Abstract
The herpes simplex virus 1 (HSV-1) multifunctional regulatory protein ICP27 shuttles between the nucleus and cytoplasm in its role as a viral mRNA export factor. Arginine methylation on glycine- and arginine-rich motifs has been shown to regulate protein export. ICP27 contains an RGG box and has been shown to be methylated during viral infection. We found by mass spectrometric analysis that three arginine residues within the RGG box were methylated. Viral mutants with substitutions of lysine for arginine residues were created as single, double, and triple mutants. Growth of these mutants was impaired and the viral replication cycle was delayed compared to wild-type HSV-1. Most striking was the finding that under conditions of hypomethylation resulting from infection with arginine substitution mutants or treatment of wild-type HSV-1-infected cells with the methylation inhibitor adenosine dialdehyde, ICP27 export to the cytoplasm occurred earlier and was more rapid than wild-type ICP27 export. We conclude that arginine methylation of the ICP27 RGG box regulates its export activity and that early export of ICP27 interferes with the performance of its nuclear functions.
Herpes simplex virus 1 (HSV-1) regulatory protein ICP27 is a multifunctional protein that plays a role in both transcriptional and posttranscriptional regulation of viral and cellular gene expression (29). Early during infection ICP27 is predominantly nuclear and undergoes a series of interactions with splicing proteins (25, 30-32), RNA polymerase II (8, 48), mRNA export factors (6, 7, 13), and viral RNA (12, 28). During infection, ICP27 has been shown to bind viral mRNA both in the nucleus and cytoplasm through an RGG box binding motif (28). At approximately 5 h postinfection, ICP27 begins to shuttle to the cytoplasm, facilitating the export of associated viral transcripts (6, 7, 12, 22, 38). Export of ICP27 to the cytoplasm requires its interaction with the nuclear export adaptor protein TAP/NXF1, and both the N and C termini of ICP27 must be intact for the interaction with TAP/NXF1 (6, 12).
Protein arginine methylation is a posttranslational modification commonly found in RNA-binding proteins that shuttle between the nucleus and cytoplasm (3, 9, 16, 17, 46). Protein arginine methylation is catalyzed by a family of enzymes known as protein arginine methyltransferases (PRMTs), for which at least nine members have been identified (1, 24). S-Adenosyl-l-methionine is used as the methyl donor in the reaction, in which the methyl group is transferred to one of the guanidinium nitrogens of arginine residues. PRMTs can be nuclear or cytoplasmic or both, and two types have been identified: type I, which catalyzes the formation of ω-NG-monomethylarginine and ω-NG,NG-(asymmetric)-dimethylarginine, and type II, which catalyzes formation of ω-NG-monomethylarginine and ω-NG,NG′-(symmetric)-dimethylarginine (1, 24). PRMT1 is the main methyltransferase in human cells, and it has a wide substrate spectrum (24). PRMT1 usually recognizes arginines within a glycine- and arginine-rich (GAR) region, a motif that is present in many RNA and DNA binding proteins. Other PRMTs have different specificities. Although the GAR motif is the preferred methylation site for PRMT1, other factors such as three-dimensional structure and accessibility are also important for substrate recognition by PRMTs (24).
Recent studies have shown that arginine methylation is important for modulating protein-protein interactions (4, 9, 11, 19, 44). Additionally, both export and import of shuttling proteins have been shown to be regulated by arginine methylation (3, 10, 19, 23, 34, 46, 47). ICP27 is a shuttling RNA-binding protein that also interacts with a number of cellular and viral proteins (29). It contains a GAR motif in the form of an RGG box RNA-binding domain in the N-terminal portion of the protein. ICP27 also contains numerous arginine residues, which are not in the context of a GAR motif, throughout the protein. ICP27 was shown by Mears and Rice (21) to be methylated on arginine residues in vivo, and these authors were not able to detect methylation in a viral mutant in which the RGG box was deleted. Using in vitro nuclear export assays, we found that the ΔRGG ICP27 protein was exported from the nucleus at a much faster rate than the wild-type ICP27 (6), suggesting that arginine methylation may regulate ICP27 export. In this report, we mapped the arginine residues that are methylated during infection by mass spectrometric analysis. Subsequently, we created viral mutants in which these residues were replaced with lysines. Mutant viral replication, gene expression, and ICP27 nuclear export kinetics were analyzed with respect to those for the wild type. The results showed that the arginine-to-lysine substitution mutants were impaired in viral replication and that they showed delayed gene expression profiles. Furthermore, export of the mutant proteins occurred earlier and faster than that for wild-type ICP27, which was also observed under conditions of hypomethylation induced by a methylation inhibitor. We conclude that the early export of ICP27 from the nucleus to the cytoplasm prevents it from carrying out its nuclear functions efficiently.
MATERIALS AND METHODS
Cells, viruses, and recombinant plasmids.
Rabbit skin fibroblasts (RSF) and Vero cells were grown in minimal essential medium (MEM) containing 8% fetal calf serum and 4% donor calf serum. The ICP27 complementing cell line 2-2 (36) was grown in MEM containing 8% fetal calf serum and 4% donor calf serum containing G418 (750 μg/ml). HeLa cells were grown in MEM containing 10% newborn calf serum. HSV-1 wild-type strain KOS 1.1 and ICP27 null mutant 27LacZ were previously described (36). ICP27 viral mutants d1-2, d2-3, d3-4, d4-5, d5-6, and n504 were generously provided by Steve Rice (14, 26, 27). N-YFP-ICP27 virus expressing enhanced yellow fluorescent protein (YFP)-ICP27 protein was constructed by homologous recombination between sequences upstream and downstream of the ICP27 coding regions derived from HSV-1 wild-type strain KOS 1.1 (L. Li and R. M. Sandri-Goldin, unpublished results). In the viral mutant 27-GFP, the ICP27 coding sequence is replaced by the coding sequence of green fluorescent protein as previously described (41). ICP27 viral mutants were propagated on 2-2 cells as described previously (36). The plasmid pGEM2-gC was cloned by inserting a 900-bp EcoRI-XbaI fragment from pUC18-gC into pGEM2 as described previously (37).
In vivo methylation assays and immunoprecipitation.
HeLa cells were grown in 100-mm2 dishes at 37°C and infected at a multiplicity of infection (MOI) of 10 with the viruses indicated in each figure legend. Metabolic labeling of methylated proteins was performed as described previously (16). At 4.5 h after infection, the medium was aspirated and replaced with medium containing 100 μg/ml cycloheximide. Cells were incubated in this medium at 37°C for 30 min to stop translation. At 5 h, the medium was replaced with MEM containing 100 μg/ml cycloheximide and 45 μCi/ml of l-[methyl-3H]methionine. Parallel control experiments were conducted to ensure that cycloheximide stopped translation efficiently. For these control experiments, MEM containing 100 μg/ml cycloheximide and 150 μCi [35S]methionine was added 5 h after infection. Cells labeled with either l-[methyl-3H]methionine or [35S]methionine were harvested at 8 h in cold phosphate-buffered saline (PBS), pH 7.2. Cellular pellets were resuspended in 1 ml cold high-salt lysis buffer (10 mM Tris pH 7.5, 1 mM EDTA, 500 mM NaCl, 0.5% NP-40, pH 7.5, 2 mM leupeptin; Roche) and 5 μg/ml Pefabloc (Roche) and were frozen at −80°C. Immunoprecipitation was performed on thawed samples as described previously (30). Extracts were precleared with rabbit anti-mouse immunoglobulin G (IgG) secondary antibody (Pierce) bound to protein A-Sepharose (Amersham Pharmacia) for 1 h. The ICP27 monoclonal antibody H1119 (15 μg/ml) (Virusys) was added, and samples were incubated at 4°C on an end-to-end rotator for 1 h. Anti-mouse IgG antibody (7.5 μg) was added, and samples were incubated at 4°C on a rotator for 1 h, after which 150 μl of a slurry of protein A-Sepharose equilibrated in high-salt lysis buffer was added and samples were incubated at 4°C on a rotator overnight. The samples were washed four times with high-salt lysis buffer and once with 1× PBS, pH 7.2. Samples were resuspended in 2× electrophoresis sample solution (ESS). Immunoprecipitated proteins were resolved on 10% polyacrylamide gels and transferred to nitrocellulose. Western blot analysis was done using ICP27 monoclonal antibody H1119 (1:5,000 dilution) as the primary and sheep anti-mouse IgG-horseradish peroxidase (HRP) (1:100,000 dilution) as the secondary. Proteins were detected with enhanced chemiluminescence (ECL). After the signal decayed, the same blots were then incubated for 10 min in 1 M salicylate-0.1 M Tris-HCl, air dried, and exposed to film for 7 to 10 days at −80°C for 3H-labeled proteins or for 30 min for 35S-labeled proteins.
For the analysis of ICP27 protein arginine methylation changes over time, HeLa cells were infected with wild-type HSV-1 KOS and samples were harvested at the times indicated in Fig. 4. Nuclear and cytoplasmic fractions were prepared and ICP27 was immunoprecipitated from the lysates as described above. Western blot analysis was performed on fractionated samples with a monoclonal antibody to ICP27, 21C7 (AbCam), which is specific for dimethylarginine, and monoclonal antibody 16B11 (AbCam), which is specific for monomethylarginine.
FIG. 4.
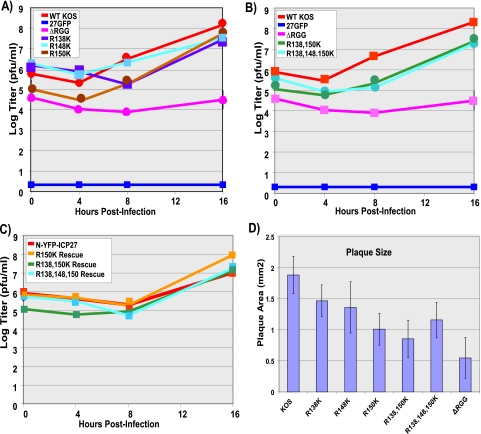
ICP27 arginine-to-lysine viral mutants exhibit a growth defect. One-step viral growth curves were performed with the ICP27 R-to-K mutants. (A and B) Vero cells were infected with wild-type (WT) HSV-1 KOS, the ICP27 null mutant 27-GFP, ΔRGG, or the ICP27 R-to-K mutants at a MOI of 1. Infected cells were harvested 0, 4, 8, and 16 h after infection. Plaque assays were performed on Vero cells. Plotted is the average titer (n = 3). (C) As described in Materials and Methods, viral mutants were rescued. Average titers are plotted (n = 3). (D) The average plaque areas of WT HSV-1 KOS virus and viral mutants were measured. Error bars represent the standard errors of the means (n = 10).
MS analysis.
ICP27 was immunoprecipitated from wild-type HSV-1 KOS 1.1-infected HeLa cells and fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide). Parallel immunoblot analysis was used to positively identify ICP27 on Coomassie-stained gels. ICP27 was excised from Coomassie-stained gels and then destained by vortexing three times for 10 min each time at room temperature in 100 μl of 25 mM NH4HCO3 and 50% CH3CN. The gel pieces were dehydrated under vacuum centrifugation and then rehydrated in either 40 μl 100 mM Tris-HCl (pH 8.0) containing Trypsin Gold (Promega; added at a 1:10 protease/substrate mass ratio) or 0.1% trifluoroacetic acid (TFA; Applied Biosystems) in water containing pepsin (Sigma; 1:3 mass ratio) or 50 mM Tris-HCl (pH 8.0), 5 mM CaCl2, and 10% (vol/vol) acetonitrile containing thermolysin from “Bacillus thermoproteolyticus” subsp. rokko (Sigma-Aldrich; 1:10 mass ratio). After 10 min at room temperature, digestion buffer lacking enzyme was added to cover the gel slice completely, followed by incubation at 37°C overnight (trypsin), 0°C for 15 min (pepsin), or 50°C for 24 h (thermolysin). Peptides were then extracted once in 50 μl water and twice in 50% CH3CN-5% TFA in water, with vortexing for 10 min each time. All extracted peptides were pooled and subjected to volume reduction under vacuum to a final volume of 5 μl. The resulting peptides were fractionated by reversed-phase nanoflow liquid chromatography (trapping, 0.3-mm-inside-diameter by 5-mm column packed with 5-μm/100-Å C18 beads; analytical, 0.1-mm-inside-diameter by 150-mm column packed with 5-μm/200-Å C18 beads). With instrumentation from LC Packings, CH3CN-isopropanol gradients (10 to 50 or 45% over 38 min) in 0.1% TFA-water were developed at 0.2-μl/min split flow rate. The column output was dosed online with α-cyano-4-hydroxycinnamic acid matrix solution (7.5 mg/ml in CH3CN-water [3:1] containing 130 μg/ml ammonium citrate, 1 mM ammonium monobasic phosphate, and trace amounts of Glu-fibrinopeptide) at a sample/matrix mixing ratio of 1:2, followed by robotic generation of 576 spots on the matrix-assisted laser desorption ionization (MALDI) target plate. The 4700 MALDI-time of flight (TOF)-TOF mass spectrometer (Applied Biosystems) was used to acquire mass spectrometry (MS) spectra in the m/z 800 to 4,000 range for all spots, followed by tandem MS (MS/MS) on all ions showing a signal/noise ratio of >30 (strongest first; maximum MS/MS value per spot = 20). SwissProt (taxonomy: other viruses) was searched against the resulting MS/MS ion peak lists via Mascot (Matrix Science Ltd.), setting precursor mass tolerance at 75 ppm, fragment mass tolerance at 0.3 Da, and variable modifications at oxidized methionine, methylarginine, and dimethylarginine. Spectra for candidate methylated peptides scoring with >95% confidence were visually inspected, comparing accepted versus alternative spectral interpretations using DeNovo Explorer (Applied Biosystems) and then confirming the presence of diagnostic fragments. In addition, the combined MS peak list for all spots of an experiment was sent to FindMod (http://ca.expasy.org/tools/findmod/) along with the ICP27 protein sequence for identification of candidate methylated peptides at the MS level (mass tolerance = ±30 ppm; ≤3 missed cleavages). Spectra for such candidates were subjected to visual inspection in DeNovo Explorer as described above.
Construction of ICP27 arginine-to-lysine viral mutants.
Site-directed mutagenesis was used to construct point mutations in the ICP27 coding sequence of pSG130/BS plasmid producing arginine-to-lysine mutations (33) with primers 5′-CCAAAGCCCAGCCTGCCAAGGGCGGACGCCGTGGGCGTCG-3′ (R138K), 5′-CGTCGCAGGGGTAAGGGTCGCGGTGGTCCC3′ (R148K), and 5′-CGCAAGGGTCGGGGTAAGGGTGGTCCCGGGGC-3′ (R150K) using the QuikChange II site-directed mutagenesis kit (Stratagene). The mutant plasmids were linearized using restriction endonucleases at BamHI and SstI sites. Linearized mutant plasmid DNA and viral DNA from 27-GFP were cotransfected into 2-2 cells with Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. Viral recombinants were screened by picking nongreen plaques, which were plaque purified at least five times. The ICP27 gene open reading frame and 5′ and 3′ untranslated regions of purified viral recombinants were sequenced to ensure maintenance of the point mutations and absence of extraneous mutations. Rescued viruses were engineered by cotransfecting the BamHI-to-SstI fragment from the N-EYFP-ICP27 coding sequence and DNA from the ICP27 arginine to lysine mutant viral recombinants into 2-2 cells to allow homologous recombination. Viral recombinants were screened by picking yellow plaques, which were plaque purified and sequenced as described above.
One-step viral growth curves.
Vero cells were seeded in six-well plates and infected the next day at a MOI of 1. Infected cells were collected immediately after virus adsorption (zero time) and 4, 8, and 16 h after infection. Viral titers were determined by plaque assay on Vero cells. Growth curves represent the means of three independent infections, each titrated in duplicate.
Microarray analysis.
HeLa cells were seeded in 150-mm2 dishes and infected the next day at a MOI of 10 with the viruses indicated in each figure. Infections were allowed to proceed at 37°C, and the cells were collected 6 h after infection by harvesting the infected cells in PBS and resuspending the cell pellet directly in 7 ml Trizol reagent (Invitrogen). RNA was extracted, biotin labeled, and purified as described previously (40, 41). Biotin-labeled cDNAs were hybridized and labeled as previously described (41). Details regarding the characteristics and construction of the HSV-1-specific oligonucleotide-based DNA microarray used in this experiment have been described previously (39, 45).
Protein expression profile.
HeLa cells were infected at a MOI of 10 with the viruses indicated in the figures. Infected cells were collected 4, 8, and 12 h after infection by harvesting directly in 2× ESS. The lysates were subjected to three rounds of freeze-thaw cycles, followed by boiling for 10 min. Proteins were resolved on 12% polyacrylamide gels and transferred to nitrocellulose. Western blot analysis was performed using monoclonal antibodies against ICP4 (1:5,000 dilution; Virusys), ICP27 (1:5,000 dilution; Virusys), ICP8 (1:2,500 dilution; AbCam), gD (1:2,500 dilution; Virusys), and β-actin (1:10,000 dilution; Sigma). The secondary antibody was sheep anti-mouse IgG-HRP (1:100,000 dilution; Jackson Immunoresearch). Proteins were detected with ECL.
Quantitative real-time PCR.
HeLa cells were infected at a MOI of 10 with the viruses indicated in each figure. Adenosine dialdehyde (AdOx) (20 μM) was added 2 h after infection as indicated in the figure legends. Cells were harvested 1 and 12 h after infection in 1 ml PBS. The cells were centrifuged at 4°C for 10 min, and cell pellets were resuspended in 500 μl of lysis buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, 1% SDS). After one freeze-thaw cycle, cell lysates were incubated at 65°C for 20 min to inactivate DNase. RNA was degraded by incubation with 0.2 mg/ml RNase A for 30 min at 37°C. Proteins were degraded by incubation with proteinase K (10 mg/ml) for 2 h at 37°C. DNA was extracted with phenol-chloroform and precipitated in ethanol. The plasmid pGEM2-gC was used for standard curve titrations. The real-time PCR mixture contained 1 ng viral DNA, 50% (vol/vol) Sybr green mixture (Bio-Rad), 1.3 μM betaine (Sigma), 1.3% dimethyl sulfoxide (Fisher), 0.32 pmol/μl forward primer 5′-GTTCACCACCGTCTCTACC-3′, and 0.32 pmol/μl reverse primer 5′-CGAGAACGTCACGGAGTC-3′.
ICP27 protein stability assay.
HeLa cells were infected at a MOI of 10 with the viruses indicated in the figures. AdOx at 20 μM was added to samples 2 h after infection. Cycloheximide at 100 μg/ml was added 5 h after infection to some of the samples. Cells were harvested directly in 200 μl 2× ESS. Proteins were resolved on 10% polyacrylamide gels and transferred to nitrocellulose. Western blot analysis was performed using primary monoclonal antibodies against ICP27 (1:5,000 dilution; Virusys) and β-actin (1:10,000 dilution; Sigma) and sheep anti-mouse IgG-HRP (1:100,000 dilution) as the secondary antibody. Proteins were detected with ECL. The relative amounts of the proteins were quantified by scanning the X-ray films with Personal FX (Bio-Rad) for quantification with Quantity One software (Bio-Rad).
Immunofluorescence.
HeLa cells were seeded on coverslips in 24-well plates and infected at a MOI of 10 with the viruses indicated in the figures. AdOx (20 μM) was added to samples 2 h after infection. Cells were fixed in 3.7% formaldehyde at the times indicated in the legends to Fig. 8 and 10. Immunofluorescence staining was performed as described previously (15, 32) with either anti-ICP4 monoclonal antibody (Virusys) at a 1:500 dilution, anti-ICP27 monoclonal antibody H1119 (Virusys) at 1:500 dilution, or anti-hnRNP A2 monoclonal antibody (23). Cells were viewed by fluorescence microscopy at 100× magnification with a Zeiss Axiovert S100 microscope. Images were pseudocolored and merged with Adobe Photoshop.
FIG. 8.
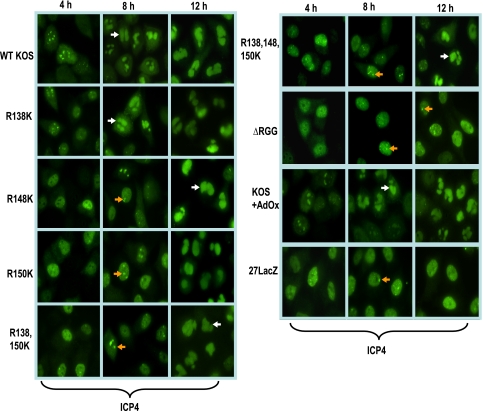
ICP27 arginine-to-lysine mutant viruses show delayed replication compartment formation. HeLa cells were infected with either wild-type (WT) HSV-1 KOS, ΔRGG, 27LacZ, or the indicated ICP27 arginine-to-lysine viral mutants. Cells were fixed 4, 8, or 12 h after infection and subsequently immunostained with anti-ICP4 monoclonal antibody. White arrows point to fully formed replication compartments, and orange arrows indicate small prereplication sites.
FIG. 10.
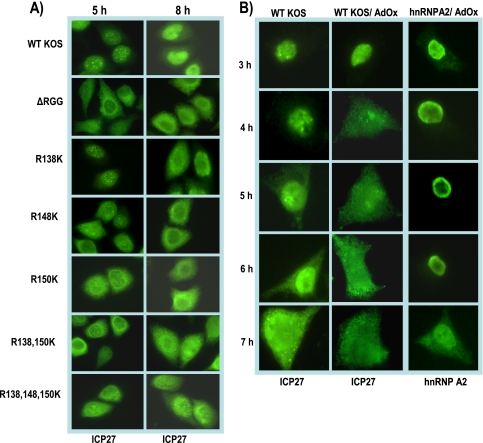
ICP27 hypomethylation affects its cellular localization. (A) HeLa cells were infected with wild-type (WT) HSV-1 KOS, ΔRGG, or the indicated ICP27 arginine-to-lysine viral mutants. At 5 and 8 h after infection, the cells were fixed and immunostained with anti-ICP27 monoclonal antibody. (B) HeLa cells were either mock infected or infected with WT HSV-1 KOS. The general methylation inhibitor AdOx was added 2 h after infection to the indicated samples. At the times indicated, cells were fixed and immunostained with anti-ICP27 or anti-hnRNPA2 monoclonal antibodies.
In vitro nuclear export assays.
RSF cells were seeded in 24-well plates and infected at a MOI of 10 with HSV-1 wild-type KOS or with an ICP27 viral mutant. Nuclear export assays were carried out 5 h after infection. Cytoplasmic membranes were permeated with digitonin and extensively washed with PBS. Export was initiated by addition of 50% rabbit reticulocyte lysate and an ATP regeneration system as described previously (7). Assays were stopped by the addition of cold transport buffer, and exported proteins were washed away by repeated ice-cold washes (6, 7). Proteins that were retained in the nucleus were suspended directly in 2× ESS. Western blot analysis was performed using anti-ICP27 (H1119) and anti-YY1 (AbCam) antibodies, as described previously (6, 7). Blots were quantified by scanning the X-ray films with Personal FX (Bio-Rad) for quantification with Quantity One software (Bio-Rad).
RESULTS
The RGG box motif is the major site of methylation for ICP27.
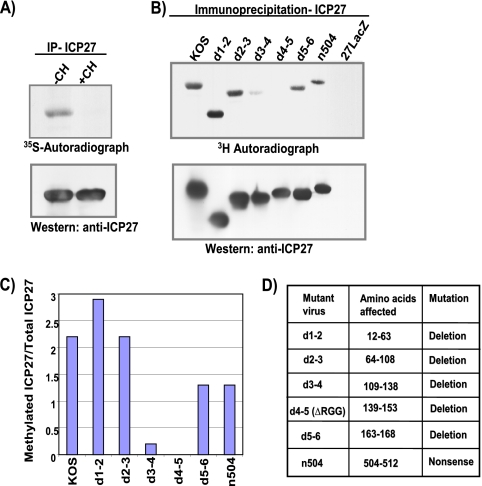
Mears and Rice (21) reported that wild-type ICP27 is methylated in vivo; however, ICP27 was not methylated when cells were infected with the mutant virus d4-5, in which the ICP27 RGG box is deleted. To determine if the RGG box, which is a GAR motif, was the major site of methylation of ICP27 in vivo, cells were infected with wild-type HSV-1 KOS and a series of viral mutants with lesions throughout the ICP27 protein (Fig. 1). A labeling procedure similar to that of Mears and Rice was used. Infected cells were labeled with l-methyl-[3H]methionine for 3.5 h between 4.5 and 8 h after infection in the presence of cycloheximide, which prevents uptake of the label by newly translated proteins. A parallel experiment was performed with [35S]methionine labeling. ICP27 was immunoprecipitated, and fractionated proteins were transferred to nitrocellulose. Immunoblot analysis was performed with anti-ICP27 antibody to determine the relative amount of protein precipitated, and autoradiography was performed to determine the amount of labeled ICP27 protein (Fig. 1A and B). Wild-type ICP27 as well mutant ICP27 protein from infections with d1-2, d2-3, d5-6, and n504 were efficiently labeled with [3H]methionine (Fig. 1B and C). In contrast, in infections with d4-5, in which the RGG box is deleted (Fig. 1D), labeled ICP27 was not detected, although equivalent amounts of ICP27 were immunoprecipitated relative to other mutants (Fig. 1B, bottom). This result confirms what was found by Mears and Rice (21). It was also observed that labeling of ICP27 with [3H]methionine in d3-4 infection was reduced at least 10-fold compared to labeling of wild-type ICP27 (Fig. 1B and C). The deletion in d3-4 is adjacent to the RGG box (Fig. 1D), and this region does not have a GAR motif. Two possible explanations for this result are as follows. First, although the GAR motif is the preferred methylation site for PRMT1, other factors such as three-dimensional structure and accessibility are important for substrate recognition (24). A deletion of the region adjacent to the RGG box may significantly affect the structure of the protein and the accessibility of the RGG box. Second, the ICP27 nuclear localization signal is deleted in d3-4 (20) and ICP27 is inefficiently imported into the nucleus. PRMT1 is predominantly nuclear. If PRMT1 is the primary methyltransferase for ICP27, a substantial fraction of ICP27 would not be available for methylation. We conclude that the major site for methylation of ICP27 is the RGG box.
FIG. 1.
The RGG box is the major site of ICP7 arginine methylation. In vivo methylation assays were performed to determine the major site of ICP27 methylation. HeLa cells were infected with either wild-type HSV-1 KOS; ICP27 viral deletion mutant d1-2, d2-3, d3-4, d4-5 (ΔRGG), or d5-6; ICP27 truncation mutant n504; or ICP27 null mutant 27LacZ (14, 26, 36). Cells were incubated from 4.5 to 8 h after infection with medium containing either [35S]methionine (A) or l-[methyl-3H]methionine (B), in the absence or presence of the translation inhibitor cycloheximide (CH). ICP27 was immunoprecipitated (IP), resolved by SDS-PAGE, and transferred to nitrocellulose. Total ICP27 protein was determined by Western blot analysis with the ICP27-specific monoclonal antibody H1119, and methylated ICP27 was determined by fluorography. (C) Densitometry was used to quantify the bands. The data shown were derived from a single experiment but are representative of several experiments. (D) The residues deleted in the mutants are shown.
Determination of which arginines are methylated by MS.
To determine which arginines are methylated during viral infection, ICP27 was immunopurified at 6 h postinfection from nuclear and cytoplasmic fractions of HSV-1 KOS-infected HeLa cell extracts. The ICP27 band was cut from Coomassie-stained gels (Fig. 2A), and the gel slice was digested in situ. Three different enzymes, trypsin, pepsin, and thermolysin, were used to digest ICP27 in parallel. Each enzyme has different intrinsic cleavage specificities, increasing the number of unique peptide fragments available for analysis by MS. Digesting ICP27 with these three enzymes created a peptide pool representing 90% of the ICP27 protein sequence, including the major methylation site, the RGG box. The peptides that resulted from these digestions were analyzed by liquid chromatography-MALDI-TOF-TOF MS as described in Materials and Methods. In the data analysis shown in Fig. 2B, the arginine at position 148 was found to be monomethylated and the arginine at position 150 was found to be dimethylated in the nuclear sample in this experiment. The arginine at position 138 was found to be dimethylated in the cytoplasmic fraction. These three arginine residues reside within the RGG box in the context of a GAR motif, as shown in Fig. 2C. The mass spectrometric analysis was performed several times using three different enzymes, trypsin, pepsin, and thermolysin, to obtain maximum coverage of the protein. The arginines at positions 138, 148, and 150 were consistently found to be methylated in these experiments. Other arginines scattered throughout the protein were also found to be methylated, but these residues were not located at the major site of methylation, the RGG box (Fig. 1B and C). The mass spectrometric results from one such experiment are shown in Fig. S1 in the supplemental material. Although these other residues are not within GAR motifs, the preferred substrate for PRMT1, -3, and -6, they may be substrates of PRMT4, -5, and -7, which are able to methylate arginines not in the context of GAR motifs (2).
FIG. 2.
Arginines 138, 148, and 150 within the ICP27 RGG box are methylated in vivo. (A) ICP27 was immunopurified from nuclear (Nuc) and cytoplasmic (Cyt) fractions of wild-type KOS-infected HeLa cell extracts. The immunoprecipitates were resolved by SDS-PAGE and visualized by Coomassie blue staining. Plus signs indicate ICP27 protein, and asterisks denote heavy-chain IgG. M, molecular mass. (B) ICP27 was cut from the Coomassie-stained gels and in-gel digested. The resulting peptides were analyzed for protein methylation by liquid chromatography-MALDI-TOF-TOF MS. WT, wild type; monoisotopic mass, the observed monoisotopic mass of the peptide; Mass Diff, the difference between the theoretical mass and the monoisotopic mass; Mod Diff, the theoretical mass of a particular posttranslational modification; MA (ppm), mass accuracy, which is Mass Diff divided by the theoretical mass in parts per million; modification, the number of methyl groups on each arginine residue at a particular position in the peptide; #MC, the number of cleavage sites that were not cleaved by the enzyme. (C) Schematic diagram of the ICP27 coding region, illustrating structural motifs, including the leucine-rich amino terminus (LRR), nuclear localization signal (NLS), RGG box RNA-binding motif (RGG), three predicted KH domains, and a zinc finger-like motif (CCHC). Specific amino acid positions of methylated arginine residues are shown within the RGG box motif.
ICP27 arginine substitution mutants are hypomethylated in vivo.
Because three arginines in the RGG box appear to be the major site of arginine methylation in ICP27, we concentrated on determining the function of methylation in the activities of ICP27. Viral recombinants in which these arginine residues were replaced with lysine, singly and in combination, were constructed. To determine if methylation of ICP27 was reduced in the substitution mutants, in vivo labeling with [3H]methionine was performed as described in the legend to Fig. 1. At 6 h after infection, ICP27 was immunoprecipitated from cell lysates. The amount of immunoprecipitated protein was determined by Western blot analysis, and the amount of labeled protein was determined by fluorography. In Fig. 3A, one such experiment is shown. This experiment was performed four times, and the relative amount of methylated protein to total immunoprecipitated protein was determined by densitometry. Figure 3B shows the average ratio for four experiments along with the standard deviations. The single substitutions R138K and R148K had relatively little effect on ICP27 methylation, while R150K produced about a twofold reduction (Fig. 3). However, the R138,150K double mutant and the R138,148,150K triple mutant showed a substantial reduction in methylation. Methylation of the d4-5 mutant (ΔRGG) was not detectable, as we saw previously. For clarity, the d4-5 mutant is referred to as ΔRGG hereafter.
FIG. 3.
The ICP27 arginine-to-lysine mutants are hypomethylated in vivo. (A) HeLa cells were infected with wild-type (WT) HSV-1 KOS, d4-5 (ΔRGG), 27LacZ, or the indicated ICP27 R-to-K viral mutants. Protein methylation was labeled in vivo from 4.5 to 8 h after infection with l-[methyl-3H]methionine. ICP27 was immunoprecipitated, resolved by SDS-PAGE, and transferred to nitrocellulose. Total ICP27 protein was determined by Western blot analysis, and methylated ICP27 was determined by fluorography. (B) Densitometry was used to quantify the bands. The average amount of methylated ICP27/total ICP27 is plotted for each virus. Error bars represent the standard errors of the means (n = 4).
ICP27 arginine-to-lysine mutants display a defect in growth.
To determine the growth properties of the RGG box substitution mutants, one-step growth experiments were performed. The single substitution mutants showed a somewhat delayed growth cycle, but final titers at 16 h were just about fivefold lower than that for wild-type HSV-1 (Fig. 4A). The R150K mutant showed the most delayed phenotype, and this mutant also had a smaller plaque size (Fig. 4D). The R138,150K double mutant and the R138,148,150K triple mutant were delayed relative to wild-type HSV-1, and final titers were about 10-fold lower (Fig. 4B). Smaller plaques were also seen for these mutants (Fig. 4D). The R150K, R138,150K, and R138,148,150K mutants were rescued, and the resulting recombinants were compared to N-YFP-ICP27 in one-step growth curves. N-YFP-ICP27 is an ICP27 N-terminally tagged viral recombinant that behaves like wild-type KOS (L. A. Johnson, L. Li, and R. M. Sandri-Goldin, unpublished data). The rescued viruses were indistinguishable from N-YFP-ICP27, indicating that the growth defects seen with the substitution mutants were due to the engineered mutations and not to mutations elsewhere in the genome.
Viral gene expression and DNA replication are decreased in infections with RGG box substitution mutants.
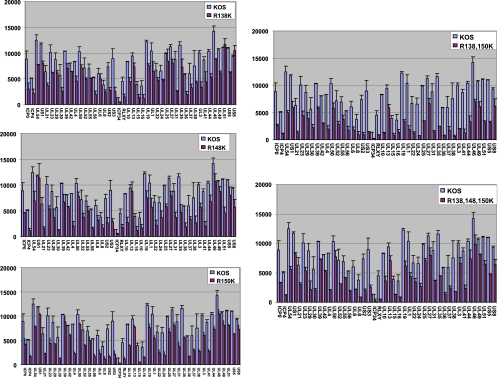
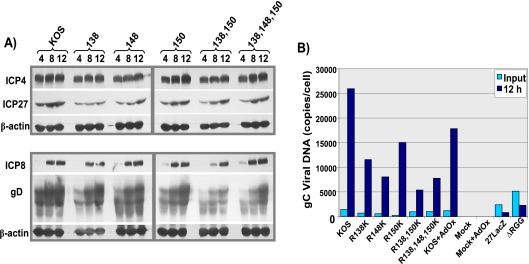
To determine the pattern of gene expression during infection with the arginine-to-lysine substitution mutants, microarray analysis was performed on total poly(A+) RNA isolated from cells infected with each of the viral mutants. Moderate decreases in the levels of all classes of viral transcripts were seen for the three single mutants (Fig. 5, left panels), and more notable decreases in some early and late transcript levels were seen for the double and triple mutants (Fig. 5, right panels). There was little effect on the expression of the immediate early protein ICP4 (Fig. 6A). ICP27 protein expression was slightly reduced compared to that of the wild type at 4 h after infection but was similar to wild-type levels by 8 and 12 h after infection for all the mutants (Fig. 6A). Expression of the early protein ICP8 appeared to be affected only during R138,150K mutant infection, in which ICP8 expression was delayed (Fig. 6A). The late protein gD was more affected than ICP27 or ICP8, in that expression was delayed and reduced, and this was most apparent in the double and triple mutants (Fig. 6A). DNA replication was also reduced relative to that for wild-type KOS, in that viral DNA copies per cell were found to be about 2.5 to 5 times lower for the mutants, as measured by quantitative real-time PCR of the glycoprotein C (gC) locus (Fig. 6B). Thus, the delayed growth kinetics observed in the one-step growth curves resulted from reduced global viral transcription and DNA replication in the substitution mutants. To determine if reduced DNA replication was due to hypomethylation, we also measured DNA copy number in cells infected with KOS in the presence of the methylation inhibitor AdOx. This inhibitor is a general inhibitor of methylation and has been shown to reduce arginine methylation to about 50% of the level seen in the absence of the inhibitor (24). DNA copy number was reduced in the presence of this drug, but not to the extent seen with the arginine substitution mutants. This does suggest, however, that the arginine substitution mutants are impaired because of hypomethylation.
FIG. 5.
Global transcription of the ICP27 arginine-to-lysine viral mutants is decreased. HeLa cells were infected with wild-type HSV-1 KOS or the indicated ICP27 arginine-to-lysine viral mutants. At 6 h after infection, total poly(A+) RNA was purified and microarray analysis was performed against an array of HSV-1 transcript-specific probes as described previously (12, 15). Each experiment was performed three times, and hybridizations were performed in duplicate for each experiment. Average values for each transcript are plotted, and error bars represent the standard errors of the means.
FIG. 6.
Protein expression profile and DNA replication. (A) HeLa cells were infected with wild-type (WT) HSV-1 KOS or the indicated ICP27 arginine-to-lysine viral mutants. Cell lysates were collected at 4, 8, and 12 h after infection. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose, and Western blot analysis was performed. (B) HeLa cells were mock infected or were infected with WT HSV-1 KOS, ΔRGG, 27LacZ, or the indicated ICP27 arginine-to-lysine viral mutants. The general methylation inhibitor AdOx was added to the indicated samples. At 1 h and 12 h postinfection, DNA was purified from cell lysates by phenol-chloroform extraction. Quantitative real-time PCR was performed with a probe for the gC gene to determine viral DNA copy number.
Hypomethylation of ICP27 reduces its stability.
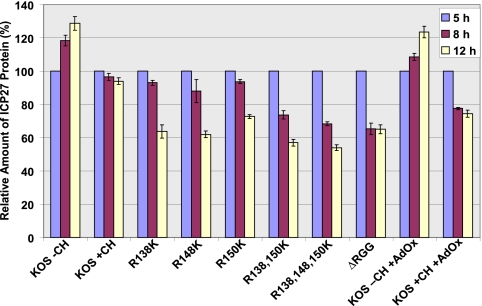
To determine if methylation affects the stability of ICP27, protein stability was measured during viral infection. Cycloheximide was added to cells infected with either HSV-1 KOS or the substitution mutants or to KOS-infected cells treated with AdOx. Protein levels were measured by Western blot analysis on samples harvested 5 h after infection, before the addition of cycloheximide, and 8 and 12 h after infection. The experiment was performed three times, and protein levels were quantified by densitometry. In the absence of cycloheximide, ICP27 levels increased during the course of the experiment, as expected (Fig. 7). In KOS-infected cells treated with cycloheximide, it was found that ICP27 was very stable over the course of the experiment. In contrast, the arginine substitution mutants showed reduced stability, with more-pronounced reductions seen with the R138,150K and R138,148,150K mutants. Stability of the ΔRGG mutant ICP27 was also reduced. Similarly, ICP27 stability was reduced in the presence of AdOx, suggesting that the decreased stability was due to hypomethylation and that arginine methylation of ICP27 may enhance its stability. Importantly, we saw no effect on the stability of the cellular protein actin, which was used as a loading control and which is not methylated (data not shown); thus, the effect of AdOx was specific and not an indirect effect.
FIG. 7.
ICP27 arginine-to-lysine mutant proteins are less stable than wild-type (WT) HSV-1 ICP27. HeLa cells were infected with WT HSV-1 KOS, ΔRGG, or the indicated ICP27 arginine-to-lysine viral mutants. The general methylation inhibitor AdOx was added 2 h after infection, and the translation inhibitor cycloheximide (CH) was added 5 h after infection to the indicated samples. Cell lysates were collected 5 h after infection, and proteins were resolved by SDS-PAGE and transferred to nitrocellulose. ICP27 and β-actin were identified by Western blot analysis using monoclonal antibodies. Densitometry analysis was used to quantify the bands, and the relative amount of ICP27 was normalized against the loading control β-actin. The amount of ICP27 present 5 h after infection was set to 100%. Plotted are the averages of three experiments. Error bars represent standard errors of the means.
Replication compartment formation is delayed during infection with ICP27 arginine substitution mutants.
Because viral gene expression and DNA replication were reduced in infections with the arginine substitution mutants, we monitored the formation of viral replication compartments by immunofluorescence staining for ICP4, a marker of viral replication compartments (8, 18, 42). Whereas replication compartments were clearly seen to form by 8 h after infection with HSV-1 KOS (Fig. 8), full-blown replication compartments were not seen until 12 h after infection with the R148K, R150K, R138,150K, and R138,148,150K mutants. Smaller prereplication sites were formed at 8 h with these mutants (Fig. 8), similar to what is observed with the ICP27 null mutant 27LacZ. In infections with ΔRGG, replication compartments were mostly restricted to prereplication sites, even at 12 h after infection. While some cells infected with wild-type KOS in the presence of AdOx were found to have only prereplication sites, several cells in each field observed did show replication compartment formation by 8 h after infection. This may reflect the fact that methylation is reduced by only about 50% in the presence of this inhibitor.
Arginine methylation of ICP27 occurs primarily in the nucleus and increases during infection.
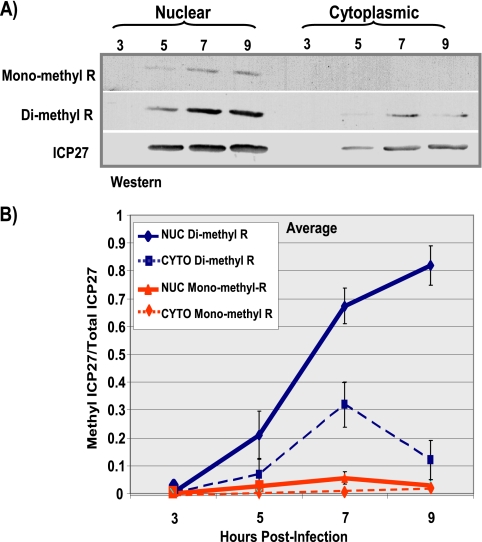
To monitor ICP27 methylation during infection, a time course experiment was performed. ICP27 was immunoprecipitated from nuclear and cytoplasmic fractions from KOS-infected cells at different times after infection (Fig. 9). Western blots were probed with monoclonal antibodies specific for monomethylarginine and dimethylarginine (Fig. 9A). The ratio of methylated ICP27 to the total amount of ICP27 was greater in the nuclear fractions than in the cytoplasmic fractions, and the amount of methylated ICP27 increased over the course of infection (Fig. 9B). This further suggests that PRMT1, a nuclear methyltransferase with a strong preference for GAR motifs (2), may be the major enzyme responsible for methylation of ICP27.
FIG. 9.
ICP27 protein-arginine methylation changes over time. HeLa cells were infected with wild-type HSV-1 KOS virus. Infected-cell lysates were collected 3, 5, 7, and 9 h after infection, and ICP27 was immunoprecipitated. Samples were resolved by SDS-PAGE and transferred to nitrocellulose. (A) Western blot analysis was performed using monoclonal antibodies to ICP27, the monoclonal antibody 21C7 (AbCam), which is specific for dimethylarginine, and monoclonal antibody 16B11 (AbCam), which is specific for monomethylarginine. (B) Densitometry analysis was performed to quantify the bands. Plotted are the average ratios of methylated ICP27/total ICP27 as a function of time postinfection. Error bars represent standard errors of the means (n = 3). NUC, nuclear; CYTO, cytoplasmic.
Hypomethylation of ICP27 affects its cellular localization.
To determine if hypomethylation of ICP27 affects its export to the cytoplasm, we performed immunofluorescence assays on cells infected with either HSV-1 KOS or the arginine substitution mutants and on cells infected with KOS in the presence of AdOx (Fig. 10). In wild-type KOS-infected cells, cytoplasmic ICP27 begins to be seen by 5 h after infection. ICP27 export occurs even earlier in ΔRGG-infected cells, such that ICP27 is predominantly cytoplasmic by 5 h (Fig. 10). Pronounced cytoplasmic fluorescence was also seen with the arginine substitution mutants, which appeared to be similar to ΔRGG. Hypomethylation of ICP27 in the presence of AdOx also resulted in much more rapid export of ICP27, with considerable cytoplasmic fluorescence seen by 4 h after infection. A similar result was seen for the cellular shuttling protein hnRNP A2 in the presence of AdOx, which has been previously reported (23). This result was reproduced as a control in the experiment shown in the right panels of Fig. 10B.
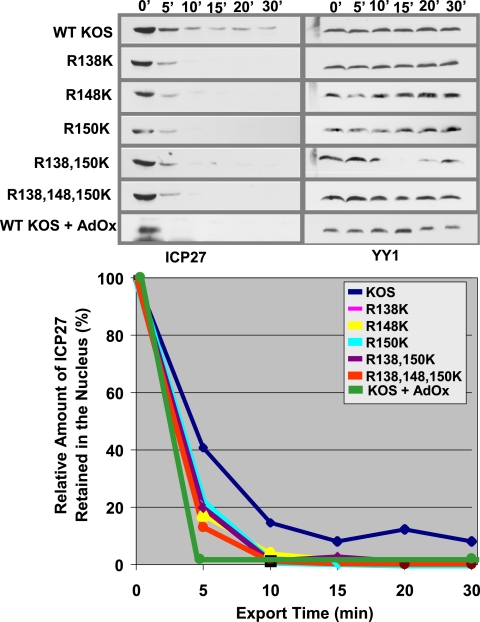
To confirm that export of ICP27 is affected by hypomethylation and to measure the approximate rate of export relative to that for the wild-type protein, in vitro nuclear export assays were performed 5 h after infection as we described previously (6, 7). In these assays, we monitor the amount of nuclear ICP27 that remains after the initiation of the export assay. Wild-type ICP27 was exported efficiently, with about 90% of the protein exiting the nucleus by 15 min after the initiation of the assay (Fig. 11). This result is similar to what we have reported previously (6, 7). Export of the arginine substitution mutant proteins was even more rapid, with nearly 100% of ICP27 having left the nucleus by 10 min after the start of the assay (Fig. 11). ICP27 from KOS-infected cells treated with AdOx was exported completely within the first 5 min. These results confirm that arginine methylation regulates the export of ICP27 and that, under conditions of hypomethylation, ICP27 is rapidly exported to the cytoplasm.
FIG. 11.
ICP27 arginine-to-lysine mutant proteins are exported from the nucleus to the cytoplasm faster than wild-type (WT) ICP27. RSF cells were infected with either WT HSV-1 KOS or the indicated ICP27 arginine-to-lysine viral mutants. In vitro nuclear export assays were performed 5 h after infection as described previously (6, 7). The assays were stopped at the indicated times. Western blot analysis was performed with anti-ICP27 and anti-YY1 monoclonal antibodies. Western blots were scanned and quantified by densitometry. The export assays were performed three times, and a representative experiment is shown.
DISCUSSION
Protein arginine methylation is a posttranslational modification that has been shown to affect a protein's ability to recognize its binding partners and that may regulate its activities (2). For shuttling RNA binding proteins, arginine methylation has been shown to modulate nuclear-cytoplasmic transport (2, 3, 9, 16, 17, 19, 23, 34, 47). For example, many of the RNA binding proteins in Saccharomyces cerevisiae that contain GAR motifs shuttle between the nucleus and cytoplasm. Two of these proteins, Npl3 and Hrp1, have been shown to be trapped in the nucleus in cells lacking the primary type of PRMT1 in yeast, which is called Hmt1 (34).
Here we showed that HSV-1 shuttling protein ICP27 is methylated primarily on three arginine residues within its RGG box RNA binding motif. Mutation of these residues to lysine, which, while a conservative change with respect to charge, is not one that results in a substrate for arginine methyltransferases, resulted in hypomethylation of ICP27. Hypomethylation of ICP27 had two major effects, decreased protein stability (Fig. 7) and an earlier and more rapid export of ICP27 from the nucleus to the cytoplasm (Fig. 11). The same effects were observed with the methylase inhibitor AdOx. These results strongly suggest that it is the hypomethylation of ICP27, rather than structural changes that may have occurred in the RGG box associated with mutating arginines to lysines, that is responsible for the reduced stability and more-rapid nuclear export of ICP27. This is further supported by results from nuclear magnetic resonance studies on the RGG box region of ICP27. In studies to be published elsewhere, we found no discernible structural differences in the triple-substitution mutant compared to wild-type ICP27 (K. Corbin-Lickfett, S. K. Souki, M. Cocco, and R. M. Sandri-Goldin, unpublished data).
It is interesting that a phenotype of more-rapid export to the cytoplasm was seen when ICP27 was hypomethylated, in contrast to what has been seen in yeast for the proteins Npl3 and Hrp1, which become confined to the nucleus. At present we cannot explain the difference with ICP27, but it might be due to the effect of hypomethylation on the protein interactions that ICP27 undergoes. Disrupting these interactions may prevent ICP27 from remaining in the nucleus. It has also been shown that the Epstein-Barr virus protein EBNA1 is methylated in vivo by PRMT1 (35). EBNA1 is important for replication and mitotic segregation of Epstein-Barr virus genomes in latently infected cells and also for the activation of the transcription of several viral latency genes. Hypomethylation of EBNA1 either by substitution mutations or treatment with AdOx altered its localization such that EBNA1 was seen to form rings around the nucleoli (35). Thus, it is possible that the disruption of some protein interactions caused by hypomethylation results in the altered cellular localization of proteins that undergo methylation.
The present study also showed that viral recombinants harboring the arginine mutations were impaired in virus replication and gene expression. Replication compartment formation was also delayed. The more rapid export of ICP27 from the nucleus may mean that it is unable to engage in a number of its nuclear protein interactions, which contribute to viral gene expression, and it may also mean that at least some of ICP27's protein interactions are regulated by methylation. We are currently investigating ICP27's ability to interact with the cellular splicing factor serine-arginine protein kinase 1 (SRPK1) and the cellular export adaptor protein Aly/REF, because these interactions have been mapped to the region encompassing the RGG box (7, 32). Results to be published elsewhere indicate that the interaction of ICP27 with SRPK1 and Aly/REF was altered under hypomethylation conditions. Studies are also ongoing to determine if methylation of the RGG box affects RNA binding by ICP27 during viral infection. For example, arginine methylation has been shown to alter the DNA binding specificity of some cellular transcription factors (5, 43).
These studies have shown that arginine methylation of ICP27 serves to regulate its export from the nucleus and also affects its biological activities. Under conditions of ICP27 hypomethylation, viral replication and gene expression are impaired, which may result from earlier and faster ICP27 nuclear export. Arginine methylation may serve as a regulatory switch for at least some of ICP27's numerous activities.
Supplementary Material
Acknowledgments
We thank Steve Rice for providing viral mutants, William Rigby for hnRNP A2 antibody, and Gayathri Devi-Rao for assistance with the microarray experiments.
This work was supported by National Institute of Allergy and Infectious Diseases grants AI21515 and AI61397.
Footnotes
Published ahead of print on 25 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bedford, M. T. 2007. Arginine methylation at a glance. J. Cell Sci. 1204243-4246. [DOI] [PubMed] [Google Scholar]
- 2.Bedford, M. T., and S. G. Clarke. 2009. Protein arginine methylation in mammals: who, what and why. Mol. Cell 331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford, M. T., and S. Richard. 2005. Arginine methylation: an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, F.-M., J. Cote, M.-C. Boulanger, P. Cleroux, F. Bachand, C. Auxtexier, and S. Richard. 2002. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 159957-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boisvert, F.-M., A. Rhie, S. Richard, and A. J. Doherty. 2005. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 41834-1841. [DOI] [PubMed] [Google Scholar]
- 6.Chen, I. B., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 793949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, I. B., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the export factor Aly/REF to direct herpes simplex virus 1 intronless RNAs to the TAP export pathway. J. Virol. 7612877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai-Ju, J. Q., L. Li, L. A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 803567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, D. M., K. A. Marfatia, E. B. Crafton, X. Zhang, and A. H. Corbett. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 2777752-7760. [DOI] [PubMed] [Google Scholar]
- 10.Iacovides, D. C., C. C. O'Shea, J. Oses-Prieto, A. Burlingame, and F. McCormick. 2007. Critical role for arginine methylation in adenovirus-infected cells. J. Virol. 8113209-13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansson, M., S. T. Durant, E. Cho, S. Sheahan, M. Edelmann, B. Kessler, and N. B. La Thange. 2008. Arginine methylation regulates the p53 response. Nat. Cell Biol. 101431-1439. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, L. A., and R. M. Sandri-Goldin. 2009. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 831184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lengyel, J., C. Guy, V. Leong, S. Borge, and S. A. Rice. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 7611866-11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, L., L. A. Johnson, J. Q. Dai-Ju, and R. M. Sandri-Goldin. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS ONE 3e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, Q., and G. Dreyfuss. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 152800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukong, K. E., and S. Richard. 2004. Arginine methylation signals mRNA export. Nat. Struct. Mol. Biol. 11914-915. [DOI] [PubMed] [Google Scholar]
- 18.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 712390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride, A. E., J. T. Cook, E. A. Stemmler, K. L. Ruteledge, K. A. McGrath, and J. A. Rubens. 2005. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 28030888-30896. [DOI] [PubMed] [Google Scholar]
- 20.Mears, W. E., V. Lam, and S. A. Rice. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 707445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between the nucleus and cytoplasm. Virology 242128-137. [DOI] [PubMed] [Google Scholar]
- 23.Nichols, R. C., X. W. Wang, J. Tang, B. J. Hamilton, F. A. High, H. R. Herschman, and W. F. C. Rigby. 2000. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256522-532. [DOI] [PubMed] [Google Scholar]
- 24.Pahlich, S., R. Zakaryan, and H. Gehring. 2006. Protein arginine methylation: cellular functions and methods of analysis. Biochim. Biophys. Acta 17641890-1903. [DOI] [PubMed] [Google Scholar]
- 25.Phelan, A., M. Carmo-Fonseca, J. McLauchlan, A. I. Lamond, and J. B. Clements. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 909056-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 641704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, S. A., V. Lam, and D. M. Knipe. 1993. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J. Virol. 671778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandri-Goldin, R. M. 1998. ICP27 mediates herpes simplex virus RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandri-Goldin, R. M. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 135241-5256. [DOI] [PubMed] [Google Scholar]
- 30.Sandri-Goldin, R. M., and M. K. Hibbard. 1996. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum and an the C terminus appears to be required for this interaction. J. Virol. 70108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of HSV-1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 696063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciabica, K. S., Q. J. Dai, and R. M. Sandri-Goldin. 2003. ICP27 interacts with SRPK1 to mediate HSV-1 inhibition of pre-mRNA splicing by altering SR protein phosphorylation. EMBO J. 221608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekulovich, R. E., K. Leary, and R. M. Sandri-Goldin. 1988. The herpes simplex virus type 1 α protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J. Virol. 624510-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shire, K., P. Kapoor, M. Ng, T. Hing, N. Sivachandran, T. Nguyen, and L. Frappier. 2006. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 805261-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, I. L., M. A. Hardwicke, and R. M. Sandri-Goldin. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 18674-86. [DOI] [PubMed] [Google Scholar]
- 37.Smith, I. L., and R. M. Sandri-Goldin. 1988. Evidence that transcriptional control is the major mechanism of regulation for the glycoprotein D gene in herpes simplex virus type 1-infected cells. J. Virol. 621474-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 719188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 749916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, A., G. Devi-Rao, M. K. Rice, L. W. Gary, D. C. Bloom, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2004. The TATGARAT box of the HSV-1 ICP27 gene is essential for immediate early expression but not critical for efficient replication in vitro or in vivo. Virus Genes 29335-343. [DOI] [PubMed] [Google Scholar]
- 41.Sun, A., G. V. Devi-Rao, M. K. Rice, L. W. Gary, D. C. Bloom, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2004. Immediate-early expression of the herpes simplex virus type 1 ICP27 transcript is not critical for efficient replication in vitro or in vivo. J. Virol. 7810470-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor, T. J., E. E. McNamee, C. Day, and D. M. Knipe. 2003. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology 309232-247. [DOI] [PubMed] [Google Scholar]
- 43.Valentini, S. R., V. H. Weiss, and P. A. Silver. 1999. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA 5272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie, B., C. F. Invernizzi, S. Richard, and M. A. Wainberg. 2007. Arginine methylation of the human immunodeficiency virus type Tat protein by PRMT6 negatively affects Tat interactions with both cyclin T1 and Tat transactivation region. J. Virol. 814226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, W. C., G. V. Devi-Rao, P. Ghazal, E. K. Wagner, and S. J. Triezenberg. 2002. General and specific alterations in programming of global viral gene expression during infection by VP16 activation-deficient mutants of herpes simplex virus type 1. J. Virol. 7612758-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, M. C., F. Bachand, A. E. McBride, S. Komili, J. M. Casolari, and P. A. Silver. 2004. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 182024-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yun, C. Y., and X. D. Fu. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150707-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, C., and D. M. Knipe. 2002. Association of herpes simplex virus 1 ICP8 and ICP27 with cellular RNA polymerase II holoenzyme. J. Virol. 765893-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.