Abstract
During cell culture isolation experiments to recover Dobrava hantavirus from a suspension of liver from a striped field mouse (Apodemus agrarius), an unknown virus was coisolated. Atypically for hantaviruses, it had extensive cytopathic effects. Using a random PCR approach, it was identified as a novel murine adenovirus, MAdV-3 (for MAdV type 3). A plaque-purified virus clone was prepared and further characterized. The complete genome sequence of MAdV-3 was determined to be 30,570 bp in length. Sequence comparisons to other adenovirus species revealed highest similarity to MAdV-1, the representative of the murine adenovirus A species. However, substantial differences were found in the E1, E3, and E4 genomic regions. The phylogenetic distance of MAdV-3 amino acid sequences for pVIII, protease, polymerase, and hexon from MAdV-1 is markedly higher than 0.1 exchange per position, and, based on our cross-neutralization experiments, MAdV-3 and MAdV-1 can be regarded as different serotypes. Therefore, we propose to classify MAdV-3 as the first isolate of a novel adenovirus species, designated murine adenovirus C (MAdV-C). The novel MAdV-3 virus is not only genetically and serologically distinct from MAdV-1 but also shows a unique organ tropism in infected mice. In contrast to MAdV-1, the virus was not detectable in brain but predominantly infected heart tissue. Thus, infection of mice with cardiotropic MAdV-3 might be an interesting animal model of adenovirus-induced myocarditis.
Adenoviruses (AdVs) belong to the family Adenoviridae and are nonenveloped, icosahedral viruses with a wide range of hosts from all main vertebrate groups. AdVs have linear, double-stranded DNA genomes 26 to 45 kbp in size. The genomes are characterized by an inverted terminal repeat (ITR) ranging in size from 36 to over 200 bp, and the 5′ ends are linked to a terminal protein. In phylogenetic analyses, AdVs can be divided into five major clades which correspond to one putative and four recognized virus genera. AdVs of mammals form the genus Mastadenovirus (8).
Two AdVs have been so far isolated from mice: FL (14) and K87 (15, 29), which have meanwhile been designated as MAdV-1 (for murine AdV type 1) and MAdV-2 and represent distinct species MAdV-A and MAdV-B, respectively. MAdV-2 grows very inefficiently in cell cultures (13), and experimental infection of mice leads to diarrhea (29). Sequence information on MAdV-2 is limited to short partial sequence encoding the structural hexon protein (GenBank accession no. AY618467).
In contrast, the complete genome sequence of MAdV-1 is available (GenBank accession no. NC_000942) and an infectious clone of the virus has been constructed (21). Experimental infection with MAdV-1 causes encephalomyelitis in newborn or immunodeficient mice as well as in several strains of adult mice (11, 16, 31). In accordance with the clinical symptoms, the virus load is high in the central nervous system but very limited in lung and spleen (11). In addition, endocarditis and adrenalitis after MAdV-1 infection have also been reported where endothelial cells are the main target of the virus.
Human AdVs often cause respiratory diseases: less frequent complications are conjunctivitis, diarrhea, hemorrhagic cystitis, and meningoencephalitis. Moreover, there are studies in which human AdVs are discussed as putative etiological agents of myocarditis and cardiac dysfunction (9, 19, 24). However, due to the high host specificity of AdVs, no animal models are currently available for studies on the pathogenesis and persistence of human AdVs. Novel AdVs of murine rodents might allow the filling of this gap. In addition, nonhuman AdVs could be used as vectors for human gene therapy and vaccination; advantages would be the absence of preexisting antibodies to nonhuman AdVs in the human population as well as the possibility to test the safety of the vectors in relevant animal models.
Here we report on the genetic, serological, and biological characterization of a novel MAdV accidentally coisolated from liver tissue of a striped field mouse (Apodemus agrarius) during hantavirus isolation attempts (18). The novel virus is not only genetically and serologically different from the yet known MAdVs but also shows different organ tropism and genome organization.
MATERIALS AND METHODS
Virus isolation.
Liver tissue of one A. agrarius animal was used for attempts to isolate Dobrava hantavirus (18). A 10% tissue suspension was prepared in Dulbecco's medium supplemented with 0.2% bovine serum albumin. The tissues were triturated in a closed mechanical blender FastPrep Instrument (BIO 101 Systems, Carlsbad, CA). Triturated tissues were briefly centrifuged at low speed to remove larger tissue fragments and inoculated (0.4 ml/flask) onto cultures of confluent Vero E6 cells in 25-cm2 flasks. Virus was then allowed to adsorb at 37°C. The cell culture medium (minimal essential medium plus 10% fetal calf serum, l-glutamate, penicillin, and streptomycin) was changed for the first time after 90 min and then weekly. At 2- to 3-week intervals, cells, detached by trypsin treatment, were passaged into new culture flasks with the addition of the same amount of fresh uninfected cells according to the protocol described for isolation of hantaviruses (20).
Plaque purification and detergent treatment.
For plaque purification, 10-fold serial dilutions of viral stock were inoculated into six-well plates with a nearly confluent monolayer of Vero E6 cells. After an adsorption period for 1 h at 37°C, the cells were overlaid with a mixture of 1% agarose and Eagle's basal medium containing neutral red (0.1%) (Biochrom KG, Berlin, Germany). Plates were then incubated for 10 days. Virus picked from visible plaques by a micropipette tip was then transferred on fresh uninfected cells and further passaged. To inactivate coisolated hantavirus (enveloped virus), infected cells were incubated with medium containing the detergent Triton X-100. Different concentrations of Triton X-100 have been tested (0.0125% to 10%). Only the lowest concentration, 0.0125%, was not toxic for the cells but inactivated the coisolated hantavirus.
PAN-PCR.
In a first approach, cloning of subgenomic fragments from the unknown virus was done by particle-associated nucleic acid-PCR (PAN-PCR) as described before (28). Briefly, isolation of particles was done by centrifuging supernatant at 4,000 × g for 30 min, passing it through a 0.22-μm-pore sterile filter, and further purifying it by ultracentrifugation through a 30% sucrose-phosphate-buffered saline (PBS) cushion for 3 h at 30,000 rpm. Resuspended pellets were treated with DNase before isolation of PANs, which were further used in a random PCR amplification. In a second attempt, we substituted for the random amplification of PAN-PCR with an adapter ligation step and subsequent PCR amplification following a modified protocol described previously (1). In detail, purified DNA was digested by MseI and subsequently precipitated by ethanol in the presence of 1 μg glycogen. Adapter ligation was done for 1 h at 4°C and 6 h at 16°C in a total volume of 10 μl containing the MseI-digested DNA, 400 U T4 DNA ligase and T4 DNA ligase buffer (1×, both New England Biolabs), and 20 pmol adapter composed of the hybridized oligonucleotides NBam24 (AGGCAACTGTGCTATCCGAGGGAG) and NCsp11 (TACTCCCTCGG). Two microliters of ligation mixture was used for PCR amplification in a total volume of 25 μl containing 1.25 U Platinum Taq polymerase (Invitrogen), 0.2 mM each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, and the primer NBam24 (1 mM). After an initial denaturation step of 95°C for 2 min, 30 cycles of 95°C for 30 s and 72°C for 2 min were done followed by 10 min at 72°C. Products were visualized on an agarose gel and used directly for construction of a library cloned into pCRII (Invitrogen). In both experiments, colonies were picked and tested directly by PCR as described previously (28). Different PCR products were sequenced and analyzed for homologies to viral sequences by a nucleotide-nucleotide (BLAST) and a translated BLAST search (BLASTx) at the NCBI website.
Quantification of the novel AdV in supernatants from cell culture and original tissue samples by qPCR.
For quantification of virus in supernatants and animal tissues, we established SYBR green-based real-time PCRs for two of the generated clones. One set of primers was generated to amplify a sequence located within the pV gene of the novel virus: BBKVD01s1 (AATGCAACTCCGTCCTTTGTG) and BBKVD01a1 (TCAACGCCTATCCTATTCCCA). A second set was located within the open reading frame (ORF) for DNA binding protein: BBKVD06s1 (CTGTTACAGCAAAAGATGGTTCC) and BBKVD06a1 (GGACACATCCAAATCTACATGGT). The sensitivity of both PCRs was five copies/reaction. No unspecific SYBR green-positive PCR products were obtained even in the presence of 100 ng of genomic carrier DNA. For isolation of viral DNA from cell culture supernatants, 100 μl clarified supernatant was expanded to 200 μl with PBS. To ensure that only DNA from viral particles is quantified by PCR, a DNase digestion step was performed by addition of 1 μl MgCl2 (1 M) and 0.25 μl DNase I (10 mg/ml) and incubation at 37°C for 30 min. Release and purification of packaged viral DNA from the virus particles were done by means of the QiaAmp DNA blood minikit (Qiagen), as recommended by the manufacturer, and 1 μl was taken for quantitative PCR (qPCR). We assume that DNase treatment degraded viral DNA which was not protected by the virus particle. Therefore, we use the term “particle-associated genome equivalent” (pgeq) to express the number of detected DNA molecules in qPCR. If DNase treatment was not done, the detected DNA molecules are designated as genome equivalents (geq).
For purification of viral genomes from animal tissue of the original mouse, we used the QiaAmp DNA minikit (Qiagen) as recommended by the manufacturer. Five microliters (out of 100 μl) of the eluted DNA was taken for qPCR, which was performed on a Rotor-Gene 3000 (Corbett Research) using the QuantiTect SYBR green PCR kit (Qiagen) in a total volume of 20 μl with 4 pmol of each sense and antisense primer, respectively. After an initial activation of the polymerase for 15 min at 95°C, 40 cycles of 95°C (30 s), 55°C (30 s), and 72°C (40 s) and acquisition of the SYBR green fluorescence at 78°C for 5 s were done.
Sequencing of the viral genome.
We sequenced both strands of overlapping PCR fragments from regions located between the subgenomic fragments generated by PAN-PCR using the Accu Prime high-fidelity Taq polymerase (Invitrogen). PCR primers were designed to ensure a sequence overlap of more than 200 bp between adjacent PCR products. For generation of PCR products between the ITRs and the most outward-located PAN-PCR fragments, we used one primer located within that fragment and a second one derived from the ITR sequence of MAdV-1 under low-stringency conditions (MAV-1 ITR1, ATACAGTTAGCAAAAAATGGCG). All PCR products were sequenced by primer walking in both directions.
Furthermore to get the exact sequence of genomic ends, we directly sequenced the viral DNA. For this purpose we concentrated 20 ml of cell culture supernatant by polyethylene glycol precipitation (21) and subsequent ultracentrifugation through a 30% sucrose cushion to a volume of 200 μl and extracted DNA by means of the DNA blood minikit (Qiagen). From this highly concentrated viral DNA, we used about 1010 genomic copies for each sequencing reaction. For sequencing of the 5′-ITR, we used three primers, and for sequencing of the 3′-ITR, we used two different primers (data not shown) complementary to regions adjacent to the ITRs of each genomic end, respectively. All sequences were assembled with the software Contig Express from the Vector NTI 6 Suite (InforMax/Invitrogen).
ORF prediction and determination of splice sites.
Several online bioinformatics tools were used for ORF prediction. First, a translated BLAST search (BLASTx at http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was used to identify present ORFs. The following additional online tools were then used for precise predictions of ORFs and putative splicing sites: SpliceView (http://l25.itba.mi.cnr.it/∼webgene/wwwspliceview.html), GenomeScan (http://genes.mit.edu/genomescan/), and GeneMark (http://opal.biology.gatech.edu/GeneMark/). Presented predictions represent a consensus of results obtained from these methods and a manual inspection of the sequence.
To verify splice sites predicted by the bioinformatics approach and eventually find additional splice sites, we sequenced cDNAs generated from cellular transcripts. Total RNA was purifiedwith the RNeasy kit (Qiagen) from cells 4, 6, 10, 12, 16, 20, 34, and 36 h after infection with MAdV-3. Residual DNA was degraded by RNase-free DNase (Ambion). Nine microliters of RNA solution was used for cDNA synthesis with the Thermoscript reverse transcriptase system (Invitrogen) and a 24-mer primer with random sequence at 55°C for 1 h. PCR amplification was done by different sets of primers located within predicted exons of E1A, E1B, terminal protein, and polymerase (data not shown). PCR products shorter than those expected from unspliced mRNA were extracted from an agarose gel and sequenced directly with the primers used for PCR.
Phylogenetic analysis.
Amino acid sequences were aligned using CLUSTALW (30). Maximum likelihood (ML) trees were calculated using TREE-PUZZLE 5.2 (25). PHYLIP 3.67 software package (10) was used to calculate neighbor-joining (NJ) and Fitch-Margoliash (FM) phylogenetic trees. The Dayhoff PAM (PHYLIP, TREE-PUZZLE) and VT (TREE-PUZZLE) evolutionary models were used for phylogenetic distance calculations. Bootstrap analysis with 10,000 replicates was performed to evaluate the statistical significance of the topology for the NJ and FM trees.
Generation of antisera.
Antisera against MAdV-1 and MAdV-3 for cross-neutralization assays were produced with two C57B/6 mice for each virus. Prime immunization occurred by intravenous injection of crude supernatant from cell culture diluted in sterile PBS (about 108 pgeq as quantified by qPCR). To avoid intense immune responses to components of the producer cell line (Vero E6), boost immunization after 21 days was done by subcutaneous footpad application of heat-inactivated particles (3 × 109 pgeq) purified by ultracentrifugation through a 30% sucrose cushion. Seven days after boosting, animals were sacrificed and sera were collected and pooled for each virus.
Neutralization assays.
Antisera against MAdV-1 and -3 were tested in neutralization assays against both viruses. For MAdV-3, a plaque reduction neutralization test (PRNT) was performed. Mouse antisera were first diluted serially in twofold steps starting from 1:20, mixed with an equal volume containing 30 to 80 PFU, incubated for 1 h at 37°C, and then inoculated into six-well plates with a nearly confluent monolayer of Vero E6 cells. After an adsorption period for 1 h at 37°C, the cells were overlaid with a mixture of 1% agarose and Eagle's basal medium. After 7 days of incubation, cells were fixed by methanol and stained using crystal violet dye to improve the visibility of the virus plaques. An at least 80% reduction in the number of plaques was considered as the criterion for virus neutralization. However, MAdV-1 did not plaque on NIH 3T3 cells. As an alternative, an “immunofluorescence-focus reduction neutralization assay” was developed in which infected cells were visualized by immunofluorescence staining using the Imagen AdV kit (Oxoid Ltd.). The test was performed analogously to the MAdV-3 PRNT, with the only differences being that 24-well plates with NIH 3T3 cells and about 20 focus-forming units of the virus per well were used.
Organ tropism of MAdV-1 and MAdV-3.
For both viruses, six female C57BL/6N mice (Janvier, Le Genest-St.-Isle, France) were infected by intravenous injection of about 108 pgeq into one of the lateral tail veins. Quantification of viral loads in cell culture supernatants was done by qPCR as described above, followed by dilution to the desired concentration in physiological NaCl solution to a volume of 100 μl. The used virus dose of 108 pgeq corresponds to 1.5 × 105 50% tissue culture infectious dose (TCID50) units of MAdV-1 and 4 × 105 TCID50 units of MAdV-3, as determined on A549 and Vero E6 cells, respectively. As a negative control, the same amount of heat-inactivated particles (2 h at 65°C) was applied in one mouse for each virus, respectively. After 1 week, animals were sacrificed and organs were taken. To monitor clinical signs of illness, we determined the animal's body weight on days 0, 1, 3, 5, and 7 after infection. Purification of DNA was done by means of the QIAamp DNA minikit as recommended by the manufacturer (Qiagen). Quantification of viral loads in organs was done by qPCR as described above using 5 μl of purified mouse DNA.
Nucleotide sequence accession numbers.
The complete genome sequence of MAdV-3 has been deposited into GenBank under accession no. EU835513. The GenBank accession numbers for the AdV sequences used in this study for comparisons and phylogenetic analysis are as follows: complete genomes of HAdV-12, X73487; HAdV-11, AY163756; SAdV-21, AR101858; HAdV-2, J01917; HAdV-5, M73260; HAdV-17, AF108105; SAdV-25, AR101859; HAdV-40, L19443; BAdV-2, AF252854; PAdV-5, AF289262; BAdV-3, AF030154; CAdV-1, Y07760; CAdV-2, U77082; PAdV-3, AF083132; MAdV-1, NC_000942; TSAdV-1, AF258784; DAdV-1, Y09598; OAdV-7, U40839; BAdV-4, AF036092; additional hexon amino acid sequences of HAdV-1, AP_000512; HAdV-6, AAR26226; HAdV-3, P36849; HAdV-7, AP_000548; HAdV-21, AAR26233; HAdV-35, AP_000585; HAdV-9, P36853; HAdV-19, ABA00002; HAdV-37, ABA00016; HAdV-48, AAB17439; HAdV-4, AAD03654; HAdV-41, CAA36079; and EAdV-2, AAB88060.
RESULTS
Virus isolation and identification by PAN-PCR.
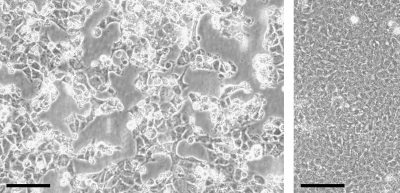
Liver tissue of a hantavirus seropositive and reverse transcription-PCR (RT-PCR)-positive A. agrarius mouse was successfully used to isolate Dobrava hantavirus (18). However, obvious plaques were seen that are atypical of hantaviruses. Cytopathic effects leading to massive cell death occurred after further passaging of the infected cells, showing that the infectious agent was enriched over the passages (Fig. 1). To identify the causative agent, PANs were isolated from cell culture supernatant (from the seventh cell culture passage since the start of the isolation attempt) and amplified by random PCR, a procedure previously termed “PAN-PCR” (28).
FIG. 1.
Cytopathic effect caused by MAdV-3 in Vero E6 cells. (Left panel) Cells 4 days after infection with MAdV-3; (right panel) uninfected cells after 4 days of incubation. The scale bar indicates 100 μm.
A total of 32 cloned DNA fragments prepared by PAN-PCR were sequenced and analyzed by BLAST search. Nine of them showed no similarity to known sequences in a nucleotide/nucleotide search (blastn) but showed significant similarities to AdV sequences when using a translated BLAST search (blastx). These sequences represented four different subgenomic fragments of the putative novel virus. An additional PAN-PCR assay was then performed using a modified protocol in which 14 out of 27 cloned fragments showed similarities to 11 different AdV subgenomic loci. In each case, the highest similarity scores were found to MAdV-1 and ranged from 47 to 98%, whereas identity scores were in the range of 34 to 96%.
The remaining sequences did not show any similarity to viral sequences. Few similarities were found to bacterial and human sequences, whereas the remaining ones could not be identified at all.
To prepare a primary virus stock of the putative novel AdV, a virus clone (no. 2) was prepared by plaque purification. Unfortunately, even after plaque purification, the culture medium was still hantavirus positive by RT-PCR. Therefore, clone 2 was then treated with the detergent Triton X-100 (0.0125%) to inactivate the present Dobrava hantavirus. To prove that the virus stock does not contain residual infectious hantavirus, it was further passaged four times but remained hantavirus negative in nested RT-PCR and the immunofluorescence test (data not shown).
Source of MAdV-3.
To ensure that the recovered AdV is not a laboratory contamination but really originates from the mouse liver tissue, qPCR was used to trace back the virus in cell culture supernatants from different stages of the isolation procedure as well as in tissues of the original donor mouse (A. agrarius). Supernatant obtained 7 days after inoculation with the liver suspension showed a load of ∼4 × 104 pgeq per ml, which rose to ∼1 × 108 pgeq/ml 32 days after inoculation. We could also detect viral genomes in stored liver and lung tissue from the original donor mouse and found ∼54 geq per μg DNA in lung and ∼15 geq/μg DNA in liver tissue. Typical viral loads of current supernatants of infected Vero E6 cells after 7 days of incubation are about 109 pgeq/ml.
Genomic organization.
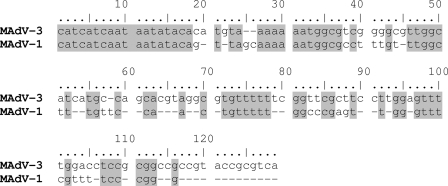
The complete genomic sequence of MAdV-3 was determined to be 30,570 bp in length and to have a GC content of 47.21%, which is very similar to that of MAdV-1 (47.78%). Most of the sequence was determined by multiple bidirectional sequencing of overlapping subgenomic PCR fragments. The ends of the double-stranded DNA linear genome were determined by direct sequencing of the purified viral DNA. ITRs at both ends were found to be 126 bp in size and to contain the consensus motif CATCATCAAT but differ substantially from the 93-bp ITR sequence of MAdV-1 (Fig. 2).
FIG. 2.
Pairwise alignment of ITR sequences of MAdV-3 and MAdV-1. Identical nucleotides are shaded.
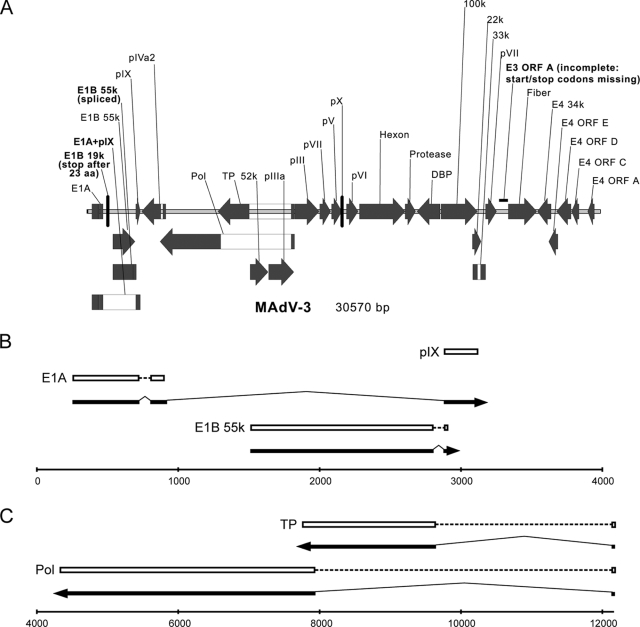
Analysis by a combination of bioinformatic approaches showed that the basic genomic organization is rather similar to that of MAdV-1 (Fig. 3 and Table 1). In contrast to other mastadenoviruses, but similar to MAdV-1 (22), we found an almost completely deleted E3 region, which in other mastadenoviruses encodes proteins with immune regulatory functions. Most ORFs of MAdV-1 were also completely present within the genome of the novel virus. However, the ORF for putative E3 glycoprotein (designated as E3 ORFA in MAdV-1 genome annotation AC_000012) of the novel virus lacked the correct start and stop codons. This seems to be in agreement with findings of Beard et al. (5) indicating that only part of this ORF is an exon in MAdV-1 (start and stop codons are therefore irrelevant) belonging to the E3 class 3 unique region. Nevertheless, this finding for the novel virus is only based on sequence analysis and needs confirmation by transcription mapping and cDNA sequencing. Moreover, ORFs designated in NC_00942 as “MAdVAgp27” encoding “tropoelastin fragment” (designated as E4 ORFB in genome annotation AC_000012), “MAdVAgp26,” and “30kD” seem to be absent in the novel virus: at least, it was not possible to identify them by any of the bioinformatic approaches. In the case of E1B 21k, we repeatedly found a stop codon 23 amino acids (aa) downstream of the predicted start codon.
FIG. 3.
(A) Genomic organization of MAdV-3. Filled arrows and boxes, ORFs; open boxes, introns. Major differences between MAdV-1 and MAdV-3 are highlighted in boldface. (B and C) Splicing patterns confirmed by sequencing. The line at the bottom represents the DNA genome, numbered according to GenBank entry EU835513; arrows indicate direction of transcription/translation; dashed lines indicate continuous ORFs located on distinct exons; and bold lines indicate exons that were sequenced.
TABLE 1.
ORF predictions for MAdV-3 in comparison with MAdV-1a
| Gene product or ORF | MAdV-1
|
MAdV-3
|
% Amino acid similarityb | % Amino acid identityb | Note | ||
|---|---|---|---|---|---|---|---|
| Nucleotide positiona | Length (aa) | Nucleotide positiona | Length (aa) | ||||
| E1A | join(281..752,829..951,2885..2892) | 200 | join(255..720,807..916,2878..3138) | 187 | 59.4 | 46.0 | Confirmed by sequencing |
| join(281..752,829..1118)c | 253 | join(255..720,807..904) | 187 | 47.5 | 36.1 | Confirmed by sequencing | |
| E1B 21k (19k)c | 1127..1654 | 175 | 1124..1648 | 23 | 65.0 | 51.4 | Stop codon after 23 aa |
| E1B 55k | join(1522..2815,2885..2892) | 433 | join(1516..2806,2878..2885) | 432 | 78.6 | 66.2 | Confirmed by sequencing |
| 1522..2865c | 448 | 1516..2814 | 432 | 75.9 | 64.1 | ||
| Early E1A | 2897..3175 | 92 | 2878..3138 | 87 | 50.5 | 41.4 | |
| pIXc | 2885..3175 | 97 | 2878..3138 | 87 | 51.5 | 42.4 | |
| pIVa2 | complement(3254..>4333) | 359 | complement(join(3205..4345,4649..4659)) | 384 | 84.6 | 75.9 | |
| complement(join(3254..4563,4878..4890))c | 441 | 78.2 | 69.8 | ||||
| Pol | complement(join(4372..8004,12387..12398))c | 1215 | complement(join(4323..7919,12302..12313)) | 1203 | 83.0 | 71.8 | Confirmed by sequencing |
| TP | complement(join(7833..9731,12387..12398))c | 637 | complement(join(7751..9622,12302..12313)) | 628 | 82.4 | 69.3 | Confirmed by sequencing |
| 52k | 9775..10854 | 360 | 9677..10756 | 360 | 90.5 | 84.1 | |
| pIIIa | 10885..12369 | 495 | 10787..12283 | 499 | 87.4 | 75.8 | |
| pIII | 12420..13889 | 490 | 12335..13807 | 491 | 89.2 | 80.7 | |
| pVII | 13986..14585 | 200 | 13847..14476 | 210 | 53.7 | 45.5 | |
| 30kD | complement(14457..15245) | 262 | Not found in MAdV-3 | ||||
| pV | 14625..15311 | 229 | 14522..15139 | 206 | 65.1 | 52.3 | |
| pX | 15347..15571 | 75 | 15171..15395 | 75 | 89.2 | 865 | |
| pVI | 15632..16345 | 238 | 15436..16116 | 227 | 80.8 | 72.9 | |
| Hexon | 16432..19161 | 910 | 16193..18922 | 910 | 92.5 | 86.6 | |
| Protease | 19166..19780 | 205 | 18925..19536 | 204 | 84.4 | 72.2 | |
| DBP | complement(19870..21255)c | 462 | complement(19622..21007) | 462 | 78.0 | 70.1 | |
| 100k | 21296..23527 | 744 | 21065..23230 | 722 | 76.5 | 66.0 | |
| 33k | join(23244..23523,23738..24039)c | 194 | join(22956..23226,23426..23715) | 187 | 66.5 | 53.6 | |
| 22k | 23244..23756c | 171 | 22926..23444 | 173 | 58.3 | 45.0 | |
| pVIII | 24047..24694 | 216 | 23724..24380 | 219 | 84.4 | 69.7 | |
| Putative E3glycoprotein (E3 ORFA)c | 24864..25349 | 162 | Not found in MAdV-3 | ||||
| Fiber | 25412..27253 | 614 | 25066..26769 | 568 | 73.5 | 59.6 | |
| E4 34kc | complement(27337..28137)c | 267 | complement(26811..27608) | 266 | 74.0 | 59.1 | |
| MAdVAgp26 | complement(join(27337..28176,29248..29277)) | 289 | Not found in MAdV-3 | ||||
| E4 ORFE | complement(27995..28501)c | 169 | complement(27478..28014) | 179 | 59.8 | 49.7 | |
| E4 ORFD | complement(28431..29210)c | 260 | complement(27950..28804) | 285 | 58.4 | 43.4 | |
| E4 ORFC | complement(29274..29711)c | 146 | complement(28850..29281) | 144 | 71.7 | 63.4 | |
| Tropoelastin fragment (E4 ORFB)c | complement(29726..30076) | 117 | Not found in MAdV-3 | ||||
| E4 ORFA | complement(30066..30464)c | 133 | complement(29805..30194) | 130 | 42.6 | 31.6 | |
Given in GenBank format (http://www.ebi.ac.uk/embl/Documentation/FT_definitions/feature_table.html).
Similarity and identity values were calculated using the Bioedit software package (12).
Bioinformatic predictions of genome annotation AC_00012 for those ORFs where positions in MAdV-1 genome annotation NC_000942 were not given exactly, were missing, or less resembled the predictions for MAdV-3.
Differences were found also in lengths of the predicted proteins. The most pronounced dissimilarities were found in the cases of fiber (difference of 46 aa), pV (23 aa), and 100k (22 aa), where the MAdV-3 proteins were predicted to be shorter than those of MAdV-1. On the other hand, some proteins such as E4 ORFD (difference of 25 aa), pIVa2 (25 aa), or pVII (10 aa) were predicted to be longer in MAdV-3 (Table 1). Large variations were found also in similarity and identity values of the predicted proteins, which varied between 42.6 and 92.5% and 31.6 and 86.6%, respectively. The highest similarity and identity values were found for the hexon (92.5% and 86.6%), 52k (90.5%, 84.1%), and pX (89.2%, 86.5%) proteins. The lowest similarity and identity values were found for the E4 ORFA (42.6%, 31.6%), E1A (47.5%, 36.1%), and pIX (51.5%, 42.4%) proteins (Table 1).
Alternative RNA splicing.
Bioinformatic predictions indicated the occurrence of mRNA splicing in the E1A, polymerase (Pol), terminal protein (TP), and 33k genes (Table 1). Splicing of these genes appears also in the genome annotation of the MAdV-1 (NC000942). Nevertheless, in the case of the MAdV-3 E1A, E1B, TP, and Pol genes, direct sequencing of cDNAs derived from mRNAs was undertaken to evaluate the predictions. In all cases, cDNA sequences of spliced RNAs confirmed our theoretical predictions. For E1A, in addition to the predicted joining of exons 1 and 2 we found an additional spliced transcript where a third exon, coding for the pIX protein, was joined. However, the ORF of the pIX protein was separated from the ORF of E1A located on the first two exons by a stop codon (Table 1). The resulting mRNA therefore encodes the protein of the same length and sequence as the one from the originally predicted spliced mRNA for E1A. A similar structure with a third exon coding for the entire pIX protein has been described also for MAdV-1 (2) but is not found in primate AdVs.
In addition, we found a splice variant also for E1B. We could not detect the unspliced transcript predicted by our bioinformatic approaches (and predicted also for MAdV-1 by Davison et al. [8]). Instead we found an mRNA in which a short intron (72 bp) starting a few nucleotides upstream from the predicted stop codon had been spliced out of the primary transcript (Table 1). The resulting ORF is complete and codes for a variant of the E1B 55k protein which is similar in amino acid length to the predicted unspliced version but in which the last 2 aa, GR, are substituted for by DD. This splicing pattern has also been found experimentally for E1B 55k of MAdV-1 (2).
Phylogenetic analysis.
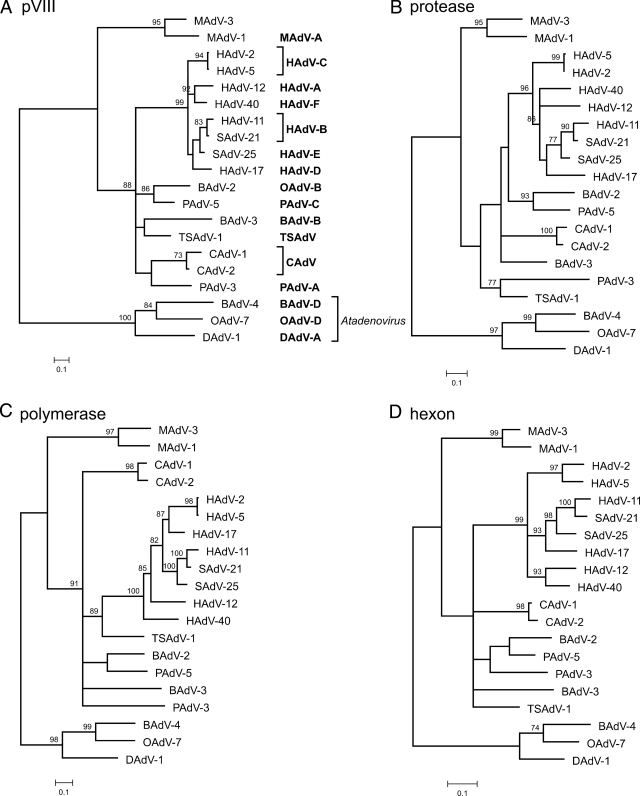
Phylogenetic analysis of the novel virus was performed for the four taxonomically relevant genes as proposed by the International Committee on Taxonomy of Viruses: the genes coding for hexon, polymerase, protease, and pVIII (6). ML (Tree-Puzzle) as well as distance-based methods (NJ and FM) were used to construct phylogenetic trees. All mastadenoviruses for which complete amino acid sequences of these four genes were available were included in the data sets. In all analyses, MAdV-3 clustered with MAdV-1 and both together formed a sister group to all other mastadenoviruses (Fig. 4). In the protease gene data set, TSAdV-1 and PAdV-3 formed a sister group to the MAdV-3 and MAdV-1 cluster when distance-based methods (NJ and FM) were used, but this clustering was not statistically supported (data not shown).
FIG. 4.
Phylogenetic analysis of MAdV-3 (top of each tree) using amino acid sequences of four taxonomically relevant genes coding for the following proteins: pVIII (A), protease (B), polymerase (C), and hexon (D). ML phylogenetic trees calculated with Tree-Puzzle software package (Dayhoff PAM model) are shown. Similar trees were obtained also when NJ and FM methods implemented in PHYLIP 3.67 software package were used. Atadenoviruses BAdV-4, OAdV-7, and DAdV-1 were used as the outgroup. The values at the tree branches represent the PUZZLE support values. The scale bar indicates an evolutionary distance of 0.1 substitution per position in the sequence. In the abbreviated virus names, the letter in front of AdV indicates the origin of the virus: B, bovine; C, canine; D, duck; H, human; M, murine; O, ovine; P, porcine; S, simian; and TS, tree shrew. Taxonomic species abbreviations are given in boldface.
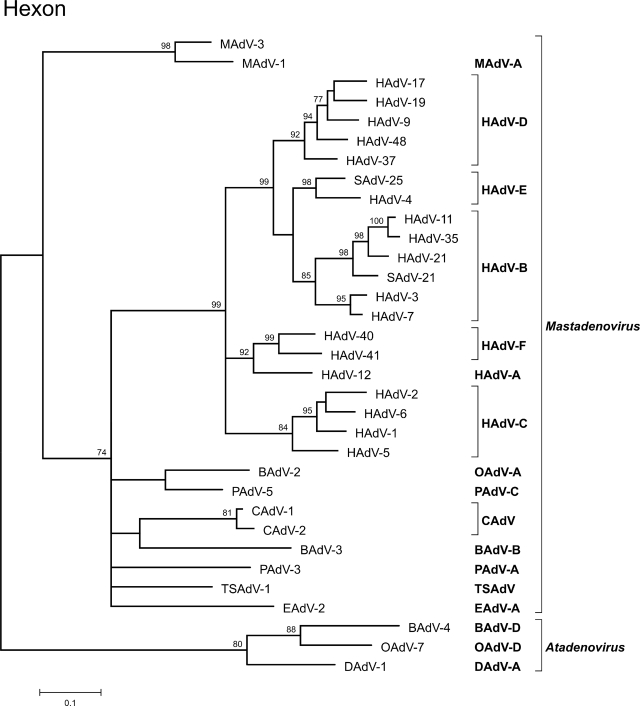
In contrast to polymerase, protease, and pVIII genes, for the hexon gene even more complete gene sequences from additional AdVs were found to be available in GenBank. Therefore, to better characterize the phylogenetic placement of MAdV-3 within mastadenoviruses, an extended data set containing all available complete hexon sequences was used to construct the trees (Fig. 5). In these analyses, the position of MAdV-3 remained unchanged; it clustered together with MAdV-1 and formed an outgroup to all other analyzed mastadenovirus sequences. Since only a very short sequence was available for the hexon gene of MAdV-2 (84 aa), a proper analysis of all three currently known MAdVs was not possible. However, sequence comparison of the short MAdV-2 fragment showed very low sequence identity values of 67.8% and 70.2% to MAdV-3 and MAdV-1, respectively. In provisional phylogenetic analysis, MAdV-2 did not even cluster with MAdV-3 and MAdV-1 into one monophyletic group (data not shown).
FIG. 5.
ML phylogenetic tree (Tree-Puzzle, Dayhoff PAM model) based on all currently available mastadenovirus complete hexon protein amino acid sequences. Atadenoviruses BAdV-4, OAdV-7, and DAdV-1 were used as the outgroup. The values at the tree branches represent the PUZZLE support values. The scale bar indicates an evolutionary distance of 0.1 substitution per position in the sequence. For the abbreviated virus names, see the legend to Fig. 4. EAdV-A, equine AdV-A.
One of the species demarcation criteria in the Mastadenovirus genus is that “properly calculated phylogenetic distance” must be higher than 0.1 (10%) (6). Therefore, we calculated the phylogenetic distance using different evolutionary models and software programs. In all cases, including the observed sequence difference (p-distance), the distance between MAdV-3 and MAdV-1 in all four relevant genes was higher than 0.1 and varied between 0.135 and 0.468, suggesting that MAdV-3 and MAdV-1 should be defined as different species (Table 2).
TABLE 2.
Genetic distance between MAdV-3 and MAdV-1 calculated for the four taxonomically relevant genes by different evolutionary models and software programs
| Protein | Genetic distance calculated by:
|
|||
|---|---|---|---|---|
| Dayhoff model
|
VT model (Tree-Puzzle software) | No modela (Bioedit software) | ||
| Protdist software | Tree-Puzzle software | |||
| pVIII | 0.468447 | 0.36270 | 0.34771 | 0.303 |
| Protease | 0.430186 | 0.35666 | 0.32742 | 0.283 |
| Polymerase | 0.434361 | 0.36061 | 0.32794 | 0.282 |
| Hexon | 0.175666 | 0.14820 | 0.14249 | 0.135 |
In the case of “no model,” the observed sequence difference (p-distance) is indicated as a proportion of positions in which the two sequences differ.
Cross-neutralization with MAdV-1.
In order to determine whether MAdV-3 and most closely related MAdV-1 represent different serotypes, a cross-neutralization study was performed. For this purpose, two C57B/6 mice per virus were immunized with MAdV-1 and MAdV-3, respectively. The neutralizing antibody titers against MAdV-3 are given as reciprocal end-point titers, as determined by PRNT. The neutralizing antibody titers against MAdV-3 for the anti-MAdV-3 and anti-MAdV-1 antisera were 640 and 40, respectively. The neutralizing antibody titers against MAdV-1 are given as reciprocal end-point titers, as determined by immunofluorescence focus reduction neutralization test. The neutralizing antibody titers against MAdV-1 for the anti-MAdV-3 and anti-MAdV-1 antisera are <20 and 1,280, respectively. Note that in neutralization assays with MAdV-3, the anti-MAdV-3 serum pool showed a 16-fold-higher neutralizing antibody titer than the anti-MAdV-1 serum pool. In the neutralizing test with MAdV-1, the anti-MAdV-3 serum did not show any cross-neutralization at all, whereas the anti-MAdV-1 serum pool reached a neutralizing antibody titer of 1,280 as described above. These results show that MAdV-3 and MAdV-1 can be regarded as different serotypes (6).
Organ tropism of MAdV-1 and -3.
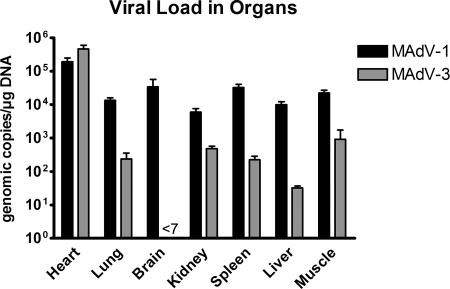
To investigate potential differences in the biology of the two viruses, two groups of six mice each were infected with 108 pgeq of MAdV-1 and -3. The viral load was then determined by qPCR 7 days after infection in various organs. Consistent with previous reports (11, 16, 29), MAdV-1-infected animals showed high viral loads in all analyzed organs. Values ranged from ∼6 × 103 (kidney) to ∼2 × 105 (heart) geq/μg of DNA (Fig. 6). The highest viral loads were found in heart muscle tissue, followed by brain, spleen, and skeletal muscle.
FIG. 6.
Organ tropism of MAdV-1 and MAdV-3. Shown are numbers of geq of each virus per μg mouse DNA 7 days after inoculation of 108 geq into six C57Bl/6N mice per group as determined by qPCR. Note that viral loads for MAdV-3 in brain were below the detection limit, which was 7 geq/μg DNA.
For MAdV-3, the organ distribution of viral loads was completely different. Whereas most organs (lung, kidney, spleen, liver, and skeletal muscle) showed (as compared to MAdV-1) low or moderate viral loads of ∼3 × 101 to ∼9 × 102 geq/μg DNA, two organs exhibited particular differences. First, we could not detect any MAdV-3 DNA in the brain tissue. Second, the myocardium showed the dominant presence of MAdV-3 at 5 × 105 geq/μg DNA, which was even higher than the load for MAdV-1 and was more than 100 times higher than MAdV-3 loads in any other organ. Control mice inoculated with the same amount of heat-inactivated viruses did not show detectable levels of MAdV-1 or MAdV-3 DNA, indicating that the detected DNAs from noninactivated viruses resulted from active replication in the animals. Although both viruses obviously replicated in their host, neither MAdV-1- nor MAdV-3-infected mice exhibited visible clinical signs of infection (loss of body weight, ruffled fur, and changes in behavior).
Altogether, MAdV-1 and MAdV-3 clearly differ in their organ tropisms. While MAdV-1 is rather homogenously distributed in all organs, including the brain, MAdV-3 shows a strong preference for myocardial tissue and is undetectable in the brain.
DISCUSSION
Two MAdVs have been described so far. MAdV-1, which has been genetically completely characterized, represents the MAdV-A species. Due to very limited sequence information, MAdV-2 is only tentatively classified as a separate species, MAdV-B (6). In this study, we report on genetic and serologic characterization of a third MAdV, MAdV-3.
Isolation of the virus from liver tissue and its detection in larger amounts in lung tissue of its natural host suggested that this virus might posses a different organ tropism from MAdV-1, which is predominantly found in the central nervous system (11). To address this question, we performed infection studies of C57BL/6N mice and found strong differences in organ tropism patterns between both viruses. MAdV-1 was found in comparable amounts in all investigated tissues (heart, lung, brain, kidney, spleen, liver, and skeletal muscle), with slightly increased viral loads in heart and brain. The occurrence of the virus in all organs is probably due to its predominant infection of the endothelium (17). In contrast, MAdV-3 showed a different pattern of organ tropism. Low viral loads were found in most organs, and no infection could be detected in brain tissue. On the other hand, high viral loads were found in the myocardium, indicating dominant infection of the heart. Human AdVs were frequently found in the myocardium of patients with myocarditis or myocardial dysfunctions (19, 24). Thus, due to its dominant tropism for cardiac tissue, MAdV-3 might be suitable as an animal model to study AdV infections of the heart.
The fact that the new virus is most closely related to MAdV-1 raises the question of whether it belongs to the MAdV-A species or represents a novel one. According to species demarcation criteria in the genus Mastadenovirus, the lack of cross-neutralization combined with a calculated phylogenetic distance of more than 10% (on the amino acid level) separate two serotypes into different species (6). Serologic and genetic comparison of MAdV-3 with MAdV-1 showed that both of these prerequisites were fulfilled. Both viruses could be clearly distinguished in cross-neutralization experiments. A 16-fold difference in neutralizing antibody titer against MAdV-3 was observed between anti-MAdV-3 and anti-MAdV-1 immune sera. Moreover, the anti-MAdV-3 serum pool showed no cross-neutralization of MAdV-1 at all. Our phylogenetic analysis demonstrated that in all four taxonomically relevant genes, coding for protease, pVIII, hexon, and DNA polymerase, the calculated evolutionary distance between MAdV-3 and MAdV-1 was always substantially higher than 10%.
Additional differences were found at genome level. ITRs of MAdV-3 are 33 bp longer than those of MAdV-1. Substantial differences were found also in ORF predictions. Putative E3 glycoprotein (E3 ORFA), tropoelastin fragment (E4 ORFB), MAdVAgp26, 30kD, and E1B 21k seem to be missing or nonfunctional in MAdV-3. Interesting findings were obtained by the direct sequencing of cDNA generated from viral mRNAs. In case of E1A, in addition to the two-exon transcript predicted by bioinformatic analysis, we found an additional transcript consisting of three exons. A similar three-exon transcript has also been found experimentally for E1A of MAdV-1 (2). However, the third exon containing the complete ORF for pIX protein is located downstream of a stop codon. In the case of E1B 55k, we found an alternatively spliced, two-exon transcript. This is particularly interesting because such a splicing pattern has also been found experimentally for MAdV-1 (2), although E1B 55k is considered to be single-exon gene in all human mastadenoviruses (8).
The classification of MAdV-3 as a novel virus species is supported by additional differences in biological properties. MAdV-3 has been isolated from a different host, a striped field mouse (A. agrarius). MAdV-3, in contrast to MAdV-1, is able to grow in Vero E6 cells, a unique property of the virus which led to its accidental discovery. Moreover, as discussed above, an important difference from MAdV-1 has been found as well in the organ tropism. Altogether, based on significant differences in serological and phylogenetic, as well as biological, characteristics, we propose to classify MAdV-3 as the first isolate of a novel mastadenovirus species, MAdV-C.
Nonhuman mastadenoviruses could also be exploited as animal models for the pathogenesis of human AdV infection or for development of AdV vectors for human gene therapy and vaccination approaches (26, 27). The potential of MAdV-3 to be used in these fields has to be thoroughly analyzed. However, there are preliminary hints that the virus might be useful in both. First, our C57BL/6N mice infection studies showed predominant infection of the heart and no detectable levels of the viral DNA in the central nervous system. Human AdV infection might be involved in myocarditis etiology in humans (9, 19, 24). MAdV-3-infected mice could therefore be relevant as a putative animal model for human cardiac AdV infections. Second, efficient virus replication in the Vero E6 cell line might be advantageous for the usage of this virus as a vector in gene therapy. Third, the expected absence of preexisting immunity to the nonhuman AdVs might increase the reproducibility of gene transfer efficiency in humans and avoid side effects. This is also of relevance for the use of AdV vectors in vaccination approaches. AdV vectors were shown to be potent inducers of cytotoxic T-cell responses to vector-encoded vaccine antigens in various animal models and humans (reviewed in reference 3). Preexisting immunity to the AdV vector can reduce immune responses to the antigens encoded by the AdV vector (4). In addition, a recent phase IIb human immunodeficiency virus (HIV) vaccine study indicates that immunization with an AdV vector vaccine encoding HIV antigens increases the susceptibility to HIV infection in volunteers with preexisting AdV-specific antibodies (7, 23).
In summary, we isolated and characterized a novel MAdV (MAdV-3) which we propose to classify as a member of a novel species, MAdV-C. In comparison to its closest relative, MAdV-1 (MAdV-A), the virus shows substantial differences in genome organization and antigenicity. Moreover, MAdV-3 predominantly infects myocardial tissue of the laboratory mice. Some of its unique properties suggest that the virus could be used as an animal model for human AdV infections or as a vector in gene therapy and vaccination, but this potential has to be further evaluated.
Acknowledgments
We thank Regina Buetermann and Christina Priemer for excellent technical help and Oliver Wildner for providing an in-house qPCR for MAdV-1.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. KR1293/6-1 and UE45/10-1), the Slovak Research and Development Agency (grant no. APVV-0108-06), and an internal research grant from the Medical Faculty of the Ruhr-University (FoRUM 402-2003).
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Allander, T., S. U. Emerson, R. E. Engle, R. H. Purcell, and J. Bukh. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc. Natl. Acad. Sci. USA 9811609-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, A. O., C. W. Beard, S. D. Redick, and K. R. Spindler. 1989. Genome organization of mouse adenovirus type 1 early region 1: a novel transcription map. Virology 170523-536. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H. 2006. Rational design of gene-based vaccines, J. Pathol. 208283-289. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., M. G. Pau, J. H. Custers, W. Koudstaal, S. Kostense, M. J. Havenga, D. M. Truitt, S. M. Sumida, M. G. Kishko, J. C. Arthur, B. Korioth-Schmitz, M. H. Newberg, D. A. Gorgone, M. A. Lifton, D. L. Panicali, G. J. Nabel, N. L. Letvin, and J. Goudsmit. 2004. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J. Immunol. 1726290-6297. [DOI] [PubMed] [Google Scholar]
- 5.Beard, C. W., A. O. Ball, E. H. Wooley, and K. R. Spindler. 1990. Transcription mapping of mouse adenovirus type 1 early region 3. Virology 17581-90. [DOI] [PubMed] [Google Scholar]
- 6.Benkö, M., B. Harrach, G. W. Both, W. C. Russell, B. M. Adair, E. Adam, J. C. de Jong, M. Hess, M. Johnson, A. Kajon, A. H. Kidd, H. D. Lehmkuhl, Q.-G. Li, V. Mautner, P. Pring-Akerblom, and G. Wadell. 2005. Family Adenoviridae, p. 213-228. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier, New York, NY.
- 7.Buchbinder, S. P., D. V. Mehrotra, A. Duerr, D. W. Fitzgerald, R. Mogg, D. Li, et al. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 3721881-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J., M. Benkö, and B. Harrach. 2003. Genetic content and evolution of adenoviruses. J. Gen. Virol. 842895-2908. [DOI] [PubMed] [Google Scholar]
- 9.Dennert, R., H. J. Crijns, and S. Heymans. 2008. Acute viral myocarditis. Eur. Heart J. 292073-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package), 3.67 ed. Department of Genetics, University of Washington, Seattle.
- 11.Guida, J. D., G. Fejer, L.-A. Pirofski, C. F. Brosnan, and M. S. Horwitz. 1995. Mouse adenovirus type 1 causes a fatal hemorrhagic encephalomyelitis in adult C57BL/6 but not BALB/c mice. J. Virol. 697674-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 13.Hamelin, C., C. Jacques, and G. Lussier. 1988. Genome typing of mouse adenoviruses. J. Clin. Microbiol. 2631-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartley, J. W., and W. P. Rowe. 1960. A new mouse virus apparently related to the adenovirus group. Virology 11645-647. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto, K., T. Sugiyama, and S. Sasaki. 1966. An adenovirus isolated from feces of mice. I. Isolation and identification. Jpn. J. Microbiol. 10115-125. [DOI] [PubMed] [Google Scholar]
- 16.Heck, F. C., W. G. Sheldon, and C. A. Gleiser. 1972. Pathogenesis of experimentally produced mouse adenovirus infection in mice. Am. J. Vet. Res. 842841-846. [PubMed] [Google Scholar]
- 17.Kajon, A. E., C. C. Brown, and K. R. Spindler. 1998. Distribution of mouse adenovirus type 1 in intraperitoneally and intranasally infected adult outbred mice. J. Virol. 721219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klempa, B., M. Stanko, M. Labuda, R. Ulrich, H. Meisel, and D. H. Krüger. 2005. Central European Dobrava hantavirus isolate from a striped field mouse (Apodemus agrarius). J. Clin. Microbiol. 432756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhl, U., M. Pauschinger, B. Seeberg, D. Lassner, M. Noutsias, W. Poller, and H. P. P. Schultheiss. 2005. Viral persistence in the myocardium is associated with progressive cardiac dysfunction. Circulation 1121965-1970. [DOI] [PubMed] [Google Scholar]
- 20.Nemirov, K., O. Vapalahti, Å. Lundkvist, V. Vasilenko, I. Golovljova, A. Plyusnina, J. Niemimaa, J. Laakkonen, H. Henttonen, A. Vaheri, and A. Plyusnin. 1999. Isolation and characterization of Dobrava hantavirus carried by the striped field mouse (Apodemus agrarius) in Estonia. J. Gen. Virol. 80371-379. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, T., J. Nery, S. Joseph, C. Rocha, G. Carney, K. Spindler, and L. Villarreal. 1999. Mouse adenovirus (MAV-1) expression in primary human endothelial cells and generation of a full-length infectious plasmid. Gene Ther. 61291-1297. [DOI] [PubMed] [Google Scholar]
- 22.Raviprakash, K. S., A. Grunhaus, M. A. el Kholy, and M. S. Horwitz. 1989. The mouse adenovirus type 1 contains an unusual E3 region. J. Virol. 635455-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson, M., D. Mehrotra, D. Fitzgerald, A. Duerr, D. Casimiro, J. McElrath, D. Lawrence, and S. Buchbinder. 5 February 2008, posting date. Efficacy results from the STEP study (Merck V520 protocol 023/HVTN 502): a phase II test-of-concept trial of the MRKAd5 HIV-1 Gag/Pol/Nef trivalent vaccine. http://www.hvtn.org/media/pr/BuchbinderSTEPEfficacy.pdf.
- 24.Savón, C., B. Acosta, O. Valdés, A. Goyenechea, G. Gonzalez, A. Piñón, P. Más, D. Rosario, V. Capó, V. Kourí, P. A. Martínez, J. J. Marchena, G. González, H. Rodriguez, and M. G. Guzmán. 2008. A myocarditis outbreak with fatal cases associated with adenovirus subgenera C among children from Havana City in 2005. J. Clin. Virol. 43152-157. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18502-504. [DOI] [PubMed] [Google Scholar]
- 26.Singh, N., A. Pandey, L. Jayashankar, and S. K. Mittal. 2008. Bovine adenoviral vector-based H5N1 influenza vaccine overcomes exceptionally high levels of pre-existing immunity against human adenovirus. Mol. Ther. 16965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sridhar, S., A. Reyes-Sandoval, S. J. Draper, A. C. Moore, S. C. Gilbert, G. P. Gao, J. M. Wilson, and A. V. S. Hill. 2008. Single-dose protection against Plasmodium berghei by a simian adenovirus vector using a human cytomegalovirus promoter containing intron A. J. Virol. 823822-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stang, A., K. Korn, O. Wildner, and K. Überla. 2005. Characterization of virus isolates by particle-associated nucleic acid PCR. J. Clin. Microbiol. 43716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiyama, T., K. Hashimoto, and S. Saski. 1967. An adenovirus isolated from the feces of mice. II. Experimental infection. Jpn. J. Microbiol. 1133-42. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wigand, R. 1980. Age and susceptibility of Swiss mice for mouse adenovirus, strain FL. Arch. Virol. 64349-357. [DOI] [PubMed] [Google Scholar]