouse | A | CPV only | 109 | 1.0-3.8 | 2,860 | 2,059 | 12.4 | |
| FPV-B | Rat | A | CPV and FPV | 56 | 1.7-3.7 | 1,576 | 1,126 | 14 | 18.7 |
| FPV-6 | Mouse | A | CPV and FPV | 42 | 1.2-4.2 | 3,500 | 2,520 | 18 | |
| FPV-8 | Mouse | B | CPV and FPV | 49 | 1.8-3.1 | 6,480 | 4,344 | 8.5 | 11.1 |
| FPV-15 | Mouse | B | CPV and FPV | 50 | 0.9-3.4 | 6,955 | 4,798 | 10.5 | 11.7 |
| FPV-16 | Mouse | B | CPV and FPV | 48 | 0.4-4.8 | 2,173 | 2,084 | 13 | 18.8 |
| FPV-E | Rat | B | CPV and FPV | 83 | 0.5-5.6 | 2,331 | 1,684 | 12 | |
| FPV-F | Rat | B | CPV and FPV | 93 | 1.0-7.3 | 2,450 | 1,769 | 14 |
Antigenic sites were mapped previously by cross-competition and escape mutational analyses.
Determined by where the Fourier shell correlation fell below 0.5 for the density representing the viral capsid between radii at 70 and 140 Å.
Determined by where the Fourier shell correlation fell below 0.5 for the density representing the entire viral capsid without setting to zero the density corresponding to the DNA for those capsids that were DNA filled.
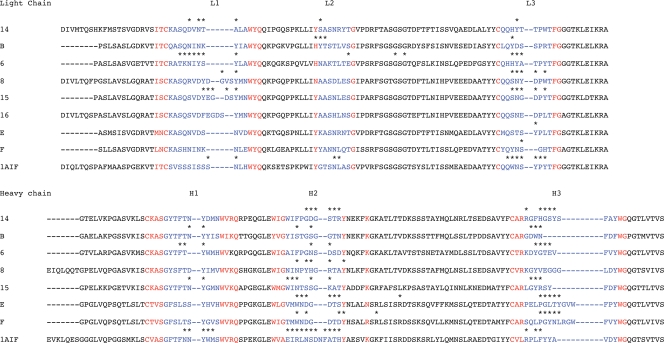
FIG. 4.
Amino acid sequences of the light and heavy chains aligned according to consensus sequences (red) and CDR loops (blue). Residues in contact with the viral surface are indicated by asterisks. Since only the light chain sequence was determined for Fab 16, the mouse antibody 1AIF Fab was used to interpret the Fab 16 density.
Sequence determination of MAb variable domains.
For each antibody, mRNA was purified from the corresponding hybridoma by use of an RNeasy purification kit (Qiagen, Valencia, CA). This mRNA was either further processed immediately or aliquoted and stored at −80°C until used. Amplified products and cDNAs were generated using a one-step reverse transcription-PCR kit (Invitrogen, Carlsbad, CA). Mouse primers were designed for variable domain amplification, using the mouse IgG database (MRC Center for Protein Engineering [http://www.mrc-cpe.cam.ac.uk]). Rat primers were designed based on degenerate nucleotide rat IgG DNA sequences obtained from the National Center for Biotechnology Information. These rat primers amplified a similar region of the IgG mRNA to that amplified by the mouse primers. PCR products were then cloned into pGEM-T Easy (Promega, Madison, WI). Plasmid inserts were then sequenced using T7 or SP6 primers. The variable sequences of MAbs 8 and 14 have been described previously (61, 64). The sequence for MAb 14 was redetermined here. The sequence of the heavy chain of antibody 16 was not determined.
Crystallization of Fab 14 and structural solution.
Fab 14 was produced and purified as described above and then concentrated to 8 mg/ml in 50 mM HEPES, 150 mM NaCl. Crystals were obtained by mixing 1 μl concentrated purified protein with 1 μl of reservoir solution containing 25% polyethylene glycol 5000 and 0.1 M HEPES, pH 7.5, and equilibrating this hanging droplet over 750 μl of reservoir solution at room temperature. Data were collected on Biocars beamline 14D at the Advanced Photon Source. The data were indexed and scaled using HKL2000 (36). The crystals belonged to space group C2, with one molecule per asymmetric unit. The structure was solved by molecular replacement using the program MOLREP (59). The immunoglobulin (Protein Data Bank [PDB] accession no. 12E8) was found to be a successful search model. Initially, the structure was refined as a rigid body by use of CNS (9), followed by model building into the electron density by use of COOT (15) between cycles of refinement with the program CNS (9). The final refinement used 2.0-Å-resolution data, which gave an Rwork of 0.256 and an Rfree of 0.293 (Table 2). Four nonglycine residues were in the disallowed area of the Ramachandran plot.
TABLE 2.
Crystallographic data collection, processing, and refinement statistics
| Parameter | Valuec |
|---|---|
| Data collection statistics | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 168.61, 39.89, 70.75 |
| β (°) | 94.6 |
| Resolution (Å) | 2.0 |
| Rmergea | 0.06 (0.47) |
| I/σ | 21.51 (3.43) |
| Completeness (%) | 99.8 (100.0) |
| Redundancy | 3.6 (3.7) |
| Refinement statistics | |
| Resolution (Å) | 20-2.0 |
| No. of reflections | 30,476 |
| Rwork/Rfreeb | 25/29 |
| No. of atoms | |
| Protein | 3,283 |
| Water | 188 |
| RMSD | |
| Bond length (Å) | 0.01 |
| Bond angle (°) | 1.31 |
| Average B factor (Å2) | |
| Main chain atoms | 39.4 |
| Side chain atoms | 41.4 |
Rmerge = (ΣhΣi|Ihi − 〈Ih〉)|/((ΣhΣiIhi), where 〈Ih〉 is the mean intensity value of I observations of symmetry-related reflections with h indices.
Rwork = Σhkl[‖F(obs)hkl| − |F(calc)hkl‖]/Σhkl|F(obs)hkl|, where |F(obs)| and |F(calc)| are the observed and calculated structure factor amplitudes. Rfree is the same as Rwork, except it is calculated for the 10% of the data that were randomly omitted from each resolution shell in the refinement (not used to calculate Rwork).
Values in parentheses are for the highest-resolution shell.
CryoEM reconstruction.
Purified Fabs were incubated with capsids at room temperature for 1 hour at a ratio of four Fab molecules per potential binding site on the virus (240 Fab molecules/capsid). Small aliquots of this mixture were applied to carbon-coated grids and vitrified in liquid ethane. Electron micrographs were recorded under minimal dose conditions (∼24 electrons/Å2) on Kodak SO-163 film, using a Phillips CM300 FEG microscope. Micrographs were digitized with a Zeiss Phodis microdensitometer at 7-μm intervals. The scans were averaged in boxes of 2 by 2 pixels, making the final pixel size 3.11 Å. The program RobEM was used to select particles and to make contrast transfer function corrections (http://cryoem.ucsd.edu/programs.shtm). The EM reconstruction processes were performed using icosahedral averaging with the programs EMPFT and EM3DR (4). The model used to initiate the reconstructions was based on the earlier X-ray crystallographic results (58). The final resolution was determined by where the Fourier shell correlation fell below 0.5 (Chuan Xiao [http://bilbo.bio.purdue.edu/∼viruswww/Rossmann_home/river_programs/]) for the density representing the viral capsid between radii 70 and 140 Å (Table 1) and by back transforming the entire map for those reconstructions of complexes in which the capsid contained DNA.
Fitting the Fab structure into the appropriate cryoEM density.
The program EMfit (45, 49) was used to calibrate the exact magnification of each of the cryoEM reconstructions by comparing it with a map derived from the X-ray crystallographically determined coordinates of CPV and FPV (PDB accession numbers 1C8D and 1FPV, respectively). A difference map was calculated for each virus-Fab complex by setting to zero the density within a radius of 3 Å surrounding each atom of the virus X-ray structure coordinates. The Fabs were fitted using the program EMfit (Table 3), using a two-step procedure (18, 65). The structure of the Fab constant domain (CL-CH) was fitted first, and the density was removed from the difference map before the variable domain Fab structure (VL-VH) was fitted into the remaining difference density. The structure used for fitting the constant domain was assumed to be similar to that for other mouse antibodies, such as the structure corresponding to PDB accession number 1AIF. For each Fab, the amino acid sequence corresponding to the variable domain was submitted to Web Antibody Modeling (WAM) (60; http://antibody.bath.ac.uk/) to obtain a three-dimensional homology model used to fit into the remaining difference density.
TABLE 3.
Statistics for fitting Fab structures into corresponding cryoEM densitye
| Fabf |
Rcrita
|
Sumfb
|
Clash (%)c
|
−Den (%)d
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fab | Con | Var | Fab | Con | Var | Fab | Con | Var | Fab | Con | Var | |
| 14 | 4.12 | 1.84 | 4.04 | 58.8 | 61.2 | 60.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| AIF | 0.76 | 3.35 | 1.39 | 55.3 | 54.7 | 58.3 | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.1 |
| 14/WAM | 3.35 | 56.4 | 0.0 | 0.0 | ||||||||
| B | 3.10 | 5.26 | 3.21 | 36.7 | 38.9 | 33.2 | 0.0 | 0.0 | 1.8 | 4.2 | 0.2 | 6.6 |
| 6 | 2.21 | 2.50 | 1.76 | 21.7 | 10.1 | 30.4 | 0.0 | 0.0 | 0.0 | 2.1 | 4.6 | 3.8 |
| 8 | 3.22 | 6.44 | 2.98 | 52.7 | 50.6 | 51.2 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 2.1 |
| 15 | 3.66 | 6.80 | 5.28 | 48.9 | 47.8 | 52.1 | 0.0 | 0.0 | 0.0 | 1.6 | 0.4 | 2.9 |
| 16 | 4.56 | 1.70 | 3.43 | 45.7 | 48.5 | 46.0 | 0.0 | 0.0 | 0.0 | 5.6 | 0.5 | 5.8 |
| E | 4.12 | 1.90 | 4.11 | 54.9 | 54.5 | 52.6 | 0.0 | 0.0 | 0.0 | 1.2 | 0.1 | 1.7 |
| F | 3.38 | 4.42 | 5.52 | 50.4 | 50.1 | 53.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 0.2 |
Rcrit = Σcriteria [(x − mean x)/(RMS from mean x)], where x is the value of each criterion.
Sumf is defined as the average value of density for all nonhydrogen atomic positions normalized by setting the highest density in the map to 100.
Clash represents the percentage of atoms in the model that have steric clashes with symmetry-related subunits.
−Den provides the percentage of atoms that are positioned in negative density.
Each Fab was fit in entirety and as separate constant (Con) and variable (Var) domains.
14 is the X-ray crystal structure. AIF is the murine Fab from virus-neutralizing antibody 730.1.4 (PDB accession no. 1AIF), and 14/WAM is the WAM computer-modeled structure of VH and VL based on the sequence of Fab 14.
Since the crystal structure of Fab 14 had been determined, it was compared to the structure of the homology model of Fab 14 obtained from the modeling service using the program HOMOlogy (43, 46, 47) in order to verify the quality of the homology modeling process used for the studies reported here. In addition, the variable and constant domains of the Fab 14 crystal structure were compared with those of another mouse Fab, 1AIF. Fab 1AIF was used for fitting Fab 16, for which the heavy chain amino acid sequence had not been determined (see Fig. 4).
Fitting of Fab 6 had a special problem in that it bound close to the icosahedral threefold axes, causing steric hindrance between threefold axis-related molecules, implying that only one of three Fabs could bind at any one of the symmetry-related sites. Thus, the fitting operation was modified by negating the effect on the fitting criterion of the atoms that were in conflict (“clash”) (Table 3) with the symmetry-related Fab molecules.
The structure of CPV (1C8D) was fitted into the corresponding virus cryoEM density by superimposing the icosahedral symmetry elements. Residues in the virus-Fab interface were identified as those in CPV or FPV that had any atoms less than 5.0 Å from any atom in the fitted Fab structure, using the CCP4 program CONTACT (14). The buried surface area was calculated using a 1.70-Å probe. Among the eight different Fabs, the surface area of contact varied from 1,238 to 2,745 Å2. The surface areas were calculated with the CCP4 programs areaimol and surface (13, 14, 30).
Nucleotide sequence accession numbers.
The sequences of the heavy chains have been deposited in GenBank under accession numbers FJ440693, FJ440688, FJ440690, FJ440695, and FJ440697, for Fabs B, 6, 15, E, and F, respectively. Sequences of the kappa chains have been deposited under accession numbers FJ440694, FJ4440689, FJ440691, FJ440692, FJ440696, and FJ440698, for Fabs B, 6, 15, 16, E, and F, respectively.
CryoEM density map accession numbers.
The EMD accession numbers are 5105, 5106, 5107, 5108, 5109, 5110, 5111, and 5112 for the cryoEM reconstructions of CPV complexed with Fab14 and FPV complexed with Fabs B, 6, 8, 15, 16, E, and F, respectively.
Protein structure accession numbers.
PDB accession numbers are as follows: 3GK8 for the X-ray crystal structure of Fab 14; and 3IY0, 3IY1, 3IY2, 3IY3, 3IY4, 3IY5, 3IY6, and 3IY7 for structures of Fabs 14, B, 6, 8, 15, 1AIF (for Fab 16), E, and F, respectively, fitted into the respective cryoEM reconstructions of the parvovirus-Fab complex.
RESULTS AND DISCUSSION
Crystal structure of Fab 14.
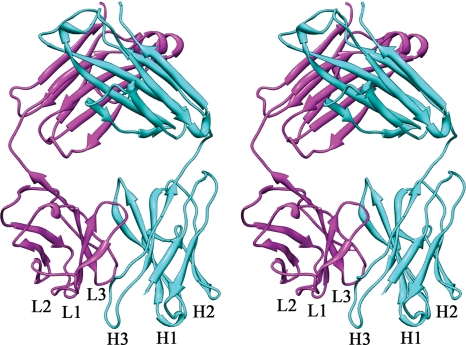
The crystal structure of Fab 14 was determined to 2.0-Å resolution and refined to a final Rwork of 25% (Rfree of 29%) (Table 2). The amino acid sequences of both the light and heavy chains were established by DNA sequencing (see Materials and Methods). All of the residues of the light (residues 1 to 212) and heavy (residues 1 to 220) chains were built into the electron density map. The secondary structure and the domain organization of the variable and constant domains of Fab 14 were typical for fragments of immunoglobulins (Fig. 2). The angle between the two pseudo-dyad rotation axes of the VLVH and CLCH domains, or elbow angle, was 176°, which is within the observed range for Fab fragment elbow angles of 127° to 225° (62). Thus, Fab 14 has an extended, nearly linear overall geometry. A Ramachandran plot of the torsion angles shows that 99% of the residues are within the generously allowed region.
FIG. 2.
The crystal structure of Fab 14 is shown in stereo as a ribbon diagram. The CDRs are labeled for the heavy chain (cyan) and the light chain (magenta).
Fitting of the Fab structures into the cryoEM density.
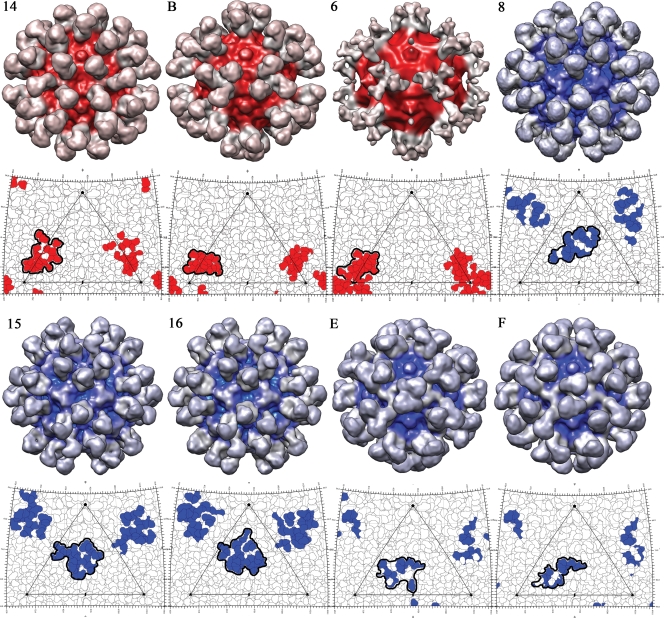
The resolutions of the reconstructions of eight different Fab-virus complexes varied from 8.5 to 18 Å and roughly correlated with the number of particles selected for each reconstruction (Fig. 3; Table 1). In seven of the reconstructions, the cryoEM density corresponding to Fab was approximately equal in magnitude to that of the virion capsid, indicating that most or all of the 60 binding sites had been occupied by a Fab molecule. However, the Fab 6 molecules bound close to the threefold axis of symmetry, such that they sterically interfered with each other, allowing only one of the threefold axis-related sites to be occupied. Thus, the density corresponding to Fab was approximately only one-third of the magnitude of the capsid density, except at the very base of the Fab. This position would always be filled, no matter which of the three possible sites were occupied (7).
FIG. 3.
Surface-rendered cryoEM reconstructions of virus complexed with each of eight fragments from neutralizing antibodies described by Nelson et al. (35). Surfaces are shown at one sigma of the cryoEM density. Density further than a 125-Å radius from the center of the virus is shown in gray. Shown directly below each reconstruction is the footprint of the Fab in the complex. Both complexes and footprints are color coded red for antibodies directed to the A site and blue for antibodies directed to the B site.
Using the known X-ray crystal structures of CPV and FPV (PDB accession no. 1C8D and 1FPV, respectively), the density corresponding to virus was removed from each cryoEM map (see Materials and Methods), producing a Fab difference density map that was used for fitting the Fab structures. For each Fab, the constant domain was fitted first, followed by fitting of the variable domain (Table 3). Although eight different Fab molecules were used in the interpretation of the Fab-virus complexes, only the crystal structure of Fab 14 was known. Comparison of the crystal structure with the homology model provided a measure of the degree of error introduced by the use of homology models. The root mean square deviation (RMSD) between equivalent C-α atoms upon superimposing the crystal structure onto the homology model was 0.8 Å between 123 C-α atoms, with the biggest difference being 5 Å in the heavy chain complementarity determining region (CDR) H3 loop. Three very similar results were obtained for the interpretation of CPV Fab 14 cryoEM density by fitting the variable domains obtained from the crystal structure of Fab 14 or from another mouse Fab structure (PDB accession no. 1AIF) or using the homology model of Fab 14 calculated from the sequence. The RMSD between equivalent C-α atoms was never greater than 1.0 Å between any of the three fitted results, with the biggest deviation being 6.3 Å in the CDR H1 loop. On using the complete Fab 14 crystal structure without any adjustment to the elbow angle, the RMSD between the C-α atoms of the variable domain differed less than 0.7 Å with respect to any of the three independently fitted variable domains, showing that the elbow angle was not affected by Fab binding to the virus or by lattice forces within the Fab 14 crystal.
Atoms located on the virus surface as well as atoms in each Fab molecule were identified if they were within 5.0 Å of the fitted virus and Fab structure. Thus, a footprint for each Fab was identified and plotted, and the Fab contacts were mapped (Fig. 3). Each of the three Fab 14 models produced nearly the same footprint, although the contact residues identified using the homology model more closely matched the footprint obtained with the Fab crystal structure (Table 4). This justified the use of homology models for the interpretation of the other Fab-capsid reconstructions where there was no Fab crystal structure (see Table S1 in the supplemental material).
TABLE 4.
List of virus surface residue contacts identified by fitting the X-ray crystal structure of Fab 14, the computer-generated homology model (WAM), and the murine Fab 730.1.4 (PDB accession no. 1AIF)
| X-ray crystal structure | WAM | Mouse |
|---|---|---|
| Asp88 | Asp88 | Asp88 |
| Met87 | ||
| Lys89 | Lys89 | |
| Ala91 | Ala91 | |
| Asn93 | Asn93 | Asn93 |
| Gly94 | ||
| Asn95 | ||
| His222 | His222 | |
| Thr223 | Thr223 | Thr223 |
| Gly224 | Gly224 | Gly224 |
| Thr225 | Thr225 | |
| Gly227 | Gly227 | |
| Pro229 | ||
| Asp305 | Asp305 | Asp305 |
| Gln309 | Gln309 | |
| Asp311 | ||
| Lys312 | ||
| Tyr324 | ||
| Leu422 | Leu422 | |
| Pro423 | Pro423 | Pro423 |
| Val424 | ||
| Thr425 | Thr425 | Thr425 |
| Asn426 | Asn426 | |
| Asp427 | Asp427 | Asp427 |
| Asn428 |
After mapping of the virus interactions to the Fab sequences, the most notable difference between the locations of contacts among the six different CDR loops was that for Fabs E and F all interactions with the virus mapped to the heavy chain, except for a single contact with CDR on loop L3. The distributions of contacts were nearly equal between the light and heavy chains for all six other Fabs (Fig. 4). The quality of docking into the cryoEM density was estimated by determining the proportion of atoms that were in steric conflict between the virus and the fitted Fab structures (see Table S1 in the supplemental material). Fewer than 6% of the interactions were between atoms less than 1.5 Å apart, and all but one of these clashes were between side chain atoms. The number of interactions between each Fab and the virus was different, as was the percentage of those residues predicted to form hydrogen bonds with the virus (see Table S1 in the supplemental material).
The antigenic surface of the virus.
The three A site and five B site footprints together covered more than 60% of the virus surface, including the threefold spike, which was thought to be the most antigenic surface area. The excluded surface area was comprised mostly of recessed regions on the capsid structure, i.e., a depressed region around the fivefold axis, the “canyon,” and a depression located at the twofold icosahedral symmetry axis, the “dimple” (Fig. 5B). These recessed areas would be less accessible to antibodies, as observed in studies of other protein antigens (3). However, the fully exposed and raised region surrounding the fivefold vertex was also excluded from binding any of the eight antibodies tested here, suggesting that merely exposure on the capsid surface is not a sufficient determinant for antigenicity.
FIG. 5.
The viral surface is shown as a stereographic projection where the polar angles φ and θ represent the latitude and longitude of a point on the viral surface, respectively (63). (A) The residues shared by the Fab footprints are defined as the common A site and common B site and outlined in red and blue, respectively. (B) All of the residues within the antibody footprints define a total antigenic surface outlined in black. The viral surface residues are colored according to the distance from the center of the virus, with red amino acids being the furthest away and blue representing depressions in the viral surface.
The region common to all of the A site antibodies consisted of the three residues previously identified by escape mutant analysis (Asn/Lys 93, His 222, and Gly 224) plus seven additional residues (Thr 223, Thr 225, Pro 423*, Thr 425*, Asn 426*, Asp 427*, and Asn 428*, where an asterisk identifies a neighboring subunit). Similarly, the region common to all of the B site antibodies consisted of the three previously identified residues (Gly 299, Ala/Gly 300, and Asn 302) plus additional residues (Glu 298, Thr 301, and Lys 387*) (Fig. 5A). This grouping of the antibodies into two binding regions is consistent with antibody competition binding experiments (40), epitope mapping (54), and peptide mapping (64).
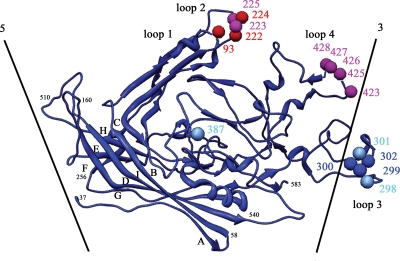
The amino acids of the CPV surface consisted of 42% polar but noncharged, 27% charged, and 32% hydrophobic amino acids. Within the A site antibody footprints, the exposed amino acid residues were 57% polar noncharged, 23% charged, and 20% hydrophobic, and within the B site antibody footprints, the surface residues were 52% polar noncharged, 25% charged, and 23% hydrophobic. Apparently, there is nothing obvious that differentiates residues in the antibody epitopes from other surface amino acids. Nevertheless, neutralizing antibodies bind either to the region common to all the A site antibodies or to the region common to all the B site antibodies, suggesting that these regions have some special property. All of the amino acids in these sites are at the very ends of loops 1, 2, and 4 in site A and at loop 3 as well as the insertion loop between beta strands G and H in site B (Fig. 6). This suggests that the common A and common B site surface regions might tolerate more mutational changes than elsewhere on the viral surface. It might also be relevant that the amino acids defining the A and B sites are at boundaries between subunits. Similar properties have been found for escape mutations to neutralizing antibodies for picornaviruses, such as human rhinovirus 14 (48), where the escape mutations are clustered into four sites at the end of loops and also situated at the boundaries between subunits. The high variability of the surface antigenic sites may also be responsible for variation in host specificity, as indicated by the overlap of the TfR receptor binding site with the antigenic sites and the known effects of some antigenic mutations on the host range for dogs (Fig. 7).
FIG. 6.
Capsid protein of CPV viewed approximately tangentially to the surface of the virus, showing the closest icosahedral five- and threefold symmetry axes. Spheres show the positions of residues previously identified as belonging to the A site (red) and B site (blue). Additional residues found in the footprints of all antibodies directed to the A site are shown in magenta, and residues common to all B site antibodies are shown in cyan. Secondary structural elements, or β strands, are labeled A to I.
FIG. 7.
The Fab footprints for the A site (red X's) and the B site (blue X's) overlap with the TfR binding site, identified in black by amino acid number (19). The antibodies 8, 15, and 16 directed to the B site show the greatest amount of overlap with the receptor binding site.
Specificity of Fab 14 binding.
Residue 93 is located within the common antibody binding region of the A site. The switching of lysine 93 in FPV to an asparagine in CPV is one of two surface changes necessary and sufficient to change the tropism of FPV to that of CPV. Of the three Fab molecules that bind to site A, Fab 14 binds only to CPV, whereas Fab 6 and B bind to both CPV and FPV. Thus, a lysine at position 93 in FPV prevents binding of Fab 14 but does not inhibit binding of the other two Fab molecules. Three hydrogen bonds might form between Fab 14 and CPV, whereas only two hydrogen bonds would be likely to form in an interaction between Fab 14 and FPV. The missing hydrogen bond in the interaction of Fab 14 with FPV is a result of changes in the structures of loop 1 and loop 2 when lysine is substituted for asparagine at residue 93 (17) (Fig. 8).
FIG. 8.
The cryoEM density of the CPV-Fab 14 complex shown as a surface-contoured mesh in a slab view at the region of the threefold spike. The icosahedral threefold axis is indicated by a dark gray axis. The heavy chain (green) and the light chain (gold) of Fab 14 are fitted into the Fab density. Both CPV (blue) and FPV (magenta) have been fitted into the viral density to show the very subtle differences between the two viruses at loops 1 and 2 in the interface of the capsid interactions with the Fab.
Mechanisms of neutralization.
Bivalent binding of an antibody to a virus requires that there is a distance of approximately 25 to 29 Å between the heavy chain C-terminal ends of symmetry-related Fabs (21, 22, 56). For the Fab complexes discussed here, the shortest distance between symmetry-related Fabs varied from 48 Å (Fab 15) to 124 Å (Fab 14) (Table 5). In none of these cases were symmetry-related Fabs close enough or in an orientation to permit bivalent binding of an intact IgG. Thus, although bivalent binding would not be possible, the orientation of the Fabs on the viral surface would allow antibodies to cross-link particles, a likely mechanism of neutralization. In contrast, many rhinovirus antibodies can neutralize virus by bivalent attachment across an adjacent icosahedral twofold axis (21, 48).
TABLE 5.
Distance from fitted Fab C-terminal end of the heavy chain to the C-terminal end of the nearest symmetry-related Fab
| Fab | Distance (Å) to nearest symmetry-related Fab
|
||
|---|---|---|---|
| Twofold | Threefold | Fivefold | |
| 14 | 113.0 | 71.3 | 124.4 |
| B | 51.5 | 81.8 | 107.1 |
| 6 | 82.9 | 54.9 | 132.0 |
| 8 | 91.0 | 142.6 | 62.8 |
| 15 | 48.8 | 161.5 | 52.9 |
| 16 | 112.8 | 157.0 | 57.5 |
| E | 51.8 | 147.8 | 107.3 |
| F | 61.6 | 148.9 | 95.2 |
Fabs B, 6, 8, 14, 15, and 16 bind such that the pseudo-dyad axis of the variable domain is approximately perpendicular (90°) to the virus surface (Fig. 3). However, Fabs E and F attach to the surface of the virus at an oblique angle by which their pseudo-dyad axes make an angle of ∼120°, increasing the area of a radial projection of Fab density onto the virus surface. Furthermore, in the case of both Fabs E and F, the Fab molecules span across twofold icosahedral axes, with the heavy chain residues making almost all of the contact with the virus surface (Fig. 3 and 4).
The cryoEM densities of the virus-Fab complexes were fully occupied by the virus and Fab structures, leaving no uninterpreted density, indicating that Fab binding did not induce any gross conformational changes in the capsid. Although each Fab footprint overlapped with the previously determined footprint of the cellular receptor TfR (Fig. 7) (19), preincubation with soluble Fab has been shown not to completely inhibit subsequent receptor binding (35). Furthermore, Fabs E and F neutralize as Fabs, suggesting that their specific interaction with the surface of the virus was the key component to the mechanism of neutralization. However, the footprints of Fabs E and F overlapped with only 5 of 46 residues in the TfR footprint, whereas the other B site Fabs overlapped with 12 or more residues (Fig. 7). Thus, competition with receptor binding is not likely to be the mechanism of neutralization. However, the occlusion of the additional viral surface area and the contact between twofold axis-related Fabs may be involved in the mechanism of neutralization. Since no escape mutants have been isolated for Fab F, and only one has been found for Fab E (54), the attachment site itself may also have a necessary function for virus infectivity.
Supplementary Material
Acknowledgments
We thank Anthony Rees and Susan Crennell of the WAM antibody modeling service for generating the homology models and Sheryl Kelly for help with the preparation of the manuscript.
This work was supported by grants AI 28385 and AI 33468 from the National Institutes of Health to C.R.P. S.H. was supported by postdoctoral fellowship AI060155 from the National Institutes of Health. M.G.R. and S.H. were supported by award AI 11219 from the National Institutes of Health.
Footnotes
Published ahead of print on 25 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abdelmagid, O. Y., L. Larson, L. Payne, A. Tubbs, T. Wasmoen, and R. Schultz. 2004. Evaluation of the efficacy and duration of immunity of a canine combination vaccine against virulent parvovirus, infectious canine hepatitis virus, and distemper virus experimental challenges. Vet. Ther. 5173-186. [PubMed] [Google Scholar]
- 2.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16155-171. [DOI] [PubMed] [Google Scholar]
- 3.Alzari, P. M., M. B. Lascombe, and R. J. Poljak. 1988. Three-dimensional structure of antibodies. Annu. Rev. Immunol. 6555-580. [DOI] [PubMed] [Google Scholar]
- 4.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 9773-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16673-685. [DOI] [PubMed] [Google Scholar]
- 7.Bowman, V. D., E. S. Chase, A. W. E. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 7612250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brioen, P., D. Dekegel, and A. Boeye. 1983. Neutralization of poliovirus by antibody-mediated polymerization. Virology 127463-468. [DOI] [PubMed] [Google Scholar]
- 9.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 10.Casal, J. I., J. P. Langeveld, E. Cortes, W. W. Schaaper, E. van Dijk, C. Vela, S. Kamstrup, and R. H. Meloen. 1995. Peptide vaccine against canine parvovirus: identification of two neutralization subsites in the N terminus of VP2 and optimization of the amino acid sequence. J. Virol. 697274-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 666858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Che, Z., N. H. Olson, D. Leippe, W. M. Lee, A. G. Mosser, R. R. Rueckert, T. S. Baker, and T. J. Smith. 1998. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryoelectron microscopy and X-ray crystallography of virus-Fab complexes. J. Virol. 724610-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chothia, C. 1975. Structural invariants in protein folding. Nature 254304-308. [DOI] [PubMed] [Google Scholar]
- 14.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 15.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 16.Fry, E. E., S. M. Lea, T. Jackson, J. W. I. Newman, F. M. Ellard, W. E. Blakemore, R. Abu-Ghazaleh, A. Samuel, A. M. Q. King, and D. I. Stuart. 1999. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 18543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govindasamy, L., K. Hueffer, C. R. Parrish, and M. Agbandje-McKenna. 2003. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J. Virol. 7712211-12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafenstein, S., V. D. Bowman, P. R. Chipman, C. M. Bator Kelly, F. Lin, M. E. Medof, and M. G. Rossmann. 2007. Interaction of decay-accelerating factor with coxsackievirus B3. J. Virol. 8112927-12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafenstein, S., L. M. Palermo, V. A. Kostyuchenko, C. Xiao, M. C. Morais, C. D. Nelson, V. D. Bowman, A. J. Battisti, P. R. Chipman, C. R. Parrish, and M. G. Rossmann. 2007. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. USA 1046585-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hangartner, L., R. M. Zinkernagel, and H. Hengartner. 2006. Antiviral antibody responses: the two extremes of a wide spectrum. Nat. Rev. Immunol. 6231-243. [DOI] [PubMed] [Google Scholar]
- 21.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 151515-1523. [PMC free article] [PubMed] [Google Scholar]
- 22.Hewat, E. A., T. C. Marlovits, and D. Blaas. 1998. Structure of a neutralizing antibody bound monovalently to human rhinovirus 2. J. Virol. 724396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewat, E. A., N. Verdaguer, I. Fita, W. Blakemore, S. Brookes, A. King, J. Newman, E. Domingo, M. G. Mateu, and D. I. Stuart. 1997. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 161492-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoelzer, K., L. A. Shackelton, C. R. Parrish, and E. C. Holmes. 2008. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J. Gen. Virol. 892280-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. R. Parrish. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 7710099-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 771718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katpally, U., C. E. Wobus, K. Dryden, H. W. Virgin IV, and T. J. Smith. 2008. Structure of antibody-neutralized murine norovirus and unexpected differences from virus-like particles. J. Virol. 822079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langeveld, J. P., J. I. Casal, E. Cortes, G. van de Wetering, R. S. Boshuizen, W. M. Schaaper, K. Dalsgaard, and R. H. Meloen. 1994. Effective induction of neutralizing antibodies with the amino terminus of VP2 of canine parvovirus as a synthetic peptide. Vaccine 121473-1480. [DOI] [PubMed] [Google Scholar]
- 29.Langeveld, J. P., J. I. Casal, C. Vela, K. Dalsgaard, S. H. Smale, W. C. Puijk, and R. H. Meloen. 1993. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J. Virol. 67765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, B., and F. M. Richards. 1971. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55379-400. [DOI] [PubMed] [Google Scholar]
- 31.Liang, X. 2008. CXCR4, inhibitors and mechanisms of action. Chem. Biol. Drug Des. 7297-110. [DOI] [PubMed] [Google Scholar]
- 32.Llamas-Saiz, A. L., M. Agbandje-McKenna, J. S. Parker, A. T. Wahid, C. R. Parrish, and M. G. Rossmann. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 22565-71. [DOI] [PubMed] [Google Scholar]
- 33.Lok, S. M., V. Kostyuchenko, G. E. Nybakken, H. A. Holdaway, A. J. Battisti, S. Sukupolvi-Petty, D. Sedlak, D. H. Fremont, P. R. Chipman, J. T. Roehrig, M. S. Diamond, R. J. Kuhn, and M. G. Rossmann. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15312-317. [DOI] [PubMed] [Google Scholar]
- 34.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 9914758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson, C. D., L. M. Palermo, S. L. Hafenstein, and C. R. Parrish. 2007. Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the Fab fragments of monoclonal antibodies. Virology 361283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276307-326. [DOI] [PubMed] [Google Scholar]
- 37.Palermo, L. M., S. L. Hafenstein, and C. R. Parrish. 2006. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 808482-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker, J. S. L., and C. R. Parrish. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 719214-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parrish, C. R. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183195-205. [DOI] [PubMed] [Google Scholar]
- 40.Parrish, C. R., and L. E. Carmichael. 1983. Antigenic structure and variation of canine parvovirus type-2, feline panleukopenia virus, and mink enteritis virus. Virology 129401-414. [DOI] [PubMed] [Google Scholar]
- 41.Parrish, C. R., L. E. Carmichael, and D. F. Antczak. 1982. Antigenic relationships between canine parvovirus type 2, feline panleukopenia virus and mink enteritis virus using conventional antisera and monoclonal antibodies. Arch. Virol. 72267-278. [DOI] [PubMed] [Google Scholar]
- 42.Pollock, R. V., and L. E. Carmichael. 1982. Maternally derived immunity to canine parvovirus infection: transfer, decline, and interference with vaccination. J. Am. Vet. Med. Assoc. 18037-42. [PubMed] [Google Scholar]
- 43.Rao, S. T., and M. G. Rossmann. 1973. Comparison of super-secondary structures in proteins. J. Mol. Biol. 76241-256. [DOI] [PubMed] [Google Scholar]
- 44.Rimmelzwaan, G. F., J. Carlson, F. G. C. M. UytdeHaag, and A. D. M. E. Osterhaus. 1990. A synthetic peptide derived from the amino acid sequence of canine parvovirus structural proteins which defines a B cell epitope and elicits antiviral antibody in BALB c mice. J. Gen. Virol. 712741-2745. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann, M. G. 2000. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D 561341-1349. [DOI] [PubMed] [Google Scholar]
- 46.Rossmann, M. G., and P. Argos. 1975. A comparison of the heme binding pocket in globins and cytochrome b5. J. Biol. Chem. 2507525-7532. [PubMed] [Google Scholar]
- 47.Rossmann, M. G., and P. Argos. 1976. Exploring structural homology of proteins. J. Mol. Biol. 10575-95. [DOI] [PubMed] [Google Scholar]
- 48.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, et al. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317145-153. [DOI] [PubMed] [Google Scholar]
- 49.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136190-200. [DOI] [PubMed] [Google Scholar]
- 50.Rossmann, M. G., Y. He, and R. J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10324-331. [DOI] [PubMed] [Google Scholar]
- 51.Rossmann, M. G., and J. E. Johnson. 1989. Icosahedral RNA virus structure. Annu. Rev. Biochem. 58533-573. [DOI] [PubMed] [Google Scholar]
- 52.Rossmann, M. G., and A. C. Palmenberg. 1988. Conservation of the putative receptor attachment site in picornaviruses. Virology 164373-382. [DOI] [PubMed] [Google Scholar]
- 53.Smith, T. J., N. H. Olson, R. H. Cheng, E. S. Chase, and T. S. Baker. 1993. Structure of a human rhinovirus-bivalently bound antibody complex: implications for viral neutralization and antibody flexibility. Proc. Natl. Acad. Sci. USA 907015-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strassheim, M. L., A. Gruenberg, P. Veijalainen, J. Y. Sgro, and C. R. Parrish. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198175-184. [DOI] [PubMed] [Google Scholar]
- 55.Taniguchi, K., Y. Morita, T. Urasawa, and S. Urasawa. 1987. Cross-reactive neutralization epitopes on VP3 of human rotavirus: analysis with monoclonal antibodies and antigenic variants. J. Virol. 611726-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thouvenin, E., S. Laurent, M. F. Madelaine, D. Rasschaert, J. F. Vautherot, and E. A. Hewat. 1997. Bivalent binding of a neutralising antibody to a calicivirus involves the torsional flexibility of the antibody hinge. J. Mol. Biol. 270238-246. [DOI] [PubMed] [Google Scholar]
- 57.Truyen, U., A. Gruenberg, S. F. Chang, B. Obermaier, P. Veijalainen, and C. R. Parrish. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 694702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, and C. R. Parrish. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 2511456-1464. [DOI] [PubMed] [Google Scholar]
- 59.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 301022-1025. [Google Scholar]
- 60.Whitelegg, N. R., and A. R. Rees. 2000. WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng. 13819-824. [DOI] [PubMed] [Google Scholar]
- 61.Wikoff, W. R., G. Wang, C. R. Parrish, R. H. Cheng, M. L. Strassheim, T. S. Baker, and M. G. Rossmann. 1994. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure 2595-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson, I. A., and R. L. Stanfield. 1994. Antibody-antigen interactions: new structures and new conformational changes. Curr. Opin. Struct. Biol. 4857-867. [DOI] [PubMed] [Google Scholar]
- 63.Xiao, C., and M. G. Rossmann. 2007. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 158182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, W., and C. R. Parrish. 2000. Comparison of two single-chain antibodies that neutralize canine parvovirus: analysis of an antibody-combining site and mechanisms of neutralization. Virology 269471-480. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 222604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.