Abstract
The mammalian interferon (IFN) signaling pathway is a primary component of the innate antiviral response. As such, viral pathogens have devised multiple mechanisms to antagonize this pathway and thus facilitate infection. Dengue virus (DENV) encodes several proteins (NS2a, NS4a, and NS4b) that have been shown individually to inhibit the IFN response. In addition, DENV infection results in reduced levels of expression of STAT2, which is required for IFN signaling (M. Jones, A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs, J. Virol. 79:5414-5420, 2005). Translation of the DENV genome results in a single polypeptide, which is processed by viral and host proteases into at least 10 separate proteins. To date, no single DENV protein has been implicated in the targeting of STAT2 for decreased levels of expression. We demonstrate here that the polymerase of the virus, NS5, binds to STAT2 and is necessary and sufficient for its reduced level of expression. The decrease in protein level observed requires ubiquitination and proteasome activity, strongly suggesting an active degradation process. Furthermore, we show that the degradation of but not binding to STAT2 is dependent on the expression of the polymerase in the context of a polyprotein that undergoes proteolytic processing for NS5 maturation. Thus, the mature form of NS5, when not expressed as a precursor, was able to bind to STAT2 but was unable to target it for degradation, establishing a unique role for viral polyprotein processing in providing an additional function to a viral polypeptide. Therefore, we have identified both a novel mechanism by which DENV evades the innate immune response and a potential target for antiviral therapeutics.
Dengue virus (DENV) is the causative agent of dengue fever, dengue hemorrhagic fever, and dengue shock syndrome (2). The virus and its arthropod vector, Aedes aegypti (21), are endemic to over 100 countries around the world including the United States. It is responsible for an estimated 50 million to 100 million infections annually, with over 24,000 deaths resulting, predominantly in children under 14 years of age (25). The virus exists in four serotypes (DEN1 to DEN4) and is grouped into the flavivirus genus along with a number of additional human pathogens including West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). These viruses have a positive-strand, nonsegmented genome of ∼11 kb (5, 55), the organization of which is highly conserved, encoding, in order, three structural proteins (C, M, and E), followed by seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5). The genome is translated as a single endoplasmic reticulum (ER)-bound polyprotein, which is co- and posttranslationally processed by both viral (NS2b and NS3) and cellular proteases (8, 44).
A critical component of the human antiviral response is the type 1 interferon (IFN) pathway, which acts to delay virus replication and to stimulate the activation of antiviral effector cells. The production of IFN is initiated upon the initial detection of virus by pattern recognition receptors such as RIG-I-like receptors and Toll-like receptors, which bind viral pathogen-associated molecular patterns including double-stranded RNA and 5′-phosphate-containing single-stranded RNA. These pattern recognition receptors, when activated, convey their signal through the transcription factors IFN regulatory factor 3, NF-κB, and AP1, which act in concert to induce the expression of IFN. Secreted IFN binds to IFN receptors found on the same cell or nearby cells and stimulates the IFN signaling pathway. The type I IFN receptor (IFNAR) is a heterodimer composed of two chains (IFNAR1 and IFNAR2) (4, 37). IFN binding stimulates receptor heterodimerization, and this triggers the activation of the Janus kinases Tyk2 and Jak1, which are associated with the IFNAR cytoplasmic tails (9, 10, 13). Phosphorylated tyrosine residues on IFNAR act as a docking site, binding STAT2 (36, 56), which in turn becomes phosphorylated on its Y-690 (22) residue. Active STAT2 recruits STAT1 (24, 43, 49), which is subsequently phosphorylated on the Y-701 residue (50). Tyrosine-phosphorylated STAT1 and STAT2 heterodimerize and complex with IFN regulatory factor 9 to form the transcription factor complex ISGF3 (17, 27). ISGF3 translocates to the nucleus and is recruited to specific genetic elements, termed IFN-sensitive response elements (ISREs), located within upstream promoter regions of IFN-stimulated genes (27). ISGF3 activation results in increased levels of expression of over 100 different proteins that function to create an antiviral state within the cell (1, 6, 41, 46), thus inhibiting viral replication.
The overall importance of the IFN signaling pathway and the ISGF3 complex in particular has been demonstrated using mice deficient in any of the three ISGF3 components. These animals manifest an increased susceptibility to viral infection. Interestingly, STAT1/STAT2 double knockouts have a more severe phenotype than either single knockout alone, suggesting that STAT2 may have additional roles in the antiviral response independent of its ISGF3-related activity (39). In the case of DENV, mice lacking a functional STAT1 protein show higher viral titers at early points in infection, although the virus is eventually cleared through STAT1-independent mechanisms, indicating an importance for IFN signaling in controlling initial virus replication and early clearance (48).
Additional evidence for the vital role of ISGF3 factors in preventing infection lies in the fact that many successful human-pathogenic RNA viruses encode proteins which target ISGF3 activity and thus block IFN-mediated signaling. Signaling antagonists come in many forms, but the majority identified thus far act directly on the ISGF3 complex. The paramyxovirus family, for example, encodes several proteins, each of which inhibits the pathway using different methods. These include nuclear or cytoplasmic sequestration of STAT1, as is the case for Nipah virus W (NiV-W) and NiV-V proteins, with the concomitant cytoplasmic sequestration of STAT2 by NiV-V (40, 45). Alternatively, the V proteins of other paramyxoviruses may target STAT1 or STAT2 for degradation, as is the case for the human parainfluenza virus 5-V protein and human parainfluenza virus 2-V, respectively (12, 38). The C proteins of several paramyxoviruses have also been shown to inhibit IFN signaling through specific interactions with STATs (18, 20, 47).
In an analogous fashion, flaviviruses are known to encode multiple IFN signaling antagonists. In the case of Kunjin virus (a strain of WNV), five of the seven nonstructural proteins (NS2a, NS2b, NS3, NS4a, and NS4b) were shown to inhibit STAT translocation to the nucleus and subsequent IFN-dependent reporter activity when individually expressed in plasmid-transfected cells (32). In contrast, the NS5 proteins of JEV and TBEV were shown to block STAT1 phosphorylation (3, 30). For DENV, the NS2a, NS4a, and NS4b proteins were shown to antagonize IFN signaling when individually expressed, with NS4b being more potent and preventing STAT1 phosphorylation (35). In addition, it was also found that DENV infection results in a loss of STAT2 expression (26), but the viral factors responsible for this effect were not identified. In this report, we demonstrate that DENV NS5 binds STAT2 and inhibits IFN-dependent signaling. In addition, we also show that in the absence of any additional viral sequence, DENV NS5 targets STAT2 for proteasome-mediated degradation but only when NS5 is expressed as a proteolytically processed precursor, indicating that NS5 expression as part of a viral polyprotein is required for potent IFN signaling antagonism by DENV. The ability of multiple flavivirus proteins to block IFN signaling illustrates the major role that this pathway plays in controlling infections with these viruses.
MATERIALS AND METHODS
Cells and viruses.
293T cells, wild-type Vero (wtVero) cells, and U6A cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Clonal U6A cells stably expressing STAT2-green fluorescent protein (GFP) were generated by cotransfection of pCAGGS-STAT2-GFP and pCDNA-zeo and subsequently selecting for zeocin resistance and GFP expression. Vero cells expressing the DEN1 replicon (Western Pacific strain) (42) were grown in DMEM-10% fetal calf serum with 0.5 mg G418. A recombinant Newcastle disease virus (NDV) culture expressing GFP was grown in 10-day-old embryonated chicken eggs. High-titer stocks of DENV (DEN2 16681) were obtained by passage in C6/36 cells. All transfections were performed using Lipofectamine 2000 (Invitrogen).
Plasmids and antibodies.
All DENV protein-encoding plasmids and the tobacco etch virus (TEV) protease NIa were generated in the pCAGGS (chicken β-actin promoter) background. Primer sequences used for the generation of these constructs are available upon request. hemagglutinin (HA)-NiV-V plasmid and plasmids encoding human STAT1 and STAT2 were a kind gift from Megan Shaw. The ISRE-54-chloramphenicol acetyltransferase (CAT) reporter and plasmids pCAGGS-Firefly luciferase and pCAGGS-GFP were kind gifts from Luis Martinez-Sobrido. pCDNA-zeo was a kind gift from Ben tenOever. Plasmid pCAGGS, encoding the OTU domain of the Crimean Congo hemorrhagic fever L protein; an OTU mutant; and UBP43 were kind gifts from Natalia Frias-Staheli. An HA-ubiquitin-expressing plasmid was obtained from Domenico Tortorella. Antibodies utilized for this study include those raised against HA (Sigma), FLAG (Sigma), STAT1 (BD), STAT2 (SC476; Santa Cruz), GFP (Sigma), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Research Diagnostics Incorporated), tubulin (Sigma), β-actin (Sigma), and ubiquitin (Cell Signaling Technologies). Rabbit antibody raised against DEN2 NS5 was generated by the serial injection of the bacterially expressed and purified DEN2 NS5 protein.
IFN reporter assay.
293T cells were cotransfected with an HA-tagged plasmid encoding various viral proteins, the IFN-inducible CAT reporter (ISG54-CAT), and a plasmid constitutively expressing the firefly luciferase protein (pCAGGS-Firefly luciferase). Twenty-four hours posttransfection, cells were treated with 1,000 U/ml of universal IFN (PBL). Twenty-four hours posttreatment, cells were lysed and measured for CAT activity. Induction of the sample is calculated as the CAT activity of the treated sample normalized to the firefly luciferase value of that sample, which is then divided by the normalized value of untreated empty-HA-transfected cells. P values were calculated using a two-tailed Student's t test.
NDV-GFP bioassay.
wtVero cells or Vero cells stably expressing the DEN1 replicon were treated with the stated amounts of either universal IFN (PBL) or IFN-γ (Calbiochem). Twenty-four hours posttreatment, cells were challenged with NDV-GFP, and subsequent fluorescence images were obtained 14 h postinfection (p.i.).
IP assays.
293T cells were transfected with an HA-tagged plasmid encoding the viral protein and both STAT1-FLAG and STAT2-FLAG. Lysis, immunoprecipitation (IP), and washes were performed using buffer containing 50 mM Tris (pH 7.5), 280 mM NaCl, 0.2 mM EDTA, 2 mM EGTA, 0.5% NP-40, 10% glycerol, 1 mM dithiothreitol, and 1 mM sodium orthovanadate.
DENV infection.
Vero cells were infected with 10 multiplicities of infection (MOI) of DEN2 strain 16681 virus for 1 h. Cells were then washed and subsequently maintained in DMEM-10% fetal calf serum at 37°C. Twenty-four hours p.i., cells were lysed and analyzed via Western blotting. U6A cells stably expressing STAT2-GFP (U6A-STAT2-GFP) were infected at an MOI of 40 to compensate for abnormally high levels of STAT2-GFP. After 1 h of infection, cells were washed and maintained in DMEM at 37°C. For time course experiments, the time point of 0 h p.i. represents the point at which infectious medium is removed and DMEM is added.
STAT2 degradation experiments.
Analysis of the degradation of endogenous STAT2 was accomplished by the transfection of virus-encoded GFP-tagged plasmids into 293T cells. Twenty-four hours posttransfection, cells were sorted by fluorescence-activated cell sorter (FACS) analysis to obtain GFP-positive cells, which were then subsequently lysed for analysis. Where densitometry analyses of the levels of STAT2 and NS5 are included, values calculated are relative to the levels in lane 1 for each Western blot, with a value of 1 in the case of NS5 levels being indicative of no detection (background levels). When NS5 was expressed as a precursor, values for NS5 represent levels of the cleaved form. Analysis of the degradation of overexpressed STAT2 was performed in U6A cells (STAT2 deficient) using virus-encoded HA-tagged plasmids cotransfected with STAT2-FLAG and HA-ubiquitin. STAT1-GFP was also transfected in these experiments and was used as a negative control.
RESULTS
The nonstructural region of the DENV polyprotein reduces STAT2 expression levels.
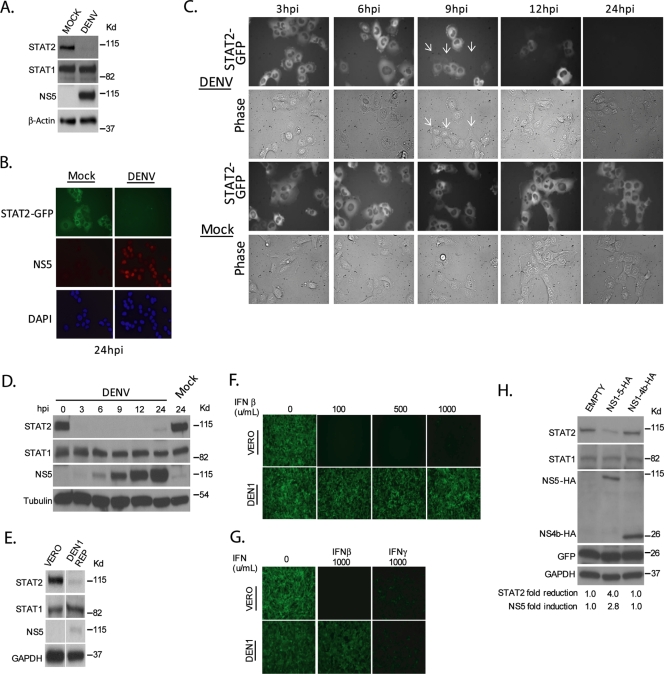
It was previously shown that DEN2 infection of K562 cells results in a decrease in levels of STAT2 expression (26). We have confirmed this finding using DEN2 virus-infected Vero cells (Fig. 1A) and infected U6A-STAT2-GFP cells (Fig. 1B). We next infected U6A-STAT2-GFP cells and monitored the loss of GFP signal over the course of the 24 h p.i. (Fig. 1C). Detectable differences in GFP intensity were observed in cells infected with DENV as early as 9 h p.i. (indicated by white arrows), and by 12 h p.i., a number of cells showed a complete loss of GFP signal. The rate of loss occurred more rapidly when we repeated the time course using Vero cells, where endogenous STAT2 levels were undetectable within 3 h p.i. (Fig. 1D). This decrease is due to the nonstructural region of the virus polyprotein, as it can be recapitulated using a DEN1 virus replicon that we have in our laboratory, which lacks the region encoding the structural proteins (Fig. 1E). We were unable to observe differences in STAT2 mRNA levels in DEN1 replicon-containing Vero cells compared to control Vero cells as measured by quantitative reverse transcription-PCR (ratio of Vero cells:DEN1 replicon without IFN treatment of 1 versus ratio of Vero cells:DEN1 replicon with IFN treatment of 7.5), suggesting that the loss of STAT2 expression is due to active degradation. In contrast, STAT1 levels were unaffected by DENV infection or in DENV replicon-containing cells (Fig. 1A, D, and E). These findings suggest both that the loss of STAT2 expression is conserved across the DENV serotypes and that it occurs independent of the structural proteins. Specificity for inhibition of STAT2 function in replicon-containing cells was evidenced after treatment with type I or type II IFN. The level of antiviral activity of type I IFN, which requires a functional STAT2, was greatly reduced in DENV replicon-containing cells, as evidenced by challenging IFN-treated cells with the IFN-sensitive virus NDV-GFP (Fig. 1F and G). However, type II IFN antiviral activity, which is mediated by STAT1 homodimers and does not require STAT2, was not dramatically affected by the presence of the DENV replicon (Fig. 1G), consistent with previous observations utilizing live virus (11).
FIG. 1.
Reduced levels of STAT2 in cells expressing the nonstructural region of the DENV polyprotein. (A) Vero cells were infected for 24 h with DENV at an MOI of 10 and subsequently lysed and examined by Western blotting. (B) U6A cells stably expressing STAT2-GFP were infected with DEN2 at an MOI of 40 for 24 h prior to fixation. Cells were then probed with antibody against NS5 and stained for DNA (DAPI [4′,6′-diamidino-2-phenylindole]). (C) U6A-STAT2-GFP cells were infected with DEN2 at an MOI of 40 and measured for a loss of GFP by live microscopy at the given time points. White arrows indicate cells in the infected samples at 9 h p.i. that have lost the GFP signal. (D) Vero cells were infected with DENV at an MOI of 10, subsequently lysed at the given time points, and examined by Western blotting. (E) Vero cells stably expressing a DEN1 replicon (NS1-5) were lysed and examined by Western blotting. (F) wtVero cells or Vero cells stably expressing a DEN1 replicon were treated with the indicated amounts of type I IFN (IFN-α/β) for 24 h. Cells were subsequently challenged with NDV-GFP and assayed via fluorescence microscopy for GFP expression at 14 h p.i. (G) Vero cells or Vero cells stably expressing a DEN1 replicon were treated with 1,000 units of type I (IFN-α/β) or type II (IFN-γ) IFN for 24 h. Cells were subsequently challenged with NDV-GFP and assayed via fluorescence microscopy for GFP expression at 14 h p.i. (H) 293T cells were cotransfected with plasmids expressing NS1-5-HA and GFP or NS1-4b-HA and GFP. Twenty-four hours posttransfection, cells were sorted by FACS for GFP-positive cells and subsequently lysed and examined via Western blotting. Densitometry analysis of the levels of STAT2 and NS5 are included at the bottom, and levels were calculated relative to the levels in lane 1, with the value of 1 in the case of NS5 levels being indicative of no detection (background levels).
DENV NS5 is required for reduced STAT2 expression.
To determine what viral polypeptide(s) within the DENV polyprotein was responsible for the reduced levels of STAT2 expression, we constructed a plasmid expressing the nonstructural region of the DEN2 polyprotein containing an HA tag at its carboxy terminus for easier detection (NS1-5-HA). This plasmid expresses the same nonstructural viral polypeptides as a DENV replicon; i.e., the presence of the DEN2 protease in the nonstructural polyprotein (NS2b-3) results in the proper processing of NS1-5-HA into NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5-HA polypeptides. However, in contrast to the replicon, the mRNA expressed by the NS1-5-HA plasmid is not replicated, since it lacks RNA signals required for transcription by the viral polymerase. This allows us to separate replicative functions of the nonstructural viral proteins from effects on STAT2 expression, since the levels of DENV proteins expressed by the plasmid do not depend on RNA replication. As expected, the transient expression of NS1-5-HA in 293T cells resulted in a loss of STAT2 (Fig. 1H, lane 2). This loss required NS5, since the expression of the region encoding NS1-4b-HA (all the nonstructural region of the DENV polyprotein except for NS5) did not affect STAT2 levels (Fig. 1H, lane 3).
DENV NS5 binds to STAT2.
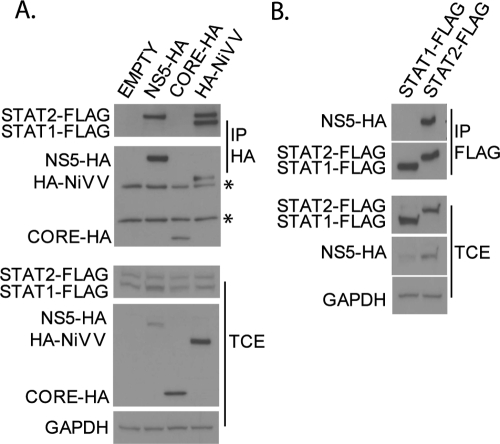
Several paramyxoviruses are known to express viral proteins that bind to STAT factors and target them for degradation (12, 19, 38, 52). We therefore tested whether DENV NS5 was able to interact with STAT2. To do this, we performed IP experiments with 293T cells using plasmids expressing FLAG-tagged STAT1 and STAT2 proteins cotransfected with DENV NS5-HA or controls (HA-tagged DENV core protein as a negative control and HA-tagged NiV-V as a positive control). IP against the HA tag on NS5 pulled down FLAG-tagged STAT2 but not STAT1 (Fig. 2A, lane 2). In contrast, both STAT1-FLAG and STAT2-FLAG are coprecipitated by the HA-NiV-V protein (Fig. 2A, lane 4), consistent with the known ability of this paramyxovirus protein to interact with both STAT1 and STAT2 (45). No interaction was detected with the negative control core-HA (Fig. 2A, lane 3). In addition, reversing the IP confirmed that NS5-HA interacted with STAT2-FLAG but not STAT1-FLAG (Fig. 2B). These results indicate that DENV NS5 specifically interacts with STAT2. Notably, analysis of levels of STAT2-FLAG in the total cell extracts suggests that NS5-HA alone fails to exert any noticeable effect on STAT2 expression levels (Fig. 2A).
FIG. 2.
DENV NS5 interacts with STAT2. (A) 293T cells were cotransfected with plasmids expressing FLAG-tagged STAT1 (STAT1-FLAG) and STAT2 (STAT2-FLAG) and empty plasmid (empty) or plasmid expressing HA-tagged DENV NS5 (NS5-HA), DENV core (CORE-HA), or NiV-V (HA-NiV-V) proteins. Lysates were then immunoprecipitated with anti-HA antibody (IP HA), and Western blotting was performed using anti-HA and anti-FLAG antibodies. Asterisks mark the heavy and light chains from the HA antibody. (B) 293T cells were cotransfected with plasmids expressing NS5-HA and either STAT1-FLAG or STAT2-FLAG. Lysates were then immunoprecipitated with anti-FLAG antibody (IP FLAG), and Western blotting was performed using anti-HA and anti-FLAG antibodies. TCE, total cell extracts were subjected to Western blotting using anti-HA, anti-FLAG, and anti-GAPDH antibodies.
Expression of NS5 as a proteolytically processed precursor results in reduced levels of STAT2.
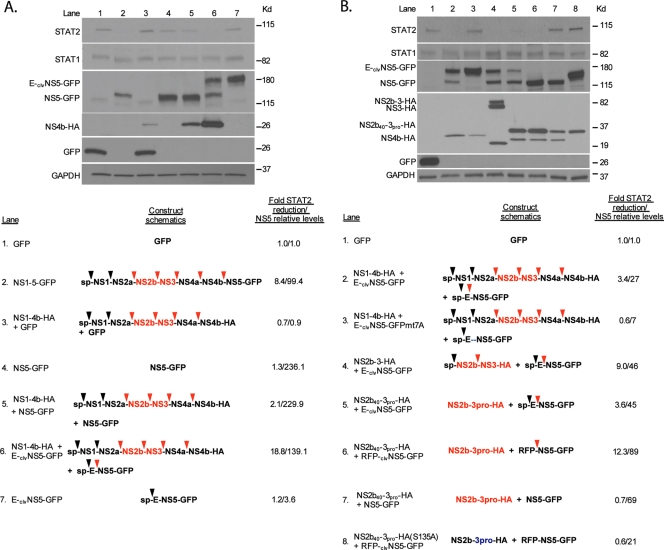
We next conducted experiments to confirm that the expression of DENV NS5 by itself does not result in reduced levels of endogenous STAT2 and to identify possible additional DENV-encoded factors participating in this reduction. For the easy detection of NS5-transfected cells, we fused GFP to the carboxy-terminal end of NS5. GFP-positive cells were then sorted by FACS analysis 24 h posttransfection and analyzed by Western blotting. Expression of the DENV NS1-5-GFP polyprotein in 293T cells resulted in reduced levels of expression of STAT2, as expected (Fig. 3A, lane 2), while the deletion of NS5 from this construct (NS1-4B-HA) eliminated the impact of this polyprotein on STAT2 expression, as described above (Fig. 1F, lane 3, and 3A, lane 3). The level of expression of NS5-GFP was also not sufficient to reduce levels of STAT2 expression, as STAT2 levels in the presence of NS5-GFP are comparable to those in the presence of GFP (Fig. 3A, lanes 1 and 4). We next attempted to complement the NS1-4b polypeptide in trans with NS5-GFP. Surprisingly, this did not result in a robust decrease in STAT2 levels (Fig. 3A, lane 5).
FIG. 3.
Expression of a precursor form of DENV NS5 cleaved by the DENV protease results in reduced STAT2 levels. (A) 293T cells were transfected with the indicated constructs. Twenty-four hours posttransfection, cells were sorted for GFP-positive cells by FACS, subsequently lysed, and examined via Western blotting using GFP-, HA-, STAT1-, STAT2-, and GAPDH-specific antibodies. Schematics of the transfected constructs are shown at the bottom. ORFs that contain an “sp” at the N terminus have a signal peptide which directs the entire polyprotein to the surface of the ER for translation. Black arrows and red arrows indicate cleavage sites for cellular and the DEN2 viral proteases, respectively. The DEN2 active protease is highlighted in red. Densitometry analysis of the levels of STAT2 and NS5 are included on the far right, and levels are calculated relative to the levels in lane 1, with a value of 1 in the case of NS5 levels being indicative of no detection (background levels). (B) Same as above (A). Mutation of the DENV protease recognition site at the N terminus of NS5 is indicated by the blue dashes. The DENV protease labeled in blue indicates a serine-to-alanine mutation within the catalytic site of the DENV protease.
One difference that may account for the loss of STAT2 seen when the DENV polypeptides are expressed by the polyprotein NS1-5-GFP but not seen when expressed by NS1-4b-HA plus NS5-GFP was that in the NS1-5-GFP construct, NS5 was expressed within the context of a polyprotein and underwent viral protease-mediated (NS2b-3) cleavage for its maturation (Fig. 3A, lane 2, third panel). We then attempted to complement NS1-4b-HA by expressing in trans NS5-GFP synthesized from a precursor cleaved by the NS2b-3 viral protease. For this purpose, we placed the DENV protease cleavage sequence present between the NS4b and NS5 open reading frames (ORFs) upstream of the NS5-GFP gene (clvNS5-GFP). We then fused clvNS5-GFP downstream of the DENV E protein (E-clvNS5-GFP). The expression of E-clvNS5-GFP together with NS1-4b-HA resulted in a matured cleaved NS5-GFP (Fig. 3A, lane 6, third panel). This coexpression also resulted in a robust decrease in levels of STAT2 expression (Fig. 3A, lane 6) similarly to the NS1-5-GFP cassette. No free matured NS5-GFP and no effects on STAT2 expression were detected when E-clvNS5-GFP was expressed in the absence of NS1-4b, suggesting that the processing of NS5-GFP by the viral protease was required for the loss of STAT2 expression (Fig. 3A, lane 7). Consistent with this, mutations in the E-clvNS5-GFP cleavage sequence that abrogated processing (E-clvNS5-GFP mt7A) prevented the reduction in STAT2 levels when the mutant protein was coexpressed with NS1-4b-HA (Fig. 3B, lane 3), indicating that cleavage of NS5 was required for the effects on STAT2. It therefore appears that the DENV viral protease is necessary for STAT2 degradation. This was confirmed by truncating the NS1-4b-HA cassette to express only NS2b-3-HA and even further truncating it to express the protease domain alone (NS2b40-3pro-HA) (7). Both forms of the protease, when coexpressed with E-clvNS5-GFP, could substitute for the NS1-4b ORF (Fig. 3B, lanes 4 and 5, respectively), indicating that the remainder of the nonstructural sequence (NS1, NS2a, NS4a, and NS4b) was not required for STAT2 degradation. However, as also expected from our previously reported data, the expression of NS5 from a cleaved precursor was required, as the coexpression of NS5-GFP with NS2b40-3pro-HA did not impact STAT2 levels (Fig. 3B, lane 7). To rule out a possible effect of E on the E-clvNS5-GFP construct, we fused clvNS5-GFP downstream of red fluorescent protein (RFP) (RFP-clvNS5-GFP). The coexpression of RFP-clvNS5-GFP with the protease domain NS2b40-3pro-HA resulted in efficient proteolytic cleavage of NS5-GFP and decreased STAT2 levels (Fig. 3B, lane 6). Active proteolytic activity was required, as the coexpression of a catalytically inactive protease [NS2b40-3pro-HA(S135A)] with RFP-clvNS5-GFP resulted in intact STAT2 protein levels (Fig. 3B, lane 8). In all cases, the impact on STAT2 levels was specific, as levels of STAT1 and GAPDH were consistently similar among all samples (Fig. 3).
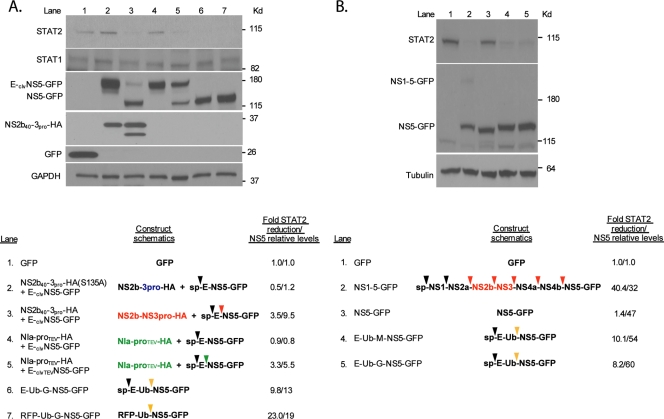
To address a potential role for the protease beyond the cleavage and maturation of NS5, we replaced the NS4b-5 cleavage signal with the TEV protease signal (clvTEVNS5-GFP) and fused this downstream of the E protein (E-clvTEVNS5-GFP). The cleavage and maturation of NS5-GFP by the TEV protease (NIa-proTEV) were sufficient to decrease the levels of endogenous STAT2, suggesting that the DENV protease does not have a role in STAT2 degradation beyond maturing NS5 (Fig. 4A, lane 5). This conclusion was confirmed by substituting the cleavage site between E or RFP and NS5-GFP by ubiquitin (E-Ub-G-NS5-GFP and RFP-Ub-G-NS5-GFP). Endogenous ubiquitin hydrolases cleave after the final two glycine residues of ubiquitin regardless of the amino acid in the P1′ position of the downstream fused protein. This results in the maturation of NS5-GFP containing an N-terminal glycine, as in the previous constructs cleaved by NS2b40-3pro or NIa-proTEV. The expression of these ubiquitin-containing fusion proteins resulted in the efficient proteolytic cleavage of NS5-GFP and in reduced STAT2 levels (Fig. 4A, lanes 6 and 7). As described above, this effect was STAT2 specific, as no impact on STAT1 and GAPDH expression levels was found (Fig. 4A).
FIG. 4.
Cleaved NS5 is sufficient for reduced STAT2 levels and does not require an N-terminal glycine. (A) 293T cells were transfected with the indicated constructs. Twenty-four hours posttransfection, cells were sorted for GFP-positive cells by FACS, subsequently lysed, subjected to 4-to-20% SDS-polyacrylamide gel electrophoresis, and examined via Western blotting using GFP-, HA-, STAT1-, STAT2-, and GAPDH-specific antibodies. The TEV protease and its cleavage site are indicated by the green text and arrow, respectively. Cleavage sequences targeted by endogenous deubiquitinases are noted by a yellow arrow. Densitometry analyses of the levels of STAT2 and NS5 are included at the bottom, and levels were calculated relative to the levels in lane 1, with a value of 1 in the case of NS5 levels being indicative of no detection (background levels). (B) Same as above (A) except that lysates were run on a 7.5% SDS-polyacrylamide gel and subsequently analyzed by Western blotting using GFP-, STAT2-, and tubulin-specific antibodies.
A clear difference between our previously described constructs expressing DENV NS5 as a precursor and the construct expressing mature NS5 is the amino-terminal residue of NS5, which, in the case of the NS5 precursors, is a glycine and not a methionine. In order to address the role of the N-terminal glycine in mediating reduced levels of STAT2, we constructed an E-Ub-M-NS5-GFP fusion protein containing a methionine at the P1′ position. The expression of this construct resulted in reduced STAT2 levels (Fig. 4B, lane 4), indicating that the proteolytic cleavage of NS5, rather than the terminal amino acid residue, is required for this effect. It should be noted that despite the fact that both proteolytically cleaved NS5-GFP from E-Ub-M-NS5-GFP and NS5-GFP are expected to have the same amino acid compositions and molecular weights, a slight difference in migration in the sodium dodecyl sulfate (SDS)-7.5% polyacrylamide gel can be seen between these two proteins (Fig. 4B). In our previous Western blots, we ran the lysates on an SDS-4-to-20%-gradient polyacrylamide gel. In order to understand whether the change in mobility seen in the SDS-7.5% polyacrylamide gel was also seen in the case of other NS5 constructs that are proteolytically cleaved and mediate reduced levels of STAT2 expression, we compared the migration patterns of NS5 in the context of the polyproteins NS1-NS5-HA, E-Ub-G-NS5-HA, RFP-Ub-G-NS5-HA, NS1-5-GFP, E-Ub-G-NS5-GFP, and RFP-Ub-G-NS5-GFP with those when expressed alone as NS5-HA and NS5-GFP. Slow migration was seen only in the case of NS5-GFP constructs associated within a polyprotein associated with the endoplasmic reticulum (ER) (data not shown). Therefore, although the reason behind the different migration patterns of the NS5-GFP constructs is unclear, these differences are unrelated to the ability of NS5 to affect STAT2 protein levels.
Impaired expression of STAT2 by DENV NS5 requires an intact proteasome and ubiquitinating activities.
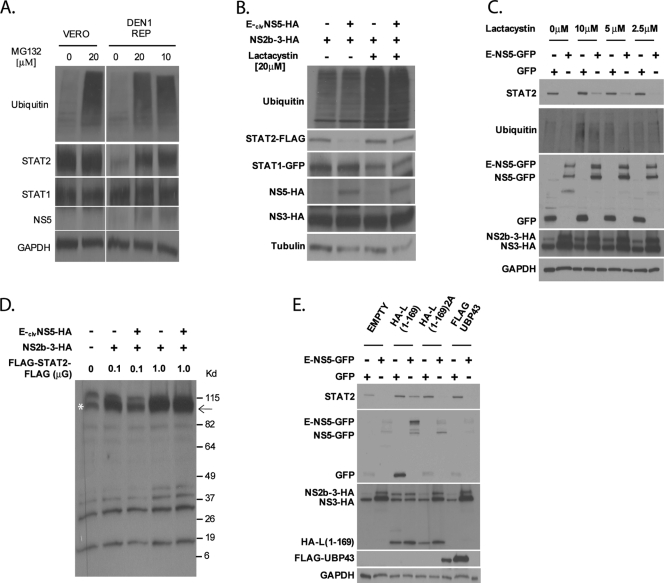
To investigate whether the DENV NS5-dependent decrease in STAT2 expression levels was mediated through the ubiquitin-proteasome pathway, we treated DEN1 replicon-containing cells with the proteasome inhibitor MG132 and monitored the level of endogenous STAT2. STAT2 levels recovered, indicating that intact proteasome activity was required (Fig. 5A). Similar results were observed in subsequent experiments using a more specific inhibitor, lactacystin. STAT2 levels were monitored in U6A cells cotransfected with HA-ubiquitin, STAT2-FLAG, NS2b-3-HA, and E-clvNS5-HA (Fig. 5B) and in 293T cells cotransfected with NS2b-3-HA and E-clvNS5-GFP (Fig. 5C). The rescue of STAT2 expression was lactacystin dose dependent (Fig. 5C). The activity of the proteasome inhibitors was monitored by the accumulation of ubiquitinated products (Fig. 5A to C). In order to determine whether STAT2 underwent a cleavage event mediated by the viral proteins, we transfected U6A cells with NS2b-3-HA, E-clvNS5-HA, and saturating amounts of a STAT2 construct containing a dual FLAG epitope at the N terminus and C terminus (Fig. 5D). When 0.1 μg of flagged STAT2 plasmid was used, coexpression with NS2b-3-HA and E-clvNS5-HA resulted in reduced levels of STAT2 expression. In contrast, when 1 μg of flagged STAT2 plasmid was used, no reduced levels were observed, which is indicative of its saturation. Since we overexposed the film to maximize the possible detection of STAT2-cleaved products, multiple low-intensity unspecific bands were detected even in the mock-transfected lane. However, no additional FLAG-specific bands were observed in the lanes transfected with NS2b-3-HA and E-clvNS5-HA compared with those transfected with NS2b-3-HA. These data, and the fact that proteasome inhibition results in the recovery of full-length STAT2 (Fig. 5A to C), indicate that STAT2 does not undergo detectable cleavage prior to its proteasome-mediated degradation. Finally, the overexpression of the OTU domain of Crimean Congo hemorrhagic fever virus L(1-169), which contains potent deubiquitinating and de-ISGylating activity (15), prevented DENV NS5-mediated STAT2 degradation. This is in contrast to the overexpression of a catalytically inactive OTU domain, L(1-169)2A, or of the de-ISGylating protein UBP43 (33) (Fig. 5E), implicating ubiquitination in STAT2 degradation. Taken together, these results indicate that a functional ubiquitin-proteasome pathway is required for STAT2 degradation.
FIG. 5.
Inhibitors of the ubiquitin-proteasome pathway prevent STAT2 degradation by DENV NS5. (A) wtVero or Vero cells stably expressing the DEN1 replicon were treated with the indicated amounts of MG132. Sixteen hours posttreatment, cells were lysed and examined for ubiquitin, STAT2, STAT1, NS5, and GAPDH levels via Western blotting. (B) STAT2-deficient U6A cells were transfected with HA-ubiquitin, STAT2-FLAG, STAT1-GFP, NS2b-3, and either E-clvNS5-HA or an empty vector plasmid. Cells were then treated with lactacystin for 8 h and subsequently lysed and examined by Western blotting using ubiquitin, STAT2, STAT1, HA, and tubulin antibodies. (C) 293T cells were cotransfected with NS2b-3-HA and the plasmids indicated at the top. Ten hours posttransfection, cells were treated with the indicated amounts of lactacystin. Twenty-four hours posttransfection, cells were sorted for GFP-positive cells by FACS, lysed, and examined by Western blotting using ubiquitin-, GFP-, HA-, and GAPDH-specific antibodies. (D) STAT2-deficient U6A cells were transfected with 0.1 or 1 μg of FLAG-STAT2-FLAG, 0.1 μg NS2b-3, and either 1 μg E-clvNS5-HA or an empty vector plasmid. Cells were then lysed and examined by Western blotting using FLAG antibody and long-term film exposure to detect any additional low-intensity bands. The arrow indicates the expected size of FLAG-STAT2-FLAG. The asterisk marks a nonspecific band running at the same mobility of FLAG-STAT2-FLAG. (E) 293T cells were cotransfected with NS2b-3 and the plasmids indicated at the top. Twenty-four hours posttransfection, cells were sorted for GFP-positive cells by FACS, lysed, and examined via Western blotting using STAT2-, GFP-, HA-, FLAG-, and GAPDH-specific antibodies.
Amino acids at the N terminus of DENV NS5 are required for STAT2 degradation but not for STAT2 binding.
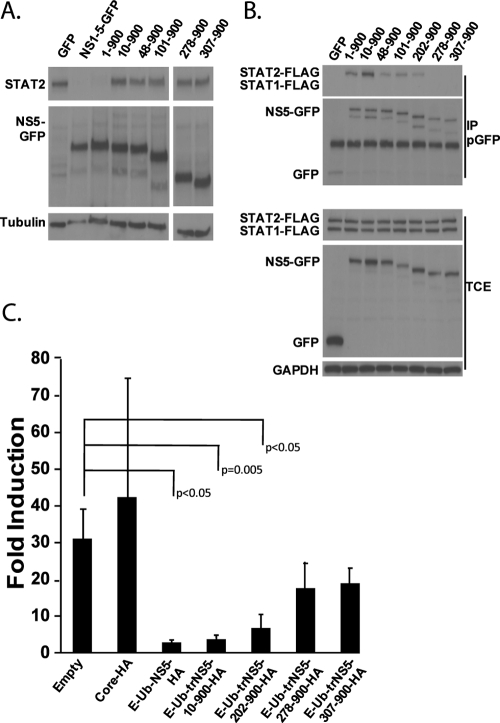
In order to preliminarily map domains in DENV NS5 required for STAT2 binding and degradation, we fused truncations of the NS5 protein downstream of the E-Ub cassette. Figure 6A demonstrates that a deletion of just the first 10 residues of NS5 results in a loss of its STAT2 degradation ability. We do not think that this abrogation is due to a gross misfolding of the truncated NS5 protein, as the deletion of up to the first 202 residues does not impact either STAT2 binding activity or IFN signaling inhibition, although a further deletion of the N-terminal 278 residues results in a significant reduction in levels of both activities (Fig. 6B and C). However, we cannot exclude the possibility that misfolded NS5 is still able to bind to STAT2 and inhibit IFN signaling. Nevertheless, our results suggest that the extreme N terminus of NS5 contains a domain required for STAT2 degradation but that the first 200 amino acids are dispensable for binding to STAT2. In addition, the binding of NS5 to STAT2 is sufficient to inhibit IFN signaling.
FIG. 6.
Truncations in the DENV NS5 protein affect its ability to associate with STAT2, decrease STAT2 levels, and inhibit IFN signaling. (A) 293T cells were transfected with the indicated constructs. Numbering refers to the glycine start position within the NS5 protein. All numbered forms of NS5 are expressed within the context of the E-Ub-NS5-GFP construct (i.e., 10-900, NS5 residues 10 to 900 fused to the C terminus of the E-Ub cassette). Twenty-four hours posttransfection, cells were sorted for GFP-positive cells using FACS, lysed, and examined by Western blotting using STAT2, GFP, and tubulin antibodies. (B) 293T cells were cotransfected with the indicated plasmids (the numbering system is identical to that described in A), STAT1-FLAG, and STAT2-FLAG. In order to detect STAT2 binding by NS5, STAT2-FLAG plasmid was transfected in excess with respect to E-Ub-NS5-GFP plasmids, resulting in a not-detectable degradation of STAT2-FLAG. Lysates were immunoprecipitated with a polyclonal GFP antibody (pGFP), and Western blots were performed using FLAG, GFP, and GAPDH antibodies. TCE, total cell extracts. (C) 293T cells were transfected with the ISRE-54-CAT reporter, a constitutively expressing firefly luciferase plasmid, and the indicated HA-tagged viral protein. Twenty-four hours posttransfection, cells were treated with 1,000 U/ml of type I IFN. Twenty-four hours posttreatment, cells were lysed and measured for CAT and luciferase activity. Data are represented with the standard deviations from three independent experiments. Samples showing P values of less than 0.05 compared with the empty control sample are indicated.
DISCUSSION
In this paper, we have identified the polymerase of DENV, NS5, as being an IFN antagonist that associates (directly or indirectly) with STAT2, a necessary component of the ISGF3 transcription complex, leading to its proteolytic processing. While the expression of DENV NS5 alone resulted in STAT2 binding, the ability of DENV NS5 to target STAT2 for degradation required the presence of a protease cleavage signal upstream of the N terminus of NS5, with the subsequent coexpression of this precursor form of NS5 with the relevant protease to target the cleavage site, mirroring the NS5 processing occurring in the context of the DENV polyprotein. The identity of the protease appears to be irrelevant, as we have used a viral serine protease (NS2b-3), a viral cysteine protease (NIa-proTEV), or a cellular cysteine protease (ubiquitin hydrolases), all of which are capable of maturing NS5 in a manner which confers the ability to degrade STAT2. We therefore conclude that a precursor form of NS5 matured via proteolytic cleavage is required for STAT2 degradation. The requirement for a cleaved form of NS5 in degrading STAT2 remains a mystery. We have tested the most obvious hypotheses and ruled them out as playing a role; NS5 does not require cleavage close to the ER for STAT2 degradation, as the processing of both E-clvNS5-GFP and RFP-clvNS5-GFP resulted in STAT2 degradation. E-clvNS5-GFP should be ER associated due to the presence of a signal sequence in front of E. The transmembrane residue of E will result in NS5 cytoplasmic localization but in close proximity to the ER. In contrast, the absence of a signal sequence in front of RFP argues that the reason why NS5, when not expressed as a precursor, does not degrade STAT2 is not due to a lack of an association with the ER. NS5 does not require a glycine residue at the N terminus, as E-UB-M-NS5-GFP was as efficient as E-Ub-G-NS5-GFP in degrading STAT2. Furthermore, the expression of NS5 without an upstream cleavage signal does not appear to grossly affect the proper folding of the polymerase, as its ability to bind STAT2 remains intact, although we do not rule out the possibility that a motif resides in the extreme N-terminal region, the folding of which may be sensitive to the mechanism by which the N terminus is generated. Indeed, the N-terminal-truncation studies appear to support this notion. It is then possible that the cleavage requirement for NS5 to degrade STAT2 is due to the presence of a motif located at the extreme N terminus of the polymerase, the tertiary structure of which may be hypersensitive to changes induced by expression with or without an upstream sequence that will subsequently be removed via cleavage. On the other hand, it is also conceivable that DENV NS5 binds to STAT2 in its precursor form and that the processing of the precursor bound to STAT2 somehow triggers the degradation of STAT2. Further experiments will continue to map the sequences within NS5 required for STAT2 binding and degradation.
The mechanism by which STAT2 degradation occurs involves ubiquitination, as STAT2 levels are restored in the presence of proteasome inhibitors and of a deubiquitinating enzyme. Although cleavage of STAT2 may occur prior to its degradation, we have been unable to detect cleaved products with the STAT2 antibody used in our studies, which recognizes the C-terminal region of STAT2, or with a FLAG antibody upon the overexpression of a STAT2 construct containing FLAG epitopes at both the N and C termini. Analysis of the primary sequence of NS5 does not immediately reveal putative motifs (i.e., RING domains and HECT domains) that would suggest that the NS5 protein is an E3 ligase. It is nevertheless possible that, analogous to the NS proteins of respiratory syncytial virus (14) or to the V protein of human parainfluenza virus type 2 (51), DENV NS5 interacts with STAT2, as shown in this paper, and with components of the ubiquitination machinery, resulting in STAT2 ubiquitination and degradation. This possibility remains to be investigated in the future. Although the binding of DENV NS5 to STAT2 was sufficient to prevent IFN signaling, the degradation of STAT2 might result in an additional advantage, since once STAT2 bound to NS5 is degraded, the NS5 molecule that was previously bound to STAT2 might now become free to perform other functions, such as RNA transcription or additional degradation of other STAT2 molecules.
The polymerase of DENV can now be added to the list of flavivirus NS5 proteins that block IFN signaling. Although the expression of JEV NS5 alone or TBEV NS5 alone appeared to be sufficient to recapitulate the phenotype observed in JEV or TBEV infection, and the inhibition of IFN signaling by these proteins did not involve binding to the STATs but upstream events in the IFN pathways (3, 30, 31), it remains to be determined whether the expression of these proteins in the context of a cleaved precursor would confer additional functions not otherwise observed outside the context of viral infection. Indeed, it has been shown that NS5 of Kunjin virus (a substrain of WNV) requires expression within the context of the NS1-5 region for efficient trans-complementation of a self replicating Kunjin minigenome that contains a catalytically dead NS5 and that the expression of NS5 alone resulted in an 100-fold decrease in replication activity despite its higher level of expression than that of the NS1-5 construct (28). We also raise the possibility that cleavage requirements may extend beyond DENV NS5 to other proteins expressed as precursors, and therefore, the expression of a viral or host protein that is proteolytically cleaved might result in additional functions beyond those exerted by intermediate products that are dependent on cleavage for proper functionality.
In order for an infecting virus to induce disease, it must mask itself from the IFN response either through preventing the initial induction of the pathway, as is the case for the influenza virus NS1 protein (53), or by blocking the IFN signaling pathway, as is the case for several flavivirus proteins including DENV NS5. While DENV NS5 targets STAT2 for degradation (this paper), DENV NS4b and, to a lesser extent, NS4a and NS2a, appear to prevent STAT1 phosphorylation (35). Multiple mechanisms to inhibit IFN signaling also apply to WNV (23, 32, 34) and may apply to other flaviviruses. This probably illustrates the need for a potent inhibition of IFN signaling for the life cycle of this group of viruses. Similar multiple mechanisms have also been described for other viruses such as paramyxoviruses and coronaviruses (16, 29, 40, 54). The knowledge gained by characterizing the various mechanisms through which viruses evade IFN paves the way for the targeted attenuation of pathogenic viruses, which may be used as potential live vaccines. In addition, small molecules that inhibit this antagonism at various points within the process (i.e., blocking the NS5-STAT2 interaction or NS5-mediated degradation) can be employed as potential therapeutics to ameliorate disease. Future experiments will focus on exploiting the antagonist function of DENV to these ends.
Acknowledgments
This work was supported by grant U54 AI57158 (to A.G.-S. and P.-Y.S.) from the National Institutes of Health and by a National Institutes of Health fellowship (to M.L.-R.).
We thank Megan Shaw, Natalia Frias-Staheli, Benjamin tenOever, and Luis Martinez-Sobrido for kindly providing plasmids. We thank Richard Cádagan for excellent technical assistance. We are also very grateful to Domenico Tortorella both for providing plasmids and for his sage advice.
Footnotes
Published ahead of print on 11 March 2009.
REFERENCES
- 1.Aebi, M., J. Fah, N. Hurt, C. E. Samuel, D. Thomis, L. Bazzigher, J. Pavlovic, O. Haller, and P. Staeheli. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 95062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburn, P. M., and C. F. Craig. 1907. Experimental investigations regarding the etiology of dengue fever. J. Infect. Dis. 4440-475. [DOI] [PubMed] [Google Scholar]
- 3.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 7912828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleary, C. M., R. J. Donnelly, J. Soh, T. M. Mariano, and S. Pestka. 1994. Knockout and reconstitution of a functional human type I interferon receptor complex. J. Biol. Chem. 26918747-18749. [PubMed] [Google Scholar]
- 5.Cleaves, G. R., and D. T. Dubin. 1979. Methylation status of intracellular dengue type 2 40 S RNA. Virology 96159-165. [DOI] [PubMed] [Google Scholar]
- 6.Clemens, M. J., and B. R. Williams. 1978. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell 13565-572. [DOI] [PubMed] [Google Scholar]
- 7.Clum, S., K. E. Ebner, and R. Padmanabhan. 1997. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 27230715-30723. [DOI] [PubMed] [Google Scholar]
- 8.Coia, G., M. D. Parker, G. Speight, M. E. Byrne, and E. G. Westaway. 1988. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J. Gen. Virol. 691-21. [DOI] [PubMed] [Google Scholar]
- 9.Colamonici, O., H. Yan, P. Domanski, R. Handa, D. Smalley, J. Mullersman, M. Witte, K. Krishnan, and J. Krolewski. 1994. Direct binding to and tyrosine phosphorylation of the α subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol. Cell. Biol. 148133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colamonici, O. R., L. C. Platanias, P. Domanski, R. Handa, K. C. Gilmour, M. O. Diaz, N. Reich, and P. Pitha-Rowe. 1995. Transmembrane signaling by the alpha subunit of the type I interferon receptor is essential for activation of the JAK kinases and the transcriptional factor ISGF3. J. Biol. Chem. 2708188-8193. [DOI] [PubMed] [Google Scholar]
- 11.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 744957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domanski, P., E. Fish, O. W. Nadeau, M. Witte, L. C. Platanias, H. Yan, J. Krolewski, P. Pitha, and O. R. Colamonici. 1997. A region of the beta subunit of the interferon alpha receptor different from box 1 interacts with Jak1 and is sufficient to activate the Jak-Stat pathway and induce an antiviral state. J. Biol. Chem. 27226388-26393. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, J., O. T. Lynch, Y. Suessmuth, P. Qian, C. R. Boyd, J. F. Burrows, R. Buick, N. J. Stevenson, O. Touzelet, M. Gadina, U. F. Power, and J. A. Johnston. 2007. Respiratory syncytial virus NS1 protein degrades STAT2 by using the elongin-cullin E3 ligase. J. Virol. 813428-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frias-Staheli, N., N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre, and H. W. Virgin IV. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2404-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 878555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 736559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcin, D., J. B. Marq, L. Strahle, P. le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295256-265. [DOI] [PubMed] [Google Scholar]
- 20.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459205-210. [DOI] [PubMed] [Google Scholar]
- 21.Graham, H. 1903. The dengue: a study of its pathology and mode of propagation. J. Trop. Med. 1903209-214. [Google Scholar]
- 22.Greenlund, A. C., M. O. Morales, B. L. Viviano, H. Yan, J. Krolewski, and R. D. Schreiber. 1995. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity 2677-687. [DOI] [PubMed] [Google Scholar]
- 23.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 791343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta, S., H. Yan, L. H. Wong, S. Ralph, J. Krolewski, and C. Schindler. 1996. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-alpha signals. EMBO J. 151075-1084. [PMC free article] [PubMed] [Google Scholar]
- 25.Halstead, S. B. 1998. Dengue viruses. WB Saunders, Philadelphia, PA.
- 26.Jones, M., A. Davidson, L. Hibbert, P. Gruenwald, J. Schlaak, S. Ball, G. R. Foster, and M. Jacobs. 2005. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 795414-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler, D. S., D. E. Levy, and J. E. Darnell, Jr. 1988. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. USA 858521-8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 7310272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 805908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 789285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 791934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malakhov, M. P., O. A. Malakhova, K. I. Kim, K. J. Ritchie, and D. E. Zhang. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2779976-9981. [DOI] [PubMed] [Google Scholar]
- 34.Muñoz-Jordán, J. L., M. Laurent-Rolle, J. Ashour, L. Martínez-Sobrido, M. Ashok, W. I. Lipkin, and A. García-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 798004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 10014333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadeau, O. W., P. Domanski, A. Usacheva, S. Uddin, L. C. Platanias, P. Pitha, R. Raz, D. Levy, B. Majchrzak, E. Fish, and O. R. Colamonici. 1999. The proximal tyrosines of the cytoplasmic domain of the beta chain of the type I interferon receptor are essential for signal transducer and activator of transcription (Stat) 2 activation. Evidence that two Stat2 sites are required to reach a threshold of interferon alpha-induced Stat2 tyrosine phosphorylation that allows normal formation of interferon-stimulated gene factor 3. J. Biol. Chem. 2744045-4052. [DOI] [PubMed] [Google Scholar]
- 37.Novick, D., B. Cohen, and M. Rubinstein. 1994. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell 77391-400. [DOI] [PubMed] [Google Scholar]
- 38.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283230-239. [DOI] [PubMed] [Google Scholar]
- 39.Park, C., S. Li, E. Cha, and C. Schindler. 2000. Immune response in Stat2 knockout mice. Immunity 13795-804. [DOI] [PubMed] [Google Scholar]
- 40.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 771501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavlovic, J., T. Zurcher, O. Haller, and P. Staeheli. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 643370-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puig-Basagoiti, F., M. Tilgner, B. M. Forshey, S. M. Philpott, N. G. Espina, D. E. Wentworth, S. J. Goebel, P. S. Masters, B. Falgout, P. Ren, D. M. Ferguson, and P. Y. Shi. 2006. Triaryl pyrazoline compound inhibits flavivirus RNA replication. Antimicrob. Agents Chemother. 501320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qureshi, S. A., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1995. Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. USA 923829-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229726-733. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 7611476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuel, C. E. 1981. Molecular mechanisms of interferon action: interferon-mediated phosphorylation of ribosome-associated protein P1 and protein synthesis initiation factor eIF-2. Tex. Rep. Biol. Med. 41463-470. [PubMed] [Google Scholar]
- 47.Shaffer, J. A., W. J. Bellini, and P. A. Rota. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315389-397. [DOI] [PubMed] [Google Scholar]
- 48.Shresta, S., K. L. Sharar, D. M. Prigozhin, H. M. Snider, P. R. Beatty, and E. Harris. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 1753946-3954. [DOI] [PubMed] [Google Scholar]
- 49.Shuai, K., C. M. Horvath, L. H. Huang, S. A. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76821-828. [DOI] [PubMed] [Google Scholar]
- 50.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 2611744-1746. [DOI] [PubMed] [Google Scholar]
- 51.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304160-166. [DOI] [PubMed] [Google Scholar]
- 52.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 776385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 7411566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wathelet, M. G., M. Orr, M. B. Frieman, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 8111620-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wengler, G., G. Wengler, and H. J. Gross. 1978. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 89423-437. [DOI] [PubMed] [Google Scholar]
- 56.Yan, H., K. Krishnan, A. C. Greenlund, S. Gupta, J. T. Lim, R. D. Schreiber, C. W. Schindler, and J. J. Krolewski. 1996. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. EMBO J. 151064-1074. [PMC free article] [PubMed] [Google Scholar]