Abstract
The respiratory syncytial virus (RSV) matrix (M) protein is localized in the nucleus of infected cells early in infection but is mostly cytoplasmic late in infection. We have previously shown that M localizes in the nucleus through the action of the importin β1 nuclear import receptor. Here, we establish for the first time that M's ability to shuttle to the cytoplasm is due to the action of the nuclear export receptor Crm1, as shown in infected cells, and in cells transfected to express green fluorescent protein (GFP)-M fusion proteins. Specific inhibition of Crm1-mediated nuclear export by leptomycin B increased M nuclear accumulation. Analysis of truncated and point-mutated M derivatives indicated that Crm1-dependent nuclear export of M is attributable to a nuclear export signal (NES) within residues 194 to 206. Importantly, inhibition of M nuclear export resulted in reduced virus production, and a recombinant RSV carrying a mutated NES could not be rescued by reverse genetics. That this is likely to be due to the inability of a nuclear export deficient M to localize to regions of virus assembly is indicated by the fact that a nuclear-export-deficient GFP-M fails to localize to regions of virus assembly when expressed in cells infected with wild-type RSV. Together, our data suggest that Crm1-dependent nuclear export of M is central to RSV infection, representing the first report of such a mechanism for a paramyxovirus M protein and with important implications for related paramyxoviruses.
The Pneumovirus respiratory syncytial virus (RSV) within the Paramyxoviridae family is the most common cause of lower-respiratory-tract disease in infants (7). The negative-sense single-strand RNA genome of RSV encodes two nonstructural and nine structural proteins, comprising the envelope glycoproteins (F, G, and SH), the nucleocapsid proteins (N, P, and L), the nucleocapsid-associated proteins (M2-1 and M2-2), and the matrix (M) protein (1, 7, 11). Previously, we have shown that M protein localizes in the nucleus at early stages of infection, but later in infection it is localized mainly in the cytoplasm, in association with nucleocapsid-containing cytoplasmic inclusions (13, 16). The M proteins of other negative-strand viruses, such as Sendai virus, Newcastle disease virus, and vesicular stomatitis virus (VSV), have also been observed in the nucleus at early stages of infection (32, 40, 48). Interestingly, the M proteins of all of these viruses, including RSV, play major roles in virus assembly, which take place in the cytoplasm and at the cell membrane (11, 12, 24, 34, 36, 39), but the mechanisms by which trafficking between the nucleus and cytoplasm occurs are unknown.
The importin β family member Crm1 (exportin 1) is known to mediate nuclear export of proteins bearing leucine-rich nuclear export signals (NES) (8, 9, 18, 19, 37, 42, 43), such as the human immunodeficiency virus type 1 Rev protein (4). In the case of the influenza virus matrix (M1) protein, binding to the influenza virus nuclear export protein, which possesses a Crm1-recognized NES, appears to be responsible for its export from the nucleus, bound to the influenza virus RNA (3).
We have recently shown that RSV M localizes in the nucleus through a conventional nuclear import pathway dependent on the nuclear import receptor importin β1 (IMPβ1) and the guanine nucleotide-binding protein Ran (14). In the present study, we show for the first time that RSV M possesses a Crm1-dependent nuclear export pathway, based on experiments using the specific inhibitor leptomycin B (LMB) (25), both in RSV-infected cells and in green fluorescent protein (GFP)-M fusion protein-expressing transfected cells. We use truncated and point-mutated M derivatives to map the Crm1-recognized NES within the M sequence and show that Crm1-dependent nuclear export is critical to the RSV infectious cycle, since LMB treatment early in infection, inhibiting M export from the nucleus, reduces RSV virion production and a recombinant RSV carrying a NES mutation in M was unable to replicate, probably because M deficient in nuclear export could not localize to areas of virus assembly, as shown in RSV-infected cells transfected to express GFP-M. This is the first report of a Crm1-mediated nuclear export pathway for a paramyxovirus M protein, with implications for the trafficking and function of other paramyxovirus M proteins.
MATERIALS AND METHODS
Cell and virus culture.
RSV strain A2, a gift from Paul Young (Department of Microbiology, University of Queensland, Australia), was used throughout the present study. Human alveolar epithelial A549 cells, and African green monkey kidney Vero cells were grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum.
Transfection.
Transfection of Vero cells was performed using Lipofectamine (Invitrogen Australia; 1:1 mix of DNA and reagent), and cells were cultured for 24 h before analysis by confocal laser scanning microscopy (CLSM; see below); when LMB treatment was used, it was added 18 h posttransfection for 6 h prior to CLSM and image analysis.
Infection and immunofluorescence.
Vero cells were grown to 80% confluence on glass coverslips before being infected at a multiplicity of infection (MOI) of 3 with RSV strain A2 (13); certain cultures had LMB added to the culture medium to a final concentration of 2.8 ng/ml for 12 h at 6 or 18 h postinfection (p.i.). The medium was then replaced with fresh medium without LMB, and cells were cultured further up to 48 h p.i. Under these treatment conditions, LMB has been shown to have no cytotoxic effects (21). At the indicated times p.i., cells were fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, and immunostained with a monoclonal antibody specific for M (MAbαM) (38) and Alexa Fluor 488-conjugated secondary antibody (Invitrogen), followed by CLSM.
CLSM and image analysis.
Fixed and live (transfected) cell samples were imaged as previously (14); single sections of 0.5 μm were obtained with Olympus ×100 oil immersion lens using a Yokogawa spinning-disk confocal with Andor Ixon EMCCD camera and IQ software. ImageJ v1.62 public domain software was used as described previously (20, 27) to determine the nuclear/cytoplasmic fluorescence ratio (Fn/c), which was determined by using the equation: Fn/c = (Fn − Fb)/(Fc − Fb), where Fn is the nuclear fluorescence, Fc is the cytoplasmic fluorescence, and Fb is the background fluorescence (autofluorescence).
Virus production.
Cell-associated RSV was estimated by using standard procedures (44). Briefly, the culture supernatant was removed, and cells were lysed in serum-free medium (218 mM sucrose, 7.1 mM K2HPO4, 4.9 mM sodium glutamate, 1% [wt/vol] bovine serum albumin), clarified by centrifugation, and stored at −80°C until required. The RSV titer was determined on Vero cells used as indicators with crystal violet staining, and the 50% tissue culture infectious dose (TCID50)/ml was calculated using the Spearman-Karber equation (33). Titers for each virus-containing cell lysate were determined three times on independent culture sets.
M expression constructs.
RSV M coding sequences containing both, only one, or neither of the proposed NES were generated by PCR from full-length RSV cDNA (23) and introduced into the Gateway system utilizing the pDONR207 entry vector and pEPI-DESTC mammalian expression destination vector as described previously (14). A QuikChange site-directed mutagenesis kit (Promega) was used to perform site-specific mutations within the putative nuclear localization sequences (NLS) and NES in the full-length M clone as previously (41). Each reaction was performed according to the manufacturer's recommendations and contained 10 ng of template DNA and 80 ng of each primer. Resultant clones were identified by using restriction analysis followed by DNA sequencing.
The plasmid pRev-GFP encoding the GFP-Rev(2-116) fusion protein containing the Crm1-recognized NES within the pEPI-DESTC mammalian expression destination vector was generated by using standard Gateway procedures (41) and used as a positive control for LMB action; GFP-Rev is largely cytoplasmic in the absence of LMB and strongly nuclear and/or nucleolar in its presence. Plasmid DNA encoding GFP (pEGFP-C1; Invitrogen) was used in all experiments as a negative control for LMB action.
All expression constructs used in the present study expressed their encoded proteins to similar levels, as determined by determination of fluorescence intensity by image analysis (NIH Image software).
Western blotting.
RSV-infected and uninfected Vero cells were collected by trypsin treatment at 6 and 24 h p.i. and fractionated into nuclear and cytoplasmic extracts by using an NE-PER reagent kit (Pierce Biotechnology) according to the manufacturer's recommendations; the total protein concentration in each fraction was estimated using Bradford reagent (Bio-Rad). The fractions were mixed with reducing Laemmli sample buffer (26), boiled, and electrophoresed on a 4 to 20% precast polyacrylamide gel (Life Gels); 25 μg of each sample was loaded per lane. The separated proteins were transferred to nitrocellulose and probed with RabbitαM (15) diluted 1:1,000 in 0.05% milk in phosphate-buffered saline (pH 7.2) containing 0.05% Tween 20 (PBST). Bound antibody was detected with alkaline phosphatase-conjugated secondary antibodies, followed by Western Blue stabilized substrate for alkaline phosphatase (Promega).
Transfection of RSV-infected cells.
RSV-infected Vero cells were transfected with indicated GFP-M expression constructs at 6 h p.i., fixed, and permeabilized as described above and then probed with polyclonal goat anti-RSV antibody (Chemicon), followed by Alexa Fluor 568-conjugated donkey anti-goat antibody. Cells were imaged for GFP and Alexa Fluor 568 using CLSM.
Generation and recovery of mutant recombinant RSV.
The shuttle vector pGEM-HX was constructed by cloning the HindIII-XhoI fragment (positions 3280 to 4482) of the RSV antigenome (6) into pGEM-7Z(−). The HindIII-NcoI cassette of GFP-M(NLSm) or GFP-M(cNES) containing the mutated NLS or cNES, respectively, was cloned into the same reading frame of pGEM-HX, creating pGEM-HX(NLSm) and pGEM-HX(cNES). These plasmids were digested with SpeI and XhoI to enable the coding sequences for the NLS and NES mutated M genes to be subcloned into vector D51 (45), predigested using the same restriction sites. Finally, the AatII-XhoI fragment of the mutant D51 constructs was cloned into the full-length antigenome (D53). The mutant D53 plasmids were used for RSV recovery as described previously (45, 47). For some attempts at recovery for the cNESm mutant, 100 ng of pTM1-M (46) was added to the initial transfection, and supernatants were added to cells transfected to express GFP-M.
Growth of mutant recombinant RSV.
Cells were infected with recombinant wild-type (rA2) or the NLS mutant (rA2NLSm) at an MOI of 0.1 or 3 or equivalent volumes of culture supernatant from the apparently lethal cNES mutant (rA2cNESm). Virus was harvested at the times indicated, and the virus titer was determined as described above.
RESULTS
M shows nuclear and cytoplasmic localization during the RSV infection cycle.
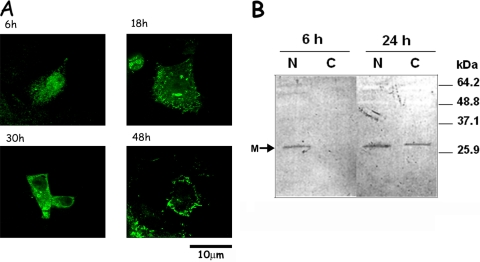
M subcellular localization during RSV infection was examined by immunofluorescence and quantitative CLSM imaging. As previously observed, it localized primarily in the nucleus of RSV-infected Vero cells, with exclusion from nucleoli, at 6 h p.i. (Fig. 1A). At 18 h p.i., M was localized in the cytoplasm as well as the nucleus, with specific staining in cytoplasmic inclusions. At 30 h p.i., most of the M protein was present in cytoplasmic inclusions, with some staining in the nucleus and at the cell membrane. The few remaining live cells at 48 h p.i. (note the characteristic cytopathic effects and changed morphology) showed M localized in the cytoplasmic inclusions and at the cell membrane, with some cells showing predominantly nuclear localization of M, a finding possibly indicative of secondary infection. These results were supported by image analysis of the digitized CLSM images (see below and Fig. 2B) and confirmed by Western blotting (Fig. 1B), where RSV-infected Vero cells were fractionated into nuclear and cytoplasmic extracts, and probed for M protein. M was found to be predominantly nuclear at 6 h p.i. but present in both the cytoplasm and the nucleus at 24 h p.i., an observation consistent with the CLSM data. That M was not observed in the cytoplasmic fraction at 6 h p.i. is not due to unequal loading in the gel since all lanes were loaded with equal amounts of total protein (25 μg) of the requisite sample. The fact that RSV-infected cells have high amounts of M in the cytoplasm at later times coincident with very low nuclear accumulation shows that the cytoplasmic localization is not due to excess M having saturated all nuclear sites and suggests that M may utilize a specific nuclear export mechanism to localize to the cytoplasm.
FIG. 1.
Dynamic localization of M protein in the nucleus and cytoplasm in RSV-infected cells. (A) Vero cells were infected with RSV, fixed, and permeabilized at the times indicated, and M localization was visualized using MAbαM and Alexa Fluor 488-coupled secondary antibodies. Fluorescence imaging was performed in the Yokogawa spinning-disk confocal with Andor Ixon EMCCD camera using a ×100 oil immersion objective lens. Scale bar, 10 μm. (B) RSV-infected Vero cells were collected at 6 and 24 h p.i., separated into nuclear and cytoplasmic fractions, and electrophoresed on a 4 to 20% polyacrylamide gel. After Western transfer, the blot was probed with RabbitαM, followed by alkaline phosphatase-conjugated secondary antibody with bound antibodies detected using Western Blue stabilized substrate for alkaline phosphatase (Promega). Molecular mass markers are indicated on the right; the position of M is shown on the left. N, nuclear fraction; C, cytoplasmic fraction.
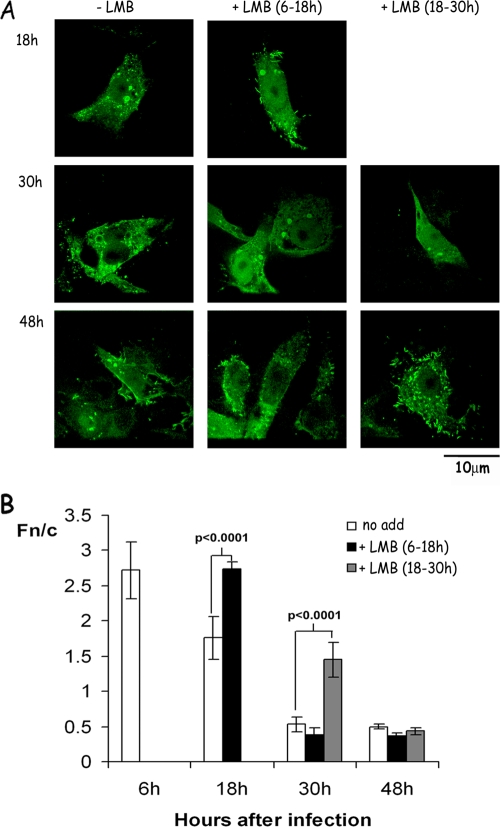
FIG. 2.
LMB treatment increases nuclear localization of M protein in RSV-infected cells. (A) RSV-infected Vero cells were treated with 2.8 ng of LMB/ml from 6 to 18 h (middle) or from 18 to 30 h (right) p.i. as indicated, fixed, and stained, and M localization was visualized by CLSM as described in Fig. 1A. Similarly stained infected untreated cells are shown on the left for direct comparison. Scale bar, 10 μm. (B) Image analysis was performed on CLSM files such as those shown in panel A to determine the Fn/c (i.e., the ratio of nuclear [Fn] to cytoplasmic [Fc] fluorescence after subtraction of the fluorescence due to background autofluorescence) as described previously (20, 28). The results represent the means ± the standard errors of the mean (SEM; n ≥ 35) from a single experiment, representative of two similar experiments, with statistically significant differences between Fn/c values in the absence or presence of LMB as indicated.
The Crm1-specific inhibitor LMB increases M nuclear accumulation in RSV-infected cells.
To assess whether M cytoplasmic localization late in infection may be attributable to Crm1-dependent nuclear export, RSV-infected cells were treated with the Crm1-specific inhibitor LMB early in infection, i.e., from 6 to 18 h p.i. (Fig. 2A, middle panels), or later in infection, i.e., from 18 to 30 h p.i. (Fig. 2A, right panels). LMB has a short half-life (3 to 6 h) in culture medium (25), and the additions as performed here resulted in short periods of Crm1 inhibition, allowing the effect of inhibiting Crm1 activity during specific time periods of infection on M localization to be assessed.
In RSV-infected cells treated with LMB from 6 to 18 h p.i., M was found to be mostly nuclear at 18 h p.i., with very few LMB-treated cells showing localization in cytoplasmic inclusions compared to untreated samples (Fig. 2A left panels; see also Fig. 1A). At 30 h p.i. M was more nuclear compared to untreated samples, although there was significant staining in cytoplasmic inclusions, and at 48 h p.i. M localization was the same as that in untreated samples. In cells treated with LMB from 18 to 30 h p.i. (Fig. 2A, right panels), M was localized primarily in the nucleus at 30 h p.i., with some cells also demonstrating localization at cytoplasmic inclusions. At 48 h p.i. the localization of M was similar to untreated cells except that there were fewer cells with predominantly nuclear localization, a finding suggestive of reduced secondary infection.
Quantitative analysis confirmed these results (Fig. 2B). In RSV-infected cells, M in the absence of LMB (columns labeled “no add”) was predominantly nuclear at 6 h p.i. with an Fn/c of ∼2.8. M became progressively more cytoplasmic with time (an Fn/c of 1.76 at 18 h and an Fn/c of 0.6 at 30 and 48 h). LMB treatment from 6 to 18 h p.i. resulted in a significant (P < 0.0001) increase in the relative nuclear levels of M at 18 h (Fn/c of 2.7) comparable to those observed at 6 h p.i. in the absence of LMB, but not at 30 or 48 h p.i. (Fn/c values of 0.38 and 0.37, respectively) compared to in its absence (Fn/c of 0.5; Fig. 2B). LMB treatment from 18 to 30 h p.i. (Fig. 2B) resulted in significantly (P < 0.0001) higher nuclear levels of M compared to untreated cells (Fn/c of 1.5) at 30 h p.i. but not at 48 h p.i. (Fn/c of 0.4).
Together, the results from Western blotting and quantitative immunofluorescence indicate that M is present in the nucleus early in infection and in cytoplasmic inclusions later. Importantly, LMB addition either early or late in infection reduces M cytoplasmic localization, indicating that M is able to shuttle into and out of the nucleus, with Crm1 a mediator of its nuclear export.
Inhibition of nuclear export impairs RSV virus production.
The importance of nuclear export of M to RSV replication was assessed by monitoring virus production in cells treated without or with LMB for the same time periods as those used above (Table 1) . LMB treatment from 6 to 18 h p.i. led to a significant decrease in the virus titer at 30 h p.i. compared to untreated cells (% TCID50/ml < 10% of untreated cells), implying that Crm1 activity, and presumably the nuclear export of M, is required for efficient virus production. At 48 h p.i., the virus titer in treated cells was still <20% of that in untreated cells. LMB treatment later in infection (18 to 30 h p.i.) resulted in a smaller decrease in the virus titer at 30 h p.i. compared to in its absence (% TCID50/ml = 74% of untreated cells); by 48 h p.i., the virus titer in treated cells had recovered to >90% of that in untreated cells. In contrast to early in infection, inhibiting Crm1-dependent nuclear export, including that of M (Fig. 2), late in infection thus appears to have only a minimal effect on virus production, probably due to the presence of cytoplasmic M already bound to nucleocapsids and envelope glycoproteins prior to LMB treatment at this time. Moreover, once LMB is removed from the culture medium, its effect appears to be reversible. That LMB treatment affects RSV virus production through direct effects on M nuclear export seems likely, since M is the only RSV structural protein that localizes in the nucleus and that has a Crm1-dependent nuclear export mechanism. Indirect host cell effects can be discounted based on the fact that LMB addition late in infection has reduced impact on virus production (Table 1) and previous work showing that LMB, as used here, does not lead to cytotoxic effects (21).
TABLE 1.
LMB treatment inhibits RSV virus production
| Time of LMB (4 μg/ml) addition | Mean log TCID50/ml ± SEa at:
|
% TCID50/mlb at:
|
||
|---|---|---|---|---|
| 30 h p.i. | 48 h p.i. | 30 h p.i. | 48 h p.i. | |
| No added LMB | 5.27 ± 0.8 | 5.4 ± 0.44 | 100 | 100 |
| 6-18 hpi | 4.24 ± 0.87 | 4.67 ± 0.46 | 9.33 | 18.6 |
| 18-30 hpi | 5.14 ± 0.78 | 5.37 ± 0.43 | 74 | 93 |
The data represent three separate experiments, each performed in triplicate.
Compared to the TCID50/ml in the absence of LMB, which was taken as 100%.
A NES within M.
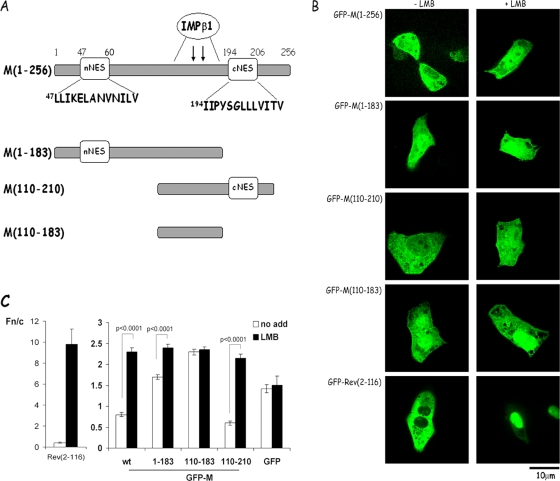
The M sequence contains several leucine/isoleucine-rich sequences that represent candidate NES, the most prominent being the nNES and cNES sequences within M residues 46 to 60 and residues 194 to 206, respectively (Fig. 3A). Full-length M and truncated GFP fusion derivatives lacking either one or both of these sequences were expressed in transfected Vero cells, and the effect of LMB on their localization was examined. GFP-Rev(2-116), containing the Crm1-recognized NES (4, 18, 25, 41) from HIV-1 Rev, was used as a control for LMB action, being largely cytoplasmic in the absence of LMB and strongly nuclear/nucleolar in its presence (Fig. 3BC). In analogous fashion, full-length M was primarily cytoplasmic 24 h after transfection but localized strongly to the nucleus (Fn/c of >2) upon LMB treatment (Fig. 3BC), confirming that M can shuttle into and out of the nucleus; Crm1-dependent nuclear export is dominant over nuclear import. The GFP-M(1-183) truncated derivative, which lacks the cNES (see Fig. 3A), showed increased nuclear accumulation in the absence of LMB compared to the full-length protein (Fig. 3B and C; Fn/c value of 1.7 compared to 0.8 for the full-length protein) and reduced nuclear accumulation in the presence of LMB, a finding consistent with reduced nuclear export activity. GFP-M(110-210) showed localization and LMB sensitivity comparable to that of the full-length protein (Fig. 3BC); this is consistent with possession of an NES. Finally, GFP-M(110-183), which lacks both nNES and cNES, localized strongly to the nucleus (Fig. 3BC) and was insensitive to LMB treatment (Fn/c of 2.4 in the absence or presence of LMB), as expected, due to lack of a NES and the fact that M residues 110 to 183 contain the IMPβ1 recognized NLS (14), used as a negative control for LMB action in this experiment. Taken together, the data suggest that the cNES is a likely candidate as the key NES.
FIG. 3.
Subcellular localization of RSV M truncated derivatives in living, transfected Vero cells. (A) Schematic diagram of the M sequence and truncated GFP-M constructs highlighting the nuclear targeting sequences, including the binding site for IMPβ1 (14) within amino acids 110 to 183, and the putative nNES and cNES sequences shown in the single-letter code. The numbers refer to the M amino acid sequence. (B) Subcellular localization of full-length and truncated M proteins in transfected Vero cells in the absence or presence of LMB. Vero cells were transfected to express the GFP-M or GFP-Rev(2-116) fusion proteins as indicated and treated either without (−LMB) or with 2.8 ng of LMB/ml (+LMB) for 6 h prior to live CLSM imaging at 24 h posttransfection using a ×100 oil immersion objective lens as described previously (41). Scale bar, 10 μm. (C) CLSM files such as those in panel B were analyzed as described in the legend to Fig. 2B. The results shown are for the means ± the SEM (n ≥ 20) determined from a single experiment, representative of two similar experiments, with significant differences between the Fn/c values in the absence or presence of LMB as indicated.
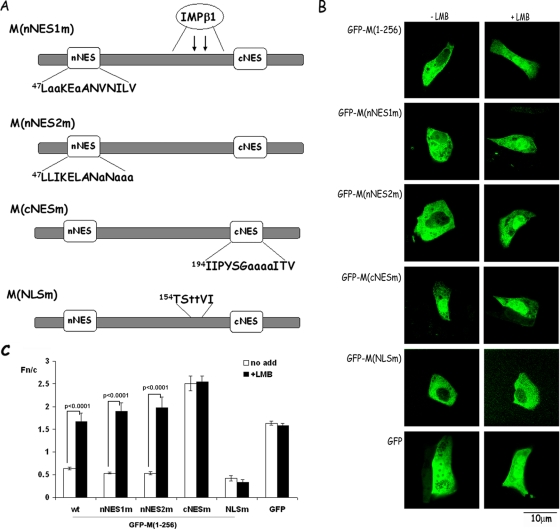
This possibility was tested directly using site-directed mutagenesis, whereby the central leucine/isoleucine residues (200 to 203) within cNES were substituted by alanine to yield M(cNESm), while comparable substitutions were made within the nNES to generate M(nNES1m) and M(nNES2m) (Fig. 4A). For comparison, a site-directed mutant was generated where the 156KK within the putative bipartite NLS were mutated to 156TT (Fig. 4A; also see reference 14). Expression constructs encoding the four mutants were transfected into Vero cells and analyzed for subcellular localization in the absence or presence of LMB (Fig. 4B and C). Both nNES mutants appeared to differ very little from wild-type M in terms of nuclear accumulation in either the absence or presence of LMB (Fig. 4BC). In contrast, however, mutation of the cNES led to strong nuclear localization even in the absence of LMB (Fig. 4B and C; Fn/c of 2.54), which was not increased in the presence of LMB (Fn/c of 2.51). The mutational analysis was thus consistent with the cNES being the key NES within M, responsible for its cytoplasmic localization in the absence of LMB by mediating Crm-1-dependent nuclear export. By comparison, the NLSm mutant was unable to localize to the nucleus either in the absence or in the presence of LMB (Fig. 4B and C; Fn/c values of 0.42 and 0.33, respectively), implying that the NLSm mutation had abolished NLS activity.
FIG. 4.
RSV M contains a Crm1-recognized NES as shown by analysis of point mutant derivatives. (A) Schematic diagram of the mutational substitutions within the nNES, cNES, and NLS sequences, with mutated residues shown in lowercase letters (see also Fig. 3A). (B) Subcellular localization of NES and NLS mutants of M protein in transfected Vero cells in the absence or presence of LMB. Vero cells were transfected to express either GFP alone or GFP-M fusion proteins as indicated and treated either without or with 2.8 ng of LMB/ml as indicated 6 h prior to live cell CLSM imaging as described for Fig. 3. Scale bar, 10 μm. (C) CLSM files such as those in panel B, were analyzed as described in the legend to Fig. 2B. The results shown are for the means ± the SEM (n ≥ 30) determined from a single experiment, representative of two similar experiments, with significant differences in the Fn/c values between the absence and presence of LMB as indicated.
Crm1-mediated nuclear export is essential for virus assembly.
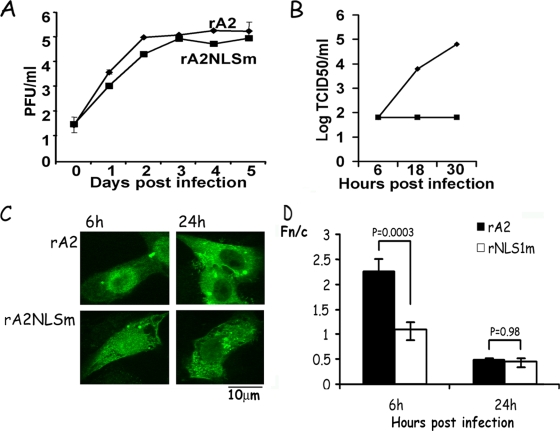
The cNESm and NLSm mutations that ablated the cNES or NLS, respectively, in the M gene were inserted into an antigenome cDNA clone of RSV. These plasmids were then used to recover M mutant recombinant RSV (rA2cNESm and rA2NLSm), using a previously described RSV reverse genetics system (6). It proved possible to recover rA2NLSm in three of three experiments, suggesting that the NLS mutation did not abolish virus production. In contrast, we were unable to recover rA2cNESm after seven attempts and multiple infection cycles over 5 days. Rescue could not be affected even using coexpression of GFP-M, the implication being that the NES mutation completely prevents virus production, which is consistent with the NES being critical to M's role in RSV infection. In all experiments, recombinant wild-type virus (rA2) was efficiently rescued. To investigate the rA2NLSm virus further, a multicycle growth (MOI = 0.1) assay was used. The rA2NLSm virus showed slower replication kinetics (Fig. 5A) relative to wt rA2 and no increase in infectious virus titer in a single step growth assay (MOI = 3) (Fig. 5B); these results are comparable to those for rA2 lacking NS2, where the virus is viable in cell culture but shows delayed growth kinetics (45). In addition, the localization of M in rA2NLSm-infected cells at early (6 h) and late (24 h) times after infection was examined by CLSM and image analysis (Fig. 5C and D) and compared to that in rA2-infected cells. M was present diffusely throughout the whole cell at 6 h in cells infected with rA2NLSm (Fn/c = 1.09 ± 0.16) but was predominantly nuclear in cells infected with rA2 (Fn/c = 2.26 ± 0.25); at 24 h after infection, M was predominantly cytoplasmic in both samples. Clearly, the NLS mutation leads to loss of nuclear accumulation of M at early times in infection.
FIG. 5.
Mutation of the cNES or NLS of M leads to loss in replication fitness. (A) Vero cells were transfected with plasmids encoding the RSV genome coding for wild-type M (rA2), nuclear-import-impaired M (rA2NLSm), or nuclear-export-deficient M (rA2cNESm). Virus was rescued from the supernatant and used to infect A549 cells; samples were collected every day for 5 days and titers of the infectious virus were determined as described in the text. rA2NLSm was recovered from all three rescued attempts, whereas rA2cNESm could not be recovered in seven attempts (data not shown). The data are shown as the logPFU/ml over time (means ± the SEM of triplicate samples). (B) Vero cells were infected with rA2 or rA2NLSm virus at an MOI of 3; samples were collected at indicated times, and titers of infectious virus were determined as described in the text. (C) Vero cells infected as in panel B were fixed, permeabilized, and analyzed for subcellular localization of M, as described in the legend to Fig. 1, at 6 h and 24 h p.i. (D) CLSM files such as those in panel C were analyzed as described in the legend to Fig. 2B. The results shown are for the means ± the SEM (n ≥ 15) determined from two similar experiments, with significant differences in the Fn/c values between the two virus infections as indicated.
Nuclear export-deficient M does not colocalize with regions of virus assembly and budding.
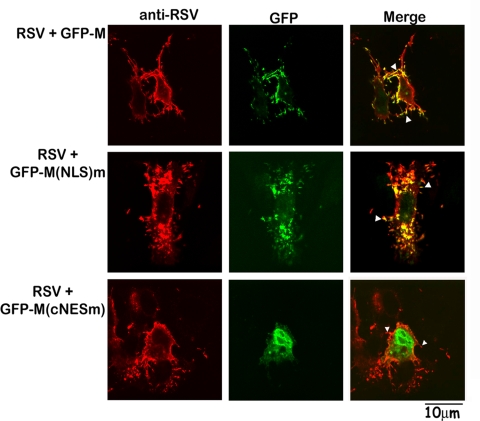
One hypothesis to explain the role of M's NES in RSV infection is that it is crucial for the targeting of M to the cytoplasm to enable its localization at sites of virus assembly. Since rA2cNESm could not be recovered, we set out to investigate this by expressing GFP-M and its GFP-NLSm and GFP-cNESm mutants in cells infected with RSV. Colocalization of GFP proteins with regions of RSV assembly and budding was analyzed 24 h later by CLSM following probing of fixed cells for RSV proteins (Fig. 6). As described previously (22), RSV can be observed budding from the cell surface; GFP-M and the nuclear import-impaired mutant (GFP-NLSm) were found to colocalize extensively with regions containing RSV proteins (yellow regions in top and middle images on the right), suggesting that they participate in virus assembly and are able to be packaged into the virus. GFP-cNESm, in contrast, did not colocalize with RSV proteins (bottom, no yellow regions in right image) and was not observed in any of the budding virus structures which have been observed by others (5, 17, 35). The clear implication is that the cNES mutation, by preventing shuttling of M to the cytoplasm, prevents its role in virus assembly. This is presumably the basis for it proving impossible to recover rA2cNESm, underlining the critical role of NES in RSV infection.
FIG. 6.
cNES mutated M cannot localize to sites of virus assembly. Vero cells were infected with RSV at MOI of 3 and transfected to express the indicated GFP-M derivatives at 6 h p.i. Cells were cultured for a further 18 h, fixed, permeabilized, and probed for RSV proteins using a goat anti-RSV antibody, followed by Alexa 568-conjugated donkey anti-goat antibody. Cells were imaged for RSV proteins (left panels) and GFP-M (middle panels) to identify regions of colocalization (yellow coloration in the merged images shown on the right); filamentous virus is indicated by arrowheads.
DISCUSSION
This study represents the first report of the nuclear export mechanism of an M protein belonging to the Paramyxovirus family, with important implications for the role of M proteins of negative-strand viruses such as those from Sendai virus, NDV, and VSV, regarding their roles in virus assembly (39) and their propensity to localize in the nucleus early in infection (32, 40, 48). Like RSV M, these proteins appear to have the ability to be exported into the cytoplasm, raising the possibility that they may also possess nuclear export mechanisms analogous to those implicated here for RSV M.
Our data show that RSV M is exported out of the nucleus in RSV-infected cells by a Crm1-dependent mechanism, through the action of the cNES (amino acids 194 to 206), which ensures that it is predominantly cytoplasmic in the transfected cell context in the absence of LMB. Mutational substitution of the central hydrophobic residues of the cNES inactivate the NES, rendering M predominantly nuclear and no longer sensitive to LMB. The clear implication is that the cNES is the dominant targeting signal under normal conditions. Without direct binding data for Crm1 association with M, we are, of course, unable to assert formally that Crm1 recognizes the cNES directly, but the fact that cNES is a classic leucine-rich NES, and targeted leucine mutations therein abolish its activity, strongly implies that this is the case. That the NES is critical to the function of M in the context of RSV is indicated by the fact that recombinant RSV carrying a mutated NES could not be recovered, in contrast to an NLS mutated virus, the clear implication being that the NES within M is essential for RSV virus production. The basis for this appears to be the critical role of the NES in directing M to the cytoplasm to enable localization to sites of virus assembly as demonstrated in the localization experiments with GFP-M derivatives in infected cells (Fig. 6). Specifically, nuclear-export-deficient GFP-M does not colocalize with sites of virus assembly in infected cells, in contrast to wild-type and nuclear-import-impaired GFP-M derivatives. Clearly, nuclear export of M is critical for RSV replication to enable M protein to coordinate the various assembly steps in the cytoplasm (15, 16, 31). Consistent with this idea, Crm1-mediated nuclear protein export at the appropriate time during infection (before 18 h p.i.) appears to be necessary for efficient virus assembly. That precisely timed localization of M at different subcellular locations is required for optimal virus assembly is also supported by the fact that rA2NLSm, which shows reduced nuclear localization of M at 6 h p.i. is impaired in virus production (Fig. 5). That the observed decrease in virus titer when RSV-infected cells were treated with LMB was likely to be due to effects on M rather than cellular factors could be concluded based on the fact that LMB addition later in infection had a reduced effect; if host cell export cargoes were the key targets of LMB in this context, the time of addition of LMB with respect to infection would not be expected to have differential effects on the kinetics of virus production. Since M is the only RSV structural protein known to localize in the nucleus of infected cells and, as shown here, possesses a Crm1-dependent nuclear export pathway, it can be postulated that the effects of LMB on RSV replication are very likely to be due directly to the observed inhibition of M nuclear export (Fig. 2).
We have previously shown that M localizes to the cytoplasm late in infection. This can now be understood in terms of M's specific ability to be exported out of nucleus by Crm1, and its ability to interact with M2-1 and associate with the cytoplasmic inclusions, where it inhibits RSV transcriptase in the later stages of infection (16, 31). This implies that a key nuclear role of M may be to ensure that viral replication and/or transcription in the cytoplasm proceed until a certain level of virus protein and RNA expression is reached. Influenza virus M1, whose nuclear export is mediated by the interacting nuclear export protein (see the introduction), is also known to inhibit virus transcriptase activity, with a similar sequestration of M1 protein away from the vRNP observed in the early stages of influenza virus infection (3). Based on our previous work showing that the nuclei of RSV-infected cells are deficient in transcription (13), together with the work of others regarding a 35% reduction in cellular transcription in RSV-infected cells (30), it seems likely that nuclear RSV M may serve to inhibit host cell transcription, as has been shown for VSV M (2). The mechanism by which M localizes in the nucleus is through the action of its IMPβ1-recognized bipartite NLS (Fig. 4B and C) (14), which appears to be the dominant targeting signal early in infection. Although the cellular signals triggering cNES-dependent relocalization of M from nucleus to cytoplasm later in infection have not been examined here, it seems likely that this process may, at least in part, be regulated by phosphorylation of M (29). Additional to the NLS, M residues 110 to 183 possess DNA-binding ability (14), which could conceivably mask the NLS in a fashion analogous to the chromatin remodeling factor SRY (10), where binding to specific DNA sequences competes with IMPβ1 binding and facilitates nuclear export.
In conclusion, we have shown here for the first time that the M protein of RSV is actively exported from the nucleus via a NES/Crm1-dependent pathway which is essential for virus assembly. Together with its IMPβ1-mediated nuclear import pathway (14), this presumably represents the basis of M's ability to be either nuclear or cytoplasmic at specific stages of the infectious cycle. Based on the fact that LMB treatment inhibits RSV virus production in infected cells, and the inability of cNES mutated RSV to be rescued, RSV M export from the nucleus may well prove to be a useful target for the development of anti-RSV therapeutics, which have thus far proved elusive.
Acknowledgments
We acknowledge the support of the National Health and Medical Research Council, Australia (fellowship 384109 and project grant 436611) and of the Monash Research Fund (Monash Small Grant).
We thank Tracy Waterhouse for the cell culture of mammalian cells, Erling Norrby for the monoclonal antibody to RSV M protein, and Minoru Yoshida for the LMB.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Ahmadian, G., P. Chambers, and A. J. Easton. 1999. Detection and characterization of proteins encoded by the second ORF of the M2 gene of pneumoviruses. J. Gen. Virol. 80(Pt. 8)2011-2016. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, M., and D. S. Lyles. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 728413-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akarsu, H., W. P. Burmeister, C. Petosa, I. Petit, C. W. Muller, R. W. Ruigrok, and F. Baudin. 2003. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 224646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askjaer, P., T. H. Jensen, J. Nilsson, L. Englmeier, and J. Kjems. 1998. The specificity of the CRM1-Rev. nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 27333414-33422. [DOI] [PubMed] [Google Scholar]
- 5.Bachi, T. 1988. Direct observation of the budding and fusion of an enveloped virus by video microscopy of viable cells. J. Cell Biol. 1071689-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, P. L., M. G. Hill, E. Camargo, H. Grosfeld, R. M. Chanock, and B. R. Murphy. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. USA 9211563-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial viruses, p. 1313-1352. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 8.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 755677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forgues, M., M. J. Difilippantonio, S. P. Linke, T. Ried, K. Nagashima, J. Feden, K. Valerie, K. Fukasawa, and X. W. Wang. 2003. Involvement of Crm1 in hepatitis B virus X protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol. Cell. Biol. 235282-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forwood, J. K., V. Harley, and D. A. Jans. 2001. The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high mobility group domain mediates nuclear import through importin β1. J. Biol. Chem. 27646575-46582. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195243-247. [DOI] [PubMed] [Google Scholar]
- 12.Garoff, H., R. Hewson, and D. J. E. Opstelten. 1998. Virus maturation by budding. Microb. Mol. Biol. Rev. 621171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghildyal, R., C. Baulch-Brown, J. Mills, and J. Meanger. 2003. The matrix protein of human respiratory syncytial virus localizes to the nucleus of infected cells and inhibits transcription. Arch. Virol. 1481419-1429. [DOI] [PubMed] [Google Scholar]
- 14.Ghildyal, R., A. Ho, K. M. Wagstaff, M. M. Dias, C. L. Barton, P. Jans, P. Bardin, and D. A. Jans. 2005. Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin β1 independent of importin α. Biochemistry 4412887-12895. [DOI] [PubMed] [Google Scholar]
- 15.Ghildyal, R., D. Li, I. Peroulis, B. Shields, P. G. Bardin, M. N. Teng, P. L. Collins, J. Meanger, and J. Mills. 2005. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J. Gen. Virol. 861879-1884. [DOI] [PubMed] [Google Scholar]
- 16.Ghildyal, R., J. Mills, M. Murray, N. Vardaxis, and J. Meanger. 2002. The respiratory syncytial virus (RSV) matrix protein associates with nucleocapsids in infected cells. J. Gen. Virol. 83753-757. [DOI] [PubMed] [Google Scholar]
- 17.Gower, T. L., M. K. Pastey, M. E. Peeples, P. L. Collins, L. H. McCurdy, T. K. Hart, A. Guth, T. R. Johnson, and B. S. Graham. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 795326-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakata, Y., M. Yamada, N. Mabuchi, and H. Shida. 2002. The carboxy-terminal region of the human immunodeficiency virus type 1 protein Rev. has multiple roles in mediating CRM1-related Rev. functions. J. Virol. 768079-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakata, Y., M. Yamada, and H. Shida. 2003. A multifunctional domain in human CRM1 (exportin 1) mediates RanBP3 binding and multimerization of human T-cell leukemia virus type 1 Rex protein. Mol. Cell. Biol. 238751-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harley, V. R., S. Layfield, C. L. Mitchell, J. K. Forwood, A. P. John, L. J. Briggs, S. G. McDowall, and D. A. Jans. 2003. Defective importin beta recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. USA 1007045-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang, B. C., J. H. Paik, H. Y. Jeong, H. J. Oh, J. W. Park, T. K. Kwon, D. K. Song, J. G. Park, S. P. Kim, J. H. Bae, K. C. Mun, M. H. Suh, M. Yoshida, and S. I. Suh. 2004. Leptomycin B-induced apoptosis is mediated through caspase activation and down-regulation of Mcl-1 and XIAP expression, but not through the generation of ROS in U937 leukemia cells. Biochem. Pharmacol. 68263-274. [DOI] [PubMed] [Google Scholar]
- 22.Jeffree, C. E., G. Brown, J. Aitken, D. Y. Su-Yin, B. H. Tan, and R. J. Sugrue. 2007. Ultrastructural analysis of the interaction between F-actin and respiratory syncytial virus during virus assembly. Virology 369309-323. [DOI] [PubMed] [Google Scholar]
- 23.Jin, H., D. Clarke, H. Z. Zhou, X. Cheng, K. Coelingh, M. Bryant, and S. Li. 1998. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251206-214. [DOI] [PubMed] [Google Scholar]
- 24.Kaptur, P. E., R. B. Rhodes, and D. S. Lyles. 1991. Sequences of the vesicular stomatitis virus matrix protein involved in binding to nucleocapsids. J. Virol. 651057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242540-547. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lam, M. H., B. Henderson, M. T. Gillespie, and D. A. Jans. 2001. Dynamics of leptomycin B-sensitive nucleocytoplasmic flux of parathyroid hormone-related protein. Traffic 2812-819. [DOI] [PubMed] [Google Scholar]
- 28.Lam, M. H., R. J. Thomas, K. L. Loveland, S. Schilders, M. Gu, T. J. Martin, M. T. Gillespie, and D. A. Jans. 2002. Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol. Endocrinol. 16390-401. [DOI] [PubMed] [Google Scholar]
- 29.Lambert, D. M., J. Hambor, M. Diebold, and B. Galinski. 1988. Kinetics of synthesis and phosphorylation of respiratory syncytial virus polypeptides. J. Gen. Virol. 69313-323. [DOI] [PubMed] [Google Scholar]
- 30.Levine, S., M. Peeples, and R. Hamilton. 1977. Effect of respiratory syncytial virus infection of HeLa-cell macromolecular synthesis. J. Gen. Virol. 3753-63. [DOI] [PubMed] [Google Scholar]
- 31.Li, D., D. A. Jans, P. G. Bardin, J. Meanger, J. Mills, and R. Ghildyal. 2008. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2-1 protein. J. Virol. 828863-8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyles, D. S., L. Puddington, and B. J. J. McCreedy. 1988. Vesicular stomatitis virus M protein in the nuclei of infected cells. J. Virol. 624387-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahy, B. W., and H. O. Kangro. 1996. Virology methods manual. Harcourt Brace, New York, NY.
- 34.Markwell, M. A., and C. F. Fox. 1980. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J. Virol. 33152-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCurdy, L. H., and B. S. Graham. 2003. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J. Virol. 771747-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagai, Y., T. Yoshida, M. Hamaguchi, M. Iinuma, K. Maeno, and T. Matsumoto. 1978. Cross-linking of Newcastle disease virus (NDV) proteins. Arch. Virol. 5815-28. [DOI] [PubMed] [Google Scholar]
- 37.Ohshima, T., T. Nakajima, T. Oishi, N. Imamoto, Y. Yoneda, A. Fukamizu, and K. Yagami. 1999. CRM1 mediates nuclear export of nonstructural protein 2 from parvovirus minute virus of mice. Biochem. Biophys. Res. Commun. 264144-150. [DOI] [PubMed] [Google Scholar]
- 38.Orvell, C., E. Norrby, and M. A. Mufson. 1987. Preparation and characterization of monoclonal antibodies directed against five structural components of human respiratory syncytial virus subgroup B. J. Gen. Virol. 683125-3135. [DOI] [PubMed] [Google Scholar]
- 39.Peeples, M. 1991. Paramyxovirus M proteins: pulling it all together and taking it on the road, p. 427-456. In D. Kingsbury (ed.), The paramyxoviruses. Plenum Press, Inc., New York, NY.
- 40.Peeples, M. E., C. Wang, K. C. Gupta, and N. Coleman. 1992. Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J. Virol. 663263-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon, I. K., C. Oro, M. M. Dias, J. P. Zhang, and D. A. Jans. 2005. A tumor cell-specific nuclear targeting signal within chicken anemia virus VP3/apoptin. J. Virol. 791339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 993944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slezak, K., M. Michalik, A. Kowalczyk, and H. Rokita. 2004. YY1 is recruited to the cytoplasm of vaccinia virus-infected human macrophages by the Crm1 system. Virus Res. 102177-184. [DOI] [PubMed] [Google Scholar]
- 44.Tannock, G. A., J. C. Hierholzer, D. A. Bryce, C. F. Chee, and J. A. Paul. 1987. Freeze-drying of respiratory syncytial viruses for transportation and storage. J. Clin. Microbiol. 251769-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teng, M. N., and P. L. Collins. 1999. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J. Virol. 73466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teng, M. N., and P. L. Collins. 1998. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J. Virol. 725707-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289283-296. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, T., Y. Nagai, S. Yoshii, K. Maeno, and T. Matsumoto. 1976. Membrane (M) protein of HVJ (Sendai virus): its role in virus assembly. Virology 71143-161. [DOI] [PubMed] [Google Scholar]