Abstract
The nucleocapsid protein (N) of vesicular stomatitis virus and other rhabdoviruses plays a central role in the assembly and template functions of the viral N-RNA complex. The crystal structure of the viral N-RNA complex suggests that the central region of the N protein interacts with the viral RNA. Sequence alignment of rhabdovirus N proteins revealed several highly conserved regions, one of which spanned residues 282 to 291 (GLSSKSPYSS) in the central region of the molecule. Alanine-scanning mutagenesis of this region suggested that replacement of the tyrosine residue at position 289 (Y289) with alanine resulted in an N-RNA template that is nonfunctional in viral genome replication and transcription. To establish the molecular basis of this defect, our further studies revealed that the Y289A mutant maintained its interaction with other N protein molecules but that its interactions with the P protein or with the viral RNA were defective. Replacement of Y289 with other aromatic, polar, or large amino acids indicated that the hydrophobic and aromatic nature of this position in the N protein is functionally important and that a larger aromatic residue is less favorable. Interestingly, we have observed that several single-amino-acid substitutions in this highly conserved region of the molecule rendered the nucleocapsid template nonfunctional in transcription without adversely affecting the replication functions. These results suggest that the structure of the N protein and the resulting N-RNA complex, in part, regulate the viral template functions in transcription and replication.
The prototypic rhabdovirus, vesicular stomatitis virus (VSV) within the family Rhabdoviridae, contains a nonsegmented negative-strand genomic RNA of 11,161 nucleotides (nt). The viral genome carries five genes, in the following order: the nucleocapsid (NC) protein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the large polymerase protein (L) genes. These genes are flanked by the leader and trailer sequences at the 3′ and 5′ termini of the genome, respectively (1). In the virion core, the viral genome is packaged in the form of a ribonucleoprotein (RNP) or NC complex, in which the genomic RNA is tightly encapsidated by the N protein. The viral RNA-dependent RNA polymerase consisting of the L and the P proteins is associated with the NC complex (1, 24). The viral NC serves as the template for transcription and replication. After virus entry and uncoating inside a cell, the polymerase (transcriptase) associated with the NC first transcribes the −ve sense genome into five subgenomic capped and polyadenylated mRNAs, which are translated to generate the viral proteins. Once sufficient viral proteins are made, the polymerase (replicase) synthesizes the full-length +ve strand RNA encapsidated with the N protein, which in turn acts as the template for synthesis of encapsidated genomic RNA (3). Previous studies (9, 28) have suggested the existence of two forms of the viral polymerase: one mediating the viral genome transcription (transcriptase) and one involved in genome replication (replicase). More recent studies have shown that the viral transcriptase contains the cellular translation elongation factor 1α, heat shock protein 60, and guanylyl transferase proteins in addition to the L and P proteins but that the replicase contains the N protein along with the L and P proteins (31).
Throughout the replication cycle, concurrent encapsidation of the nascent RNA by the N protein is necessary to generate a biologically active NC template. It is known that the N protein interaction with RNA is not sequence specific and that the N protein can readily make an N-RNA complex with cellular RNAs. However, the presence of the P protein confers specificity to encapsidate the viral RNA. Furthermore, N-P interaction is important for maintaining the N protein in a soluble and encapsidation-competent form by preventing formation of large insoluble aggregates (25). Earlier studies conducted to map a specific RNA binding domain(s) in the N protein revealed that the carboxy-terminal five amino acids are critical for encapsidation as well as genome replication (8). However, the recently identified crystal structure of the VSV N-RNA complex suggests the involvement of six positively charged amino acid residues located at a unique central cavity formed at the interface of the amino- and carboxy-terminal lobes of the N protein as well as several hydrophobic interactions for N-RNA complex formation (15). Furthermore, intermolecular interactions among adjacent N molecules along with sequestration of RNA in a ring-like structure are important for the stability of the N-RNA complex (15, 34). Structural comparison of the nucleoproteins of three negative-strand RNA viruses, Borna disease virus, rhabdovirus, and influenza A virus, suggests a very similar organizational pattern, having at least five conserved helices in the amino-terminal domain and three conserved helices in the carboxy-terminal domain that form a common structural motif for interaction with the ribose phosphate backbone of RNA in these negative-strand viruses (23).
Amino acid sequence alignment of the N protein of the members of vesiculoviruses identified two conserved central domains, of which the region comprising residues 282 to 291 (GLSSKSPYSS) is highly conserved among the members of the Rhabdoviridae family, including lyssa- and ephemeroviruses. The conserved nature of this region of the N protein suggests that this region may play an important function(s), although the definitive role of this region in N-RNA complex formation or in genome replication or transcription remains unknown. In the present study, we have examined the role of this region by generating a series of alanine-scanning mutants and testing the activity of the mutant proteins in replication, transcription, and encapsidation and also in protein-protein interactions. Our results suggest that the tyrosine residue at position 289 (Y289) is a critical amino acid for encapsidation and replication functions of the protein. Replacement of the Y289 residue with a number of other amino acids revealed that the hydrophobic and aromatic nature of this position is important for a functional N-RNA template. Our studies have also identified several other amino acids in this conserved region of the N protein, whose alterations have resulted in transcription-incompetent but replication-competent N-RNA templates. Our studies, for the first time, demonstrate differential activities of the N-RNA template with mutations in the N protein, suggesting that the N-RNA structure, in part, may play a key role in recognition of the NC template by the viral replicase and transcriptase machinery.
MATERIALS AND METHODS
Cell culture, viruses, antibodies, and reagents.
Baby hamster kidney (BHK-21) cells were maintained in Eagle's minimal essential medium (MEM) containing 5% fetal bovine serum with 100 units of penicillin, 20 units of streptomycin, and 20 units of kanamycin per ml of growth medium, as described earlier (6). Stocks of recombinant vaccinia virus (vTF7-3) expressing the bacteriophage T7 RNA polymerase (12) and the defective interfering (DI) particle of VSV were prepared in BHK-21 cells as described previously (21, 28, 29). The monoclonal antibody (MAb) against VSV N protein (10G4) and the rabbit polyclonal antibody against VSV P protein used in this study have been described previously (2, 7). Transient transfection of plasmid DNA was carried out by using Lipofectamine 2000 reagent (Invitrogen) per the manufacturer's protocol. MAbs and polyclonal antibodies against hemagglutinin (HA) and the FLAG epitope were obtained from Sigma and Santa Cruz Biotechnology.
Plasmids.
Plasmids for this study were constructed by using standard molecular cloning techniques. Since the previously described plasmid (29) encoding the N protein contained noncoding sequences after the stop codon for N, for convenience in cloning and mutagenesis, we first recreated the pN plasmid by PCR amplification of the N protein coding region from pVSVFL(+) (20) with primers N(+)For and N(−)Rev (Table 1) and subcloned the product at the EcoRI and HindIII sites in the pGEM3 vector under the T7 RNA polymerase promoter. The correctness of the nucleotide sequence of the N gene was confirmed by sequencing. The N protein expressed in cells transfected with the resulting plasmid (pN) was similar in levels of expression and activity in replication of DI particle genomes (data not shown). This plasmid (pN) encoding the wild-type (wt) N protein was used as the template for mutagenesis. The amino acid residues from 282 to 291 were individually replaced with alanine by PCR-based mutagenesis using the megaprimer method (32), and the mutants generated are shown in Fig. 1C. The PCR-based approach was also used along with primers [N(−)-Rev and EcoRI-HA-For or EcoRI-FLAG-For] given in Table 1 to incorporate HA or FLAG tags at the amino terminus of the N protein. The P protein with an HA tag at its carboxy terminus was also constructed by the PCR-based approach. All constructs were verified by nucleotide sequencing.
TABLE 1.
Primers used in this study
| Primer | Nucleotide sequencea |
|---|---|
| N(+)-For | ATATATGAATTCGCCGCCACCATGTCTGTTACAGTCAAG |
| N(−)-Rev | ATATATAAGCTTGGGTCATTTGTCAAATTCTG |
| EcoRI-HA-For | TATTGAATTCGCCACCATGTACCCATACGATGTTCCAGATTACGCTCTTTCTGTTACAGTCAAGAGAATC |
| EcoRI-FLAG-For | TATTGAATTCGCCACCATGGACTACAAGGACGACGATGACAAGTCTGTTACAGTCAAGAGAATC |
| 5HHEcoRI(+) | TATATGAATTCTCTTCGTCTGATGAGTCCGTGAGGACGAAACGGTACCCGGTACCGTCACGAAGACCACAAAACCAG |
| R2(−)-Rev | CTGTTAGTTTTTTTCATACGTACGTGATTACTGTTAAAGTTTC |
| R3(+)-For | CGTACGTATGAAAAAAACTAACAGTAATCGGCGCCTATGAAAAAAACTTTGATCCTTAAGACC |
| Le(−)-Rev | ACGAAGACAAACAAACCATTATTATC |
Primer sequences are in the 5′-to-3′ direction. Restriction enzyme sites in the primers are underlined.
FIG. 1.
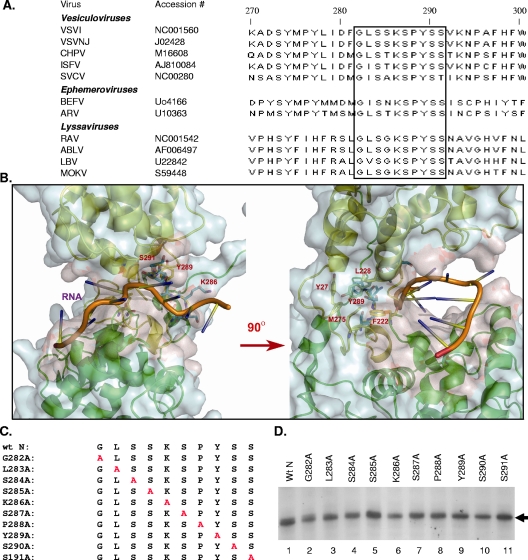
(A) ClustalW alignment of the amino acid sequences of rhabdovirus N protein presented with NCBI accession numbers. The highly conserved region comprising residues 282 to 291 (GLSSKSPYSS) of the N protein is boxed. VSVI, VSV, Indiana serotype; VSVNJ, VSV, New Jersey serotype; SVCV, spring viremia of carp virus; CHPV, Chandipura virus; ISFV, Isfahan virus; BEFV, bovine ephemeral fever virus; ARV, Adelaide river virus; RAV, rabies virus; ABLV, Australian bat lyssavirus; LBV, Lagos bat virus; MOKV, Mokola virus. (B) Structure of the VSV N-RNA complex, with a zoomed image of the conserved region identified by the sequence alignment. The two lobes of the N protein are shown as ribbons, colored green for the N-terminal lobe and yellow for the C-terminal lobe. The RNA cavity is outlined by the light red surface rendering on the surface of the N protein subunit (light blue). The conserved region spanning residues 285 to 291 is highlighted as a stick model (blue) lining the RNA cavity (left panel). The encapsidated RNA is shown with bases as rods and the ribose-phosphate backbone as a tube (orange). The cluster of hydrophobic residues around Tyr289 is shown as the stick model (yellow) sitting in the neck region between the N-terminal lobe (green) and the C-terminal lobe (yellow) (right panel, rotated 90° counterclockwise from the left panel). Residue Tyr289 and the residues in the hydrophobic cluster (Y27, F222, L228, and M275) are labeled. (C) Alanine-scanning mutants. The individual amino acid residues in the region comprising positions 282 to 291 (GLSSKSPYSS) of N are replaced with alanine (shown in red). The names of the mutants are shown on the left. (D) Expression of the alanine substitution mutants. BHK-21 cells in six-well plates were infected with vTF7-3 and then transfected with plasmids encoding either the wt or the mutant N proteins, followed by radiolabeling of the cells with [35S]methionine- and [35S]cysteine-containing medium. The radiolabeled proteins were immunoprecipitated with anti-VSV antibody and subsequently analyzed by SDS-PAGE and fluorography.
Construction of a plasmid carrying a negative-sense minigenome of VSV with eGFP as a transcription unit.
Our previous VSV minigenomes (19) encoded one or two transcription units but were of positive sense. Thus, initial replication of the positive-sense minigenomes was a prerequisite for production of the negative-sense templates for transcription. To avoid this requirement, we generated a negative-sense minigenome, pVSVmg-eGFP (see Fig. 5A). An overlapping PCR approach was used to generate the negative-sense VSV minigenome with enhanced green fluorescent protein (eGFP) as one of the transcription units. All primers used in this study for construction of various plasmids are given in Table 1. In the first step, primers 5HHEcoRI(+), containing sequences for a VSV-specific self-cleaving hammerhead ribozyme (17) at the 5′ end fused with the VSV trailer sequences, and R2(−)-Rev, with KasI and BsiWI sites, each flanked by VSV polymerase transcription initiation and termination signals, were used to amplify a 166-nt-long DNA (fragment 1) from the trailer end of pVSVFL(+). Similarly, primers R3(+)-For, containing 25 nt of complementarity to R2(−)Rev, and Le(−)-Rev, containing the VSV leader sequences, were used to amplify a 96-nt-long DNA (fragment 2) spanning the leader end of VSV. These two fragments were fused by overlapping PCR and further amplified using the 5HHEcoRI(+) and Le(−)-Rev primers, resulting in a 237-nt-long DNA (fragment 3) containing a VSV minigenome cassette. By use of overlapping PCR, fragment 3 was further fused with the hepatitis delta virus ribozyme (δ) and the T7 terminator (Tφ) sequences amplified from pDI (27) by using suitable primers with a SacI site at the 3′ end of Tφ. The resulting DNA fragment was cloned in the pGEM-3 vector at the EcoRI and SacI sites. Finally, eGFP-coding sequences released from pVSV-eGFP (6) by digestion with BsiWI were inserted in the above-mentioned construct at the same site. The resulting plasmid was named pVSVmg-eGFP.
FIG. 5.
Transcription activity of the N mutants. (A) Schematic of the plasmid carrying a negative-sense VSV minigenome with eGFP. The trailer (tr) and leader (le) regions of VSV along with the eGFP sequence in the negative-sense orientation flanked by the hammerhead ribozyme (HH) and the hepatitis delta virus ribozyme (δ) are shown. The bent arrows indicate the cleavage sites for the two ribozymes. The T7 RNA polymerase promoter (φ10) and the terminator (Tφ) are also shown in the plasmid. (B) Transcription of a negative-sense minireplicon of VSV expressing eGFP as a reporter for transcription activity. BHK-21 cells were transfected with pP, pL, and pVSVmg-eGFP and with (panel ii) or without (panel i) the plasmid encoding the wt N protein. At 24 hpt, the cells were directly observed for expression of eGFP. (C) Western blot analysis for detection of eGFP as a function of transcription activity of the minireplicon template. The cell lysates from the above-mentioned experiment were subjected to Western blot analysis by using anti-GFP antibody which detects the eGFP in the cells transfected with the plasmids expressing the P protein, the L protein, and the VSVmg-eGFP minireplicon along with the plasmid expressing (lane 2) or not expressing (lane 1) the N protein. (D) Representative fluorescent images of cells transfected with the plasmids expressing the P protein, the L protein, and the VSVmg-eGFP minireplicon along with the plasmid expressing the wt or mutant N proteins. Results for a control experiment in which wt N was not transfected (−N) are also shown. The panel numbers represent the alanine-scanning mutants corresponding to those positions in the N protein. (E) Western blot analysis for detection of eGFP as a function of transcription activity of the mutant N-RNA templates. The cell lysates from the experiment described for panel D were subjected to Western blot analysis. The upper panel shows the levels of eGFP; the lower panel shows the levels of actin determined using antiactin antibody in the Western blot. (F) Transcription activities of the N mutants per unit of the template. The ratios of average transcription activity to average replication activity (as shown in Fig. 2B) are shown for the respective N mutants. The ratio for the wt N protein is set at 1.
Metabolic labeling of RNA and analysis of RNA replication and encapsidation products.
BHK-21 cells grown in six-well tissue culture plates to 95% confluence were infected with vTF7-3 at a multiplicity of infection of 5 PFU/cell for 45 min at 37°C. Cell monolayers were washed with phosphate-buffered saline and transfected with 2 μg pDI (27) and 1.5 μg of either wt or mutant N plasmids, along with 1 μg of plasmid (pP) expressing the VSV P protein and 0.5 μg of plasmid (pL) expressing the L protein, by using Lipofectamine 2000. At 4 to 6 h posttransfection (hpt), cells were washed twice with Dulbecco's MEM containing 2% fetal bovine serum and incubated for 16 h with 0.6 ml of the same medium containing 50 μCi of [3H]uridine (Moravek Biochemicals, Brea, CA) per ml of the medium. The labeled RNAs in the form of the N-RNA complex were isolated by immunoprecipitation with anti-N or anti-VSV antibody and recovered by phenol-chloroform extraction, and subsequently, the labeled RNAs were analyzed by agarose urea gel electrophoresis and fluorography as described previously (29). RNA bands in the fluorograms were quantitated using the VersaDoc imaging system and the QuantityOne software program (Bio-Rad Laboratories).
Minigenome transcription.
For the transcription assay, BHK-21 cells in six-well tissue culture plates were infected with vTF7-3 and transfected with plasmids expressing the P protein, the L protein, and the minigenome VSVmg-eGFP along with the plasmid encoding the wt or the mutant N proteins, as described above. At 24 hpt, the transcription activity of the minigenome templates was assessed by examining the expression of eGFP in these cells. The transfected cells were visualized directly under an Olympus FV500/IX81 inverted confocal microscope, and images were captured. To determine the relative levels of transcription activity for the templates with the N mutants, cell extracts from transfected cells were analyzed by Western blotting for eGFP and quantitated by scanning of the bands in the fluorogram.
Immunoblotting and co-IP.
For N-N interaction studies, BHK-21 cells in six-well culture dishes were infected with vTF7-3 as described above and were cotransfected with 1 μg each of the plasmids encoding FLAG-tagged or HA-tagged wt N or mutant N proteins per the experimental design. At 18 hpt, cell lysates were coimmunoprecipitated by anti-FLAG or anti-HA polyclonal antibody and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis as described previously (7). The proteins were transferred to a nitrocellulose membrane and probed with MAb against the other tag per the experimental design and detected by secondary antibody conjugated with horseradish peroxidase (Millipore, Billerica, MA) and by use of an enhanced chemiluminescence detection kit (Amersham Biosciences, Pittsburgh, PA) per the manufacturer's instructions. For N-P interaction studies, BHK-21 cells were cotransfected with 1 μg each of the plasmids encoding the HA-tagged P protein and the wt N protein or the mutant N proteins. At 18 hpt, cells were washed and incubated in Dulbecco's MEM lacking methionine and cysteine for 45 min at 37°C and then labeled with 60 μCi/ml of Expre35S35S protein-labeling mix (NEN Life Sciences, MA) in 0.6 ml of the above-mentioned medium for a period of 2 h at 37°C. Cell lysates were prepared in 200 μl of coimmunoprecipitation (co-IP) buffer, and immunoprecipitation was conducted using anti-HA MAb as described previously (7). The proteins were subjected to SDS-PAGE and detected by fluorography.
Structure analysis.
Visual inspection of the N-RNA structure of VSV was carried out with the PyMol program (10), and the coordinates were obtained from the Protein Data Bank code 2GIC. The cartoon of the structure shown in Fig. 1B was also prepared by PyMol.
RESULTS
Functional significance of a highly conserved region in the N protein.
Using ClustalW software (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and the available sequence database, we identified several highly conserved regions in the N protein of the members of the vesiculovirus genus in the Rhabdoviridae family. However, when the N protein sequences for other genera (the vesiculovirus, lyssavirus, and ephemeroviurs groups) of this family were included in the analysis, the amino acid residues spanning positions 282 to 291 (GLSSKSPYSS) were found to be highly conserved (Fig. 1A). In the published crystallographic structure of the N protein (15), this region in the C-terminal lobe was found to be located in the neck connecting the two structural lobes (Fig. 1B). Five residues (S285, K286, S287, S290, and S291) line the cavity that accommodates the RNA between the two structural lobes. Residue Y289 resides in the center of a cluster of hydrophobic residues forming the core of this neck region (Fig. 1B). The location and the highly conserved nature of this region implied functional significance in viral NC template structure and function. Deletion of these 10 amino acids resulted in an N protein that did not support replication of a VSV DI RNA or transcription of a VSV minireplicon (data not shown). This deletion mutant was found to have lost its ability to encapsidate viral RNA as well as its ability to bind cellular RNA, and its interaction with the P protein was also abrogated (see Fig. 4B).
FIG. 4.
N-N and N-P interaction studies. (A) Examination of N-N interactions. BHK-21 cells were cotransfected with 1 μg of plasmid encoding HA- and/or FLAG-tagged wt or Y289A mutant N protein as shown above the panels. The cell lysates were subjected to co-IP with specific antibodies and subjected to Western blot analysis using the antibodies as shown below the panels. (B) Examination of N-P interactions. BHK-21 cells were cotransfected with the plasmid encoding P-HA and the wt or the mutant N plasmids as shown above each lane. At 16 hpt, the cells were radiolabeled with 35S-protein-labeling mixture for 2 h. The cell lysates were subjected to immunoprecipitation with anti-HA or anti-N antibody as shown below each lane and subsequently analyzed by SDS-PAGE and fluorography. The proteins detected by the co-IP assay are indicated on the right side of the panel.
To identify the important amino acid residues in this region associated with the functions of the N protein, alanine-scanning mutagenesis was employed. Each of the amino acids in this region was mutated to alanine individually (Fig. 1C). All the mutant proteins were found to be expressed in transfected cells at levels similar to that for the wt N protein (Fig. 1D), and no noticeable difference from the wt protein in terms of the stability of the mutant proteins was observed in pulse and chase experiments (data not shown). Furthermore, under denaturing and reducing gel electrophoresis conditions, the wt N protein or the mutant proteins were found as a 47-kDa monomeric protein.
The Y289A mutant is defective in the replication function of the N protein.
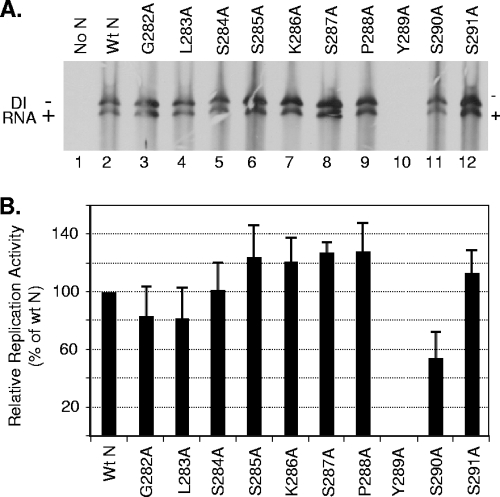
To examine if the individual alanine-scanning mutants can support replication of VSV, cells expressing the P, the L, and the wt or mutant N proteins were examined for replication of the genome of a DI particle of VSV as described previously (29). Analysis of replication products shows that all the mutant proteins, with the exception of Y289A, supported replication of DI RNA (Fig. 2A). The levels of replication with several mutant N proteins were higher than that obtained with wt N protein, whereas the replication levels supported by several other mutant proteins were either similar to or slightly lower than that of the wt N protein. The Y289A mutant protein failed to support any detectable levels of replication even after expression of larger or smaller amounts of the protein (data not shown). Quantitative analysis of results from five independent experiments (Fig. 2B) demonstrated that S285A, K286A, S287A, P288A, and S291A proteins supported consistently higher levels of replication than wt N; G282A, L283A, and S284A possessed replication activity similar to that of wt N; and S290A was 60% to 75% as active as the wt N protein. However, the Y289A mutant did not support replication to any levels in these repeat experiments.
FIG. 2.
Replication activities of the N mutants. (A) Replication of DI RNA supported by the alanine-scanning N mutants. The experiment was performed as described in Materials and Methods, and the radiolabeled RNAs were isolated, analyzed by agarose-urea gel electrophoresis, and detected by fluorography. The minus and plus signs represent genomic and antigenomic RNAs, respectively, of the DI particle. (B) Quantitative determination of the replication activity of the N mutants. The data were obtained by densitometric scanning of the fluorograms as described in Materials and Methods. The histograms represent average values from five independent experiments, with the error bars indicating standard deviations. The activity of the wt N protein is set at 100%.
Y289A is defective in encapsidation of viral RNA.
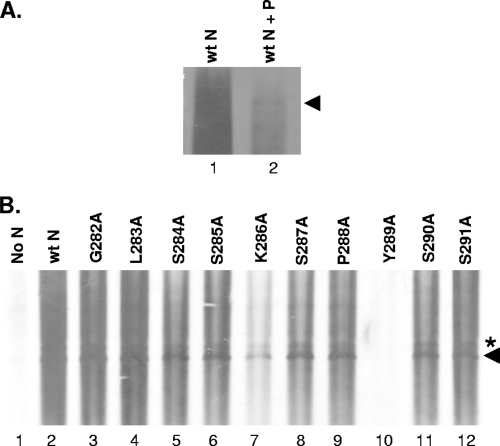
Since the N protein binds and encapsidates the viral RNA to form the NC template for replication and transcription, it is possible that the defect in replication of the Y289A mutant might have resulted from a defect in the encapsidation function of the protein. To gain insight into this possibility, we conducted an assay to measure the RNA encapsidation activity of the N mutants. BHK-21 cells infected with vTF7-3 were cotransfected with plasmids (pDI) carrying the DI genome (27) as a source of the viral genomic RNA, along with the plasmids expressing the wt or the mutant N proteins in the absence or presence of P protein. At 6 hpt, the DI RNAs were radiolabeled with [3H]uridine for 12 h and the N-RNA complexes were isolated from the cell lysates by immunoprecipitation with anti-N protein antibody. The radiolabeled RNAs recovered from the immunoprecipitated complexes were separated by electrophoresis and detected by fluorography as described previously (26). In a control experiment in which the cells were transfected with the plasmid encoding DI RNA as well as the wt N protein, we found that the N protein immunoprecipitates large amounts of cellular RNAs of heterogeneous size along with the DI RNA (Fig. 3A, lane 1) but that the amount of cellular RNAs immunoprecipitated by the N protein was significantly reduced in the presence of the P protein (Fig. 3A, lane 2) and encapsidation of DI RNA was more readily detected. The results indicate that the P protein reduces the nonspecific binding of N protein to cellular RNAs and enhances the specific encapsidation of viral RNA.
FIG. 3.
Encapsidation of DI RNA. (A) BHK-21 cells in six-well tissue culture plates were infected with vTF7-3 and transfected with the plasmids expressing the wt N proteins as well as the plasmid carrying the DI genome (pDI) in the absence (lane 1) or presence (lane 2) of P protein. At 6 hpt, the cell monolayer was radiolabeled with [3H]uridine for 16 h. The radiolabeled RNA in the form of the N-RNA complex was isolated by immunoprecipitation by using anti-N antibody, and the recovered RNAs were analyzed by agarose-urea gel electrophoresis and fluorography. The RNA band identified by the arrowhead is the cleaved full-length negative-sense DI RNA. In lane 1, the cellular RNAs of heterogeneous sizes mask the DI RNA. (B) Encapsidation of DI RNA by the mutant N proteins. The experiment was conducted as described above in the presence of the P protein. The RNA band identified by the asterisk represents the uncleaved form of the DI RNA containing the sequences of the ribozyme still attached at the 3′ end, which is detected under some conditions, whereas the RNA band identified by the arrowhead is the cleaved full-length negative-sense DI RNA.
A subsequent encapsidation experiment in the presence of P protein revealed that the DI RNA could be encapsidated by most of the mutant N proteins (Fig. 3B), with the exception of the Y289A mutant N protein, which failed to encapsidate the DI RNA in multiple repeat experiments. These results suggest that the inability of the Y289A mutant to support RNA replication is, in part, due to the loss of the encapsidation function of the protein. In addition, the Y289A mutant lost its ability to bind cellular RNAs nonspecifically, as shown by the absence of any background radioactivity with the mutant (Fig. 3B, lane 10). It is interesting to note that the K286A mutant, which supported DI RNA replication at levels greater than that of the wt protein (Fig. 2), consistently encapsidated DI RNA less efficiently, as evidenced by the relatively low levels of the RNA recovered from the immunoprecipitated complexes (Fig. 3B, lane 7), compared to those for the wt or the other mutant N proteins. This is not due to the anti-N MAb used in these studies, since the use of anti-VSV antibody also produced similar results. These results suggest that the efficiency at which the N protein encapsidates the viral RNA is not necessarily a determinant of the efficiency of replication of the NC template.
Y289A maintains N-N interaction but is defective in N-P interaction.
The crystal structure of the VSV N-RNA complex revealed that 10 molecules of N protein form a ring-like structure, in which each of the N molecules is bound to 9 nt of the RNA chain (15). Furthermore, extensive intermolecular interactions among the neighboring N molecules have also been suggested. These N-N interactions are important for a stable N-RNA complex structure. Therefore, we wanted to examine if mutation at the Y289 residue would have a negative effect on N-N interaction resulting in defects in the encapsidation functions of the protein. To determine the N-N interaction ability of the Y289A mutant, we conducted a co-IP assay using the epitope-tagged proteins. The wt N and Y289A mutant proteins tagged with an HA or FLAG epitope at the amino terminus were used in this study. Plasmids encoding HA-tagged wt N (HA-Nwt), FLAG-tagged wt N (FLAG-Nwt), HA-tagged Y289A mutant N (HA-Y289A), or FLAG-tagged Y289A mutant N (FLAG-Y289A) under the T7 RNA polymerase promoter were transfected either individually or jointly as shown in Fig. 4A into cells expressing the T7 RNA polymerase through vTF7-3 infection. At 16 hpt, cell lysates were prepared and the protein complexes were immunoprecipitated with anti-HA (lanes 1 to 3) or anti-FLAG (lanes 4 to 6) MAbs and the proteins were separated by 10% SDS-PAGE. Subsequently, the immunoprecipitated proteins were detected by Western blotting using anti-FLAG (lanes 1 to 3) or anti-HA (lanes 4 to 6) polyclonal antibodies. The results show that HA-Nwt specifically interacts with FLAG-Y289A, as evidenced by the detection of FLAG-Y289A protein in the complexes immunoprecipitated with anti-HA antibody (lane 3). The observation that similar levels of FLAG-Nwt and FLAG-Y289A proteins (lanes 2 and 3) were detected in Western blots indicated that the mutation did not have any adverse effect on N-N interaction. Similar results were obtained in a converse experiment using anti-FLAG antibody for immunoprecipitation and anti-HA antibody for Western blotting (lanes 4 to 6). Thus, our results suggest that the Y289A mutant N protein is not defective in N-N interaction.
It has been shown that the interaction of the P protein keeps the N protein in a soluble form and provides specificity to encapsidate the viral RNA (18). Since the Y289A mutant N protein maintains N-N interaction, we wanted to examine if N-P interaction is affected by the mutation. BHK-21 cells infected with vTF7-3 were cotransfected with plasmids encoding either the wt N or the mutant N proteins along with the plasmid encoding the P protein tagged with HA at its carboxy terminus (P-HA). Following radiolabeling of the proteins with a 35S-amino acid mixture, the interaction of the N protein with the P protein was examined by detection of wt or mutant N protein, using anti-HA antibody and SDS-PAGE analysis. As can be seen in Fig. 4B, the anti-HA antibody could immunoprecipitate the wt N protein (lane 6), indicating that these two proteins specifically interact with each other when coexpressed. The observation that the HA antibody did not immunoprecipitate the N protein when expressed alone (lane 1) further indicates the specificity of N-P interaction. The Y289A mutant N protein could not be detected with the anti-HA antibody (lane 7), indicating that the Y289A protein does not interact with the P protein. The deletion mutant NΔ10, which did not support DI RNA replication or DI RNA encapsidation, was also found to be defective in its ability to interact with the P protein (lane 8). These results suggest that the NΔ10 mutant as well as the Y289A mutants do not interact with the P protein, and the encapsidation defect of these mutants may, in part, be due to the loss of N-P interaction.
Transcription activity of alanine-scanning mutants.
To gain insight into how the mutations in this conserved region of the N protein affect the NC template functions in transcription, we designed an experiment to measure the transcriptional activity of a minireplicon of VSV. To carry out this experiment, we first constructed a negative-sense minigenome (minireplicon) of VSV encoding eGFP (Fig. 5A). Such a minireplicon, when assembled into an NC structure, can be transcribed by the viral RNA polymerase to generate eGFP mRNA and subsequently eGFP fluorescence even in the absence of replication of the minireplicon template. In cells transfected with the plasmid carrying this minireplicon along with the plasmids encoding the P, the L, and the wt N proteins, expression of eGFP could be readily detected by fluorescence imaging of the cells as shown (Fig.5Bii). In control experiments in which the cells were transfected with all the plasmids except the N plasmid, eGFP expression was not detected (Fig.5Bi). Furthermore, Western blotting of cell lysates with anti-GFP antibody readily detected eGFP (Fig. 5C).
With this system, we examined the transcriptional activity of all alanine-scanning N mutants by fluorescence microscopy. Representative fluorescence images for all the mutants are shown in Fig. 5D. Consistent with our finding that the Y289A mutant does not support genome replication, RNA encapsidation, or interaction with the P protein, the results showed that the Y289A mutant N protein is defective in transcription (Fig. 5D, panel 289). Interestingly, we also observed that the K286A and S290A mutants, which supported high levels of replication, were completely inactive in transcription as well (Fig. 5D, panels 286 and 290). Another mutant, L283A, showed a significant reduction in transcription activity (panel 283), while rest of the mutants showed activity to various degrees. Similar results were obtained in multiple repeat experiments. The level of eGFP in these cells was subsequently determined by Western blotting and used as a quantitative measure of transcription activity of NC templates containing the mutant N proteins (Fig. 5E). Data from three independent experiments were used to obtain a quantitative measure of average transcriptional activities of the mutant N proteins (data not shown), which revealed that the mutants exhibited transcription activities that differed widely from one another. Some mutants possessed activity similar to that of the wt N protein, while others showed significantly less activity. It is interesting to note that unlike replication in which several of the mutant N proteins supported higher levels of replication than the wt N protein, none of the mutant N proteins were hyperactive in transcription.
Since the transcription activity is dependent on the level of the NC template, we considered that the ratio of transcription activity to replication activity would provide a more accurate measure of the transcription activities of the mutants. The results (Fig. 5F) suggest that the mutant N proteins can be grouped into four categories based on their transcription activity per unit template: the G282A, S284A, and S291A mutants possessed transcription activities comparable (greater than 75%) to those for the wt N protein; the S285A and P288A mutants possessed intermediate levels (25% to 75%); the L283A and S287A mutants possessed low levels (less than 25%) of activity; and the K286A, Y289A, and S290A mutants were completely inactive in transcription.
The hydrophobic and aromatic nature of the residue at position 289 is important for N protein function.
Since our results showed that Y289 is a critical residue for the N protein functions, we investigated the nature and requirement of this residue. The residue was mutated to other aromatic amino acid residues (F and W) or charged and polar residues (K, E, and N). Using these mutants, we found that Y289F and Y289W interacted well with the P protein but that the Y289K, Y289N, or Y289E mutants did not (Fig. 6A), although very small amounts of Y289E protein could be detected (lane 6). Furthermore, Y289F supported replication consistently at levels similar to or higher than those for the wt N protein, whereas the Y289W mutant was on average about 15% as active as the wt protein (Fig. 6B). While the RNP template with the Y289F mutation demonstrated, on average, 15 to 20% transcription activity (Fig. 6B), the Y289W RNP template was completely inactive in transcription in multiple repeat experiments. The other mutants did not exhibit any measurable activity in replication or transcription (Fig. 6B). Taken together, these results suggest not only that the hydrophobic nature of this residue is important but also that an aromatic residue is critical for the functions of the N protein.
FIG. 6.
Activity of N mutants with substitutions of different amino acids at position 289. (A) Interaction of Y289 mutants of the N protein with the P protein. BHK-21 cells were transfected with plasmids encoding P-HA and the wt or mutant N proteins as shown above each lane. Interactions of the proteins were examined using anti-HA antibody as described for Fig. 4B. (B) Replication and transcription activities of Y289 mutants. Histograms represent average activities from two independent experiments. Replication activity was determined as described in the legend to Fig. 2, with the wt N protein activity set at 100%. The ratio of transcription to replication was determined as described for Fig. 5F, with the wt N protein activity set at 1.
Some mutant N proteins exhibit the ts transcription phenotype.
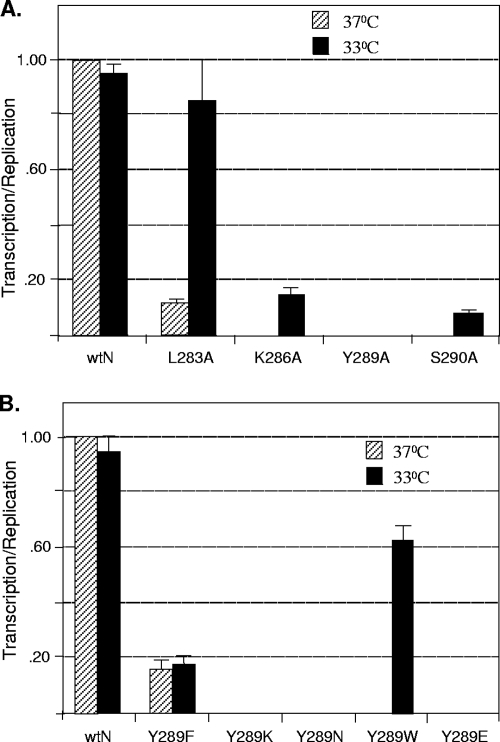
To examine if any of the mutants that are nonfunctional in transcription possess the temperature-sensitive (ts) phenotype, we determined the transcription activities of the mutant proteins at 33°C, a temperature generally considered to be permissive for the ts phenotype. Since the L283A mutant exhibited a very low level of transcription activity, we also included this mutant in our studies. Accordingly, the experiment was conducted at the lower temperature, and the replication and transcription activities of the mutants were measured as described above and compared to the activities at 37°C. Again, to provide a more accurate measure of the transcription activities of various mutants at the two temperatures, we calculated the ratio of transcription activity to replication activity at the corresponding temperature as shown in Fig. 5F. The results (Fig. 7A) show that the transcription activity of the L283A mutant is nearly fully restored at 33°C. On the other hand, the K286A and S290A mutants exhibited very low level of transcriptional activity at 33°C. The Y289A mutant was still nonfunctional at the lower temperature. Thus, our results suggest that some of the single-amino-acid substitutions in this highly conserved region of the N protein lead to the ts phenotype in the transcription of the resulting NC templates, while substitutions at several other residues can be lethal.
FIG. 7.
ts transcription phenotype of mutant N proteins. (A) Duplicate cultures of BHK-21 cells were transfected with pVSVmg-eGFP, pP, pL, and either the pN or the pN mutants, as shown at the bottom. One set of cultures was incubated at 37°C while the other was incubated at 33°C. At 24 hpt, cell lysates were prepared and subjected to Western blot analysis for detection of eGFP and further quantitated as described for Fig. 5E. The replication activities of the mutants at the two temperatures were determined by the DI replication assay as described in the legend to Fig. 2. The ratio of transcription to replication activity for the wt N protein at 37°C was set at 1, and the ratios for the wt N protein at 33°C or the mutants at the two temperatures were determined. The average values for transcription/replication from three independent experiments are plotted in the histogram, with error bars indicating standard deviations. (B) Transcription activity of Y289 substitution mutants of N. The experiment was conducted at 37°C and 33°C, and the data were obtained as described for panel A. The average values for transcription/replication from three independent experiments are plotted in the histogram, with error bars indicating standard deviations.
To determine if the other substitution mutants of Y289 exhibit the ts phenotype in transcription, we performed replication and transcription assays with the Y289F, Y289W, Y289K, Y289E, and Y289N mutants at 37°C as well as 33°C. The results (Fig. 7B) show that, although Y289F mutant exhibited low levels of transcription per unit replication at both temperatures, the Y289W mutant at 37°C exhibited about 15% replication activity but was completely inactive in transcription in multiple repeat experiments. However, when assayed at 33°C, both replication and transcription activities were readily detectable and the ratio of transcription to replication was considerably higher, suggesting that this mutant RNP possesses the ts phenotype for transcription. Other Y289 mutant RNPs were completely inactive in transcription as well as replication at both temperatures. Thus, our data indicate that replacement of Y289 with W can be conditionally defective while replacement with other residues, such as K, E, or N, is lethal for the function of the N protein in transcription.
DISCUSSION
Comparison of the amino acid sequences of the N proteins of vesiculoviruses revealed the existence of several conserved regions. The studies presented in this communication focused on a region spanning residues 282 to 291 (GLSSKSPYSS) of the VSV N protein, which is highly conserved among rhabdoviruses and is located in the central region of the molecule. By mutational analysis, we show here that this region plays critical roles in N-P interaction as well as RNA encapsidation functions of the protein. Our results further show that single- amino-acid alterations in this region have dramatic effects on transcription and replication functions of the N-RNA template, suggesting that the N protein, in part, regulates N-RNA template functions.
The crystal structure of the N-RNA complex suggests that this particular domain is located in close proximity to the central cavity of the N molecule that is formed by the amino- and carboxy-terminal lobes of the protein (15). The central cavity sequesters the viral RNA in the N-RNA complex (15). Our observation that Y289A mutant N protein does not encapsidate the viral RNA (Fig. 3) suggests that this particular residue is possibly involved in maintaining the integrity of the cavity for encapsidating the RNA. Additionally, because this mutation also resulted in the loss of binding of the P protein, it further suggests that the binding site for the P protein is in close proximity to or on the opposite side of the RNA binding cavity. Alternatively, the Y289A mutation may cause misfolding of the protein, resulting in the loss of the P binding site as well as the RNA binding. Thus, a single mutation in this highly conserved region affects the overall structure of the protein in such a manner that at least two major functions of the N protein are impaired. The structural role of the highly conserved region is also consistent with its location in the center of the neck region of the molecule. Alteration of the Y289 residue to alanine might result in exposure of nearby buried residues in the groove, thereby altering the overall conformation, leading to loss of the RNA encapsidation function of the protein. The suggestion that Y289 may be critical for holding up the RNA cavity is further strengthened by the observation that mutation of Y289 to phenylalanine or tryptophan, the two hydrophobic residues with aromatic rings, has no negative effect on RNA encapsidation or the replication or transcription functions of the protein, although replacement with the bulkier tryptophan residue resulted in a reduced or conditional (ts) phenotype for transcription. Mutants with K, E, or N substituted at this position are most likely too dramatic to fold the protein correctly and therefore do not interact with the P protein or encapsidate the RNA.
Structure of the N-RNA complex of VSV suggests involvement of at least six positively charged residues from the two lobes of the protein whose side chains directly bind the phosphate backbone of RNA (15). The comparison of the N-RNA structure of VSV with that of rabies virus supports this notion (22). Our data show that mutation of K286 to alanine resulted in a protein which could still encapsidate the RNA, albeit less efficiently (Fig. 3). These results suggest that mutation of K286 to alanine might have resulted in recruitment of additional positively charged residues located in close proximity for RNA encapsidation or that the RNA encapsidation function of the protein may not require the involvement of all the positively charged residues identified in the crystal structure (15). It would be interesting to examine how many of these positively charged residues located in the RNA binding cavity can be altered without adversely affecting the encapsidation or replication functions of the protein.
Although the K286A mutation in N resulted in less-efficient RNA encapsidation, it was interesting to see that the mutant N protein supported replication consistently at a higher level than the wt N protein. Furthermore, mutation of several residues (S285, S287, P288, and S291) in close proximity to K286 individually to alanine also supported higher levels of replication than the wt N protein (Fig. 2). The reason(s) for the hyperactivity of these mutant RNP templates in replication is not known at this time, but it is possible that mutations in this stretch of residues reduced their grips on the RNA in the RNP template such that these template RNAs are more accessible to the viral replicase machinery. However, the same mutations had varied effects on the transcription functions of the RNP template (Fig. 5). In this case, it may be possible that the viral transcriptase machinery associates with the RNA template less efficiently or not at all. The differential activity of the same RNP template for replication and transcription suggests that the viral transcription and replication machinery may recognize the RNP template distinctly and further supports the data showing that transcriptase and replicase of VSV are distinct molecular complexes (16, 31). Furthermore, our results suggest that the structures of the N protein and the resulting N-RNA template, in part, seem to play a significant role in regulating the template functions in transcription and replication. Previous studies using the polR1 mutant of VSV with a single-amino-acid substitution in the N protein (R179H) have suggested that the N-RNA template regulates the viral transcription and replication functions (4, 30). Our results presented here, and the interpretation that the structure of the N protein regulates the N-RNA template functions, are consistent with those for previous studies. It will be interesting to examine the crystal structures of the N-RNA complexes with these mutations to provide further insights into how the transcriptase and replicase recognize the same template distinctly and execute the transcription and replication activities with such varied efficiencies.
Previous biochemical and mutational studies have shown that the P binding domain in the N protein lies at the carboxy-terminal end of N, including the last 5 residues (11, 13, 14, 33). Recent structural studies also suggest that residues located in the extended loop of the carboxy-terminal lobe play a role in N-P interaction since deletion of six residues (NΔ347-352) resulted in an N protein that interacted weakly with the P protein (34). Since we were unable to detect N-P interaction using Y289A mutant in co-IP assays (Fig. 4), it is possible that the Y289A mutant does not interact with the P protein at all or interacts very weakly. The Y289A mutant may have misfolded in such a manner that the P binding site and the RNA binding cavity may have collapsed, resulting in loss of P binding and RNA encapsidation. Mutation of the Y289 residue to F or W resulted in N proteins that interacted well with the P protein, suggesting that the hydrophobic and aromatic nature of this residue is critical for N-P interaction and the N protein functions. In this context, it is noteworthy that the serine residue (S290) located adjacent to Y289 in this highly conserved region, when altered to tryptophan (S290W), resulted in an N protein that maintained its interaction with the P protein but lost its RNA encapsidation function (34). It is possible that this highly conserved region not only forms part of the RNA binding cavity of the N protein but also is critical for the overall structure of the protein such that mutations in this region have negative effects on N-P interactions as well.
Several of the N mutants that generated defective or less competent RNP templates in transcription exhibited the ts phenotype. At 33°C, the temperature used as permissive in this study, the N-RNA template with the L283A mutant showed transcription activity at a level similar to that for the wt N protein and significantly higher activity than the mutant template at 37°C, indicating that the mutant protein had recovered full activity in transcription at the lower temperature. Other mutants (K286A, Y289W, and S290A) also exhibited the ts phenotype for transcription, although the level of transcription at the permissive temperature was only about 10 to 15% of that for the wt N protein at the same temperature. These data suggest that structural alterations at the lower temperature result in RNP templates that can be readily recognized by the transcriptase complex to transcribe. It has been shown that the stability of the RNA polymerase-template RNA complex is sensitive to temperature (5). The low temperature could have restored the stability of the mutant N protein-RNA template-transcriptase complex. However, this was not observed for the Y289A mutant tested, indicating that the specific structural changes of some mutant N-RNA templates due to temperature shiftdown are more favorably recognized by the viral transcriptase machinery for transcription. It should be noted that using the VSV infectious clone (20), we have inserted the L283A and K286A mutant N protein-coding regions and have recovered these two mutant VSVs at the lower permissive temperature. Initial studies have shown that these viruses exhibit the ts phenotype for virus growth and overall RNA synthesis. More-detailed studies are being carried out to characterize these viruses and will be reported elsewhere.
Thus, from the results presented in this communication, we conclude that the highly conserved region of the N protein located near the cavity between the two lobes of the protein plays an important role in the overall structure of the protein and that the N-RNA template functions are partly regulated by the structure of the N-RNA complex. The study has identified several key residues whose alterations affect RNA encapsidation, interaction with the P protein, and the N-RNA template functions in transcription and replication.
Acknowledgments
We thank You Zhou and Terry Fangman of the microscopy core facility, Center for Biotechnology, UNL, for help with fluorescence microscopic studies and members of the Pattnaik laboratory for helpful comments on the manuscript. We also thank D. Lyles for anti-N MAb.
This investigation was supported by Public Health Service grant AI-34956 (to A.K.P.) and AI-50066 (to M.L.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 25 March 2009.
REFERENCES
- 1.Banerjee, A. K. 1987. Transcription and replication of rhabdoviruses. Microbiol. Rev. 5166-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, B. L., G. Brewer, and D. S. Lyles. 1994. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J. Virol. 68555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, B. M., M. Leppert, and D. Kolakofsky. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23837-845. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, J. L., R. L. Jackson, and J. Perrault. 1997. Isolation and characterization of vesicular stomatitis virus PoIR revertants: polymerase readthrough of the leader-N gene junction is linked to an ATP-dependent function. Virology 22957-67. [DOI] [PubMed] [Google Scholar]
- 5.Dalton, R. M., A. E. Mullin, M. J. Amorim, E. Medcalf, L. S. Tiley, and P. Digard. 2006. Temperature sensitive influenza A virus genome replication results from low thermal stability of polymerase-cRNA complexes. Virol. J. 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, S. C., D. Nayak, Y. Zhou, and A. K. Pattnaik. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 806368-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, S. C., and A. K. Pattnaik. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J. Virol. 798101-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, T., B. K. Chakrabarti, D. Chattopadhyay, and A. K. Banerjee. 1999. Carboxy-terminal five amino acids of the nucleocapsid protein of vesicular stomatitis virus are required for encapsidation and replication of genome RNA. Virology 259219-227. [DOI] [PubMed] [Google Scholar]
- 9.Das, T., A. K. Pattnaik, A. M. Takacs, T. Li, L. N. Hwang, and A. K. Banerjee. 1997. Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology 238103-114. [DOI] [PubMed] [Google Scholar]
- 10.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 11.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 845655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill, D. S., D. Chattopadhyay, and A. K. Banerjee. 1986. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc. Natl. Acad. Sci. USA 838873-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, T. J., S. Macpherson, S. Qiu, J. Lebowitz, G. W. Wertz, and M. Luo. 2000. Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J. Virol. 749515-9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, T. J., X. Zhang, G. W. Wertz, and M. Luo. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313357-360. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, A. K., D. Shaji, and A. K. Banerjee. 2003. Identification of a novel tripartite complex involved in replication of vesicular stomatitis virus genome RNA. J. Virol. 77732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. USA 1022579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard, M., and G. Wertz. 1989. Vesicular stomatitis virus RNA replication: a role for the NS protein. J. Gen. Virol. 702683-2694. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, L. N., N. Englund, and A. K. Pattnaik. 1998. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J. Virol. 721805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 924477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, T., and A. K. Pattnaik. 1997. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology 232248-259. [DOI] [PubMed] [Google Scholar]
- 22.Luo, M., T. J. Green, X. Zhang, J. Tsao, and S. Qiu. 2007. Conserved characteristics of the rhabdovirus nucleoprotein. Virus Res. 129246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, M., T. J. Green, X. Zhang, J. Tsao, and S. Qiu. 2007. Structural comparisons of the nucleoprotein from three negative strand RNA virus families. Virol. J. 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyles, D. S., and C. E. Rupprecht. 2007. Rhabdoviridae, p. 1363-1408. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 25.Masters, P. S., and A. K. Banerjee. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 622658-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pattnaik, A. K., L. A. Ball, A. LeGrone, and G. W. Wertz. 1995. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology 206760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 691011-1020. [DOI] [PubMed] [Google Scholar]
- 28.Pattnaik, A. K., L. Hwang, T. Li, N. Englund, M. Mathur, T. Das, and A. K. Banerjee. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 718167-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattnaik, A. K., and G. W. Wertz. 1990. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J. Virol. 642948-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrault, J., G. M. Clinton, and M. A. McClure. 1983. RNP template of vesicular stomatitis virus regulates transcription and replication functions. Cell 35175-185. [DOI] [PubMed] [Google Scholar]
- 31.Qanungo, K. R., D. Shaji, M. Mathur, and A. K. Banerjee. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA 1015952-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8404-407. [PubMed] [Google Scholar]
- 33.Takacs, A. M., T. Das, and A. K. Banerjee. 1993. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. USA 9010375-10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, X., T. J. Green, J. Tsao, S. Qiu, and M. Luo. 2008. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J. Virol. 82674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]