Abstract
The human adenovirus E4orf6 and E1B55K proteins promote viral replication by targeting several cellular proteins for degradation. The E4orf6 product has been shown by our group and others to form an E3 ubiquitin ligase complex that contains elongins B and C and cullin family member Cul5. E1B55K associates with this complex, where it is believed to function primarily to introduce bound substrates for degradation via proteasomes. In addition to p53, its first known substrate, the E4orf6/E1B 55-kDa complex (E4orf6/E1B55K) was shown to promote the degradation of Mre11 and DNA ligase IV; however, additional substrates are believed to exist. This notion is strengthened by the fact that none of these substrates seems likely to be associated with additional functions shown to be mediated by the E4orf6-associated E3 ubiquitin ligase complex, including export of late viral mRNAs and blockage of export of the bulk cellular mRNAs from the nucleus. In an attempt to identify new E4orf6/E1B55K substrates, we undertook a proteomic screen using human p53-null, non-small-cell lung carcinoma H1299 cells expressing either E4orf6 protein alone or in combination with E1B55K through infection by appropriate adenovirus vectors. One cellular protein that appeared to be degraded by E1B55K in combination with the E4orf6 protein was a species of molecular mass ∼130 kDa that was identified as the integrin α3 subunit (i.e., very late activation antigen 3 alpha subunit). Preliminary analyses suggested that degradation of α3 may play a role in promoting release and spread of progeny virions.
Viruses are well known to promote replication by inhibiting or enhancing endogenous cellular machinery or, in some cases, by reprogramming key cellular pathways. Human adenoviruses have developed effective ways to modulate the immune response, apoptosis, double-strand break repair, mRNA export, and translation to optimize virus replication and the spreading of progeny virions. The expression of adenovirus E1A proteins stabilizes p53 and induces apoptosis (8, 33); however, this effect is reversed in infected cells by the action of two early products: the E1B 55-kDa (E1B55K) and E4orf6 proteins (35, 36). We and others have shown that these proteins act through the formation of an E3 ubiquitin ligase complex analogous to the SCF and VBC complexes but which contains, in addition to elongins B and C and the RING protein Rbx1, the cullin family member Cul5 (18, 41, 43). This E4orf6-mediated E3 ligase complex blocks p53-induced apoptosis (35, 36) by promoting the ubiquitination of p53, followed by its degradation by proteasomes (41, 43). E4orf6 protein mediates the assembly of the complex by its interaction with elongin C through its three BC boxes (11, 41, 43). E1B55K, which appears to associate with the E4orf6 protein only when present in the ligase complex (4), is thought to function as a substrate recognition factor that brings substrates to the complex because, although both E4orf6 and E1B55K bind p53 independently, interaction of E1B55K with p53 is essential for the efficient degradation of p53 (41, 48). In addition to protecting infected cells from early lysis via p53-induced apoptosis, the E4orf6/E1B55K ligase complex performs other functions essential for virus replication. Two other substrates of the complex have been identified: a member of the MRN DNA repair complex, Mre11, and the central component of the nonhomologous end-joining DNA repair system, DNA ligase IV (2, 56). Degradation of both of these proteins prevents viral genome concatenation, which interferes with the packaging of viral DNA into virions (2, 56). E1B55K binds to p53, Mre11, and DNA ligase IV and has been demonstrated to colocalize with p53 and Mre11 in perinuclear cytoplasmic bodies termed aggresomes (1, 2, 32). More recently, we and others have obtained results that suggest that the E4orf6-associated E3 ligase complex regulates viral and cellular mRNA export (5, 66). The Cul5-based ligase activity was shown to be essential for selective viral mRNA export and the block of cellular mRNA export from the nucleus (66), thus contributing to the shutoff of cellular protein synthesis initiated by L4-100K (20). The actual substrates of the complex responsible for regulating mRNA export are currently unknown.
As discussed in detail below, our efforts to identify substrates of the E4orf6/E1B55K complex led us to consider a member of the integrin family as a potential substrate. Integrins are members of a family of surface receptors that function in several ways through the formation of cell-extracellular matrices and cell-cell interactions (reviewed in references 21, 26, and 63). Integrins are typically composed of two transmembrane glycoproteins forming heterodimers of α and β subunits each of approximately 80 to 150 kDa. There are at least 18 α subunits and 8 β subunits in mammals that can dimerize in limited combinations to form more than 20 functionally distinct integrins with different ligand specificities. Integrin heterodimers function as transmembrane receptors that link external factors to intracellular signaling pathways. In addition to roles in cell adhesion, these communication events are implicated in a large range of cellular processes, including proliferation, differentiation, translation, migration, and apoptosis. Some of these processes depend on the intracellular trafficking pathways of the integrins (reviewed in references 9, 24, 40, and 44), including the long-loop recycling pathway in which integrins present in clathrin-coated endosomes move first to the perinuclear recycling center, where some accumulate, including the β1 integrin subunit (31), before returning to the plasma membrane. The integrin α3β1 is a member of the β1 integrin subfamily in which the α3 subunit (VLA-3a) is coupled to the β1 subunit to form the very late activation antigen (VLA-3 or CD49c) (21, 59, 60). α3β1 is expressed in a wide range of tissues in which it binds a variety of extracellular matrix substrates, including fibronectin, collagen, thrombospondin 1, and laminins 1, 5, 8, 10, and 11 (13). These associations allow the integrin α3β1 to fill its primary role in cell adhesion. α3β1 also participates in intercellular adhesion through several protein-protein interactions (10, 27, 53, 55, 58), making it a major contributor in the regulation of cellular adhesion.
Human adenovirus type 5 (Ad5) particles interact with cell surface receptors to facilitate internalization into target cells. In the high-affinity interacting model (reviewed in reference 29), the viral fiber knob polypeptide binds the coxsackie adenovirus receptor (CAR) protein on the surface of cells as the primary cell binding event (primary receptor). The penton base polypeptide then binds a cell surface integrin (secondary receptor), leading to entry of the capsid into the cell by a process termed receptor-mediated endocytosis or clathrin-mediated endocytosis. Several types of integrins have been identified as being used by Ad5 to mediate virus internalization: αMβ1, αMβ2, αVβ1, αVβ3, αVβ5, and α5β1 (22, 30, 49, 65). Salone et al. have shown that α3β1 serves as an alternative cellular receptor for adenovirus serotype 5 (49). It promotes entry of the virus into cells, transduction of DNA, and mediates adenovirus infection in both CAR-positive and CAR-negative cell lines. Thus, in addition to functions related to cell adhesion, integrin α3β1 plays an important role in the adenovirus infection cycle.
To identify new targets for degradation by the E4orf6/E1B55K ubiquitin ligase, we used a proteomic screen covering most cellular proteins to look for any polypeptide that exhibited a significant decrease in amount following the coexpression from appropriate adenovirus vectors of the E4orf6 protein and E1B55K. This screen revealed several interesting candidates, including integrin α3, a species of 130 kDa that also was found to be reduced in wild-type (wt) virus infection. The degradation of α3 was seen to be dependent on the Cul5-based ligase complex driven by E4orf6 and E1B55K. We also found evidence that the E4orf6/E1B55K ligase complex appears to be involved in cell detachment from the extracellular matrix, a function that could play a role in virus spread.
MATERIALS AND METHODS
Cell lines, plasmids and viruses.
Human non-small-cell lung carcinoma H1299 cells (ATCC CRL-5803) were grown in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (HyClone) at 37°C in 5% CO2. The H1299 Cul5 knockdown cell line (H1299 Cul5 KD) and the H1299 control cell line (H1299 pCDNA3) were described previously (11) and grown as with the H1299 cells but with the addition of 1 mg of puromycin/liter.
All pCDNA plasmids, adenoviral vectors and virus mutants used have been described previously: pCDNA3-E4orf6 (43), pCDNA3-E1B55K (38), AdE1B55K (34, 42), AdE4orf6 (34, 42), AdLacZ (3), H5pg4100 (wt) (17), H5pm4154 (E4orf6−) (5), and H5pm4149 (E1B55K−; K. Kindsmuller, S. Schreiner, F. Leinenkugel, P. Groitl, and T. Dobner, unpublished data).
Antibodies and reagents.
Rabbit polyclonal antibodies to E4orf6 (1807) (7), Mre11 (catalog no. pNB 100-142; Novus Biologicals, Inc.), integrin α3 (catalog no. AB1920; Millipore), and Cul-5 H-300 (catalog no. sc-13014; Santa Cruz) were used. Monoclonal antibodies were as follows: E1B55K, mouse monoclonal 2A6 (50) or rat monoclonal 7C11 (19); E4orf3, rat monoclonal 6A11 (37); E2A DNA-binding protein, mouse monoclonal B6-8 (47); human integrin α3β1, mouse monoclonal M-KID2 (catalog no. MAB1992; Millipore); integrin α3A, mouse monoclonal 29A3 (catalog no. MAB2290; Millipore); actin, mouse monoclonal C4 (catalog no. 691001; MP Biomedicals); and normal mouse immunoglobulin G1 (IgG1) as a negative control for flow cytometry (catalog no. CBL600; Millipore). The secondary antibodies conjugated to horseradish peroxidase for detection in Western blotting were goat anti-mouse IgG, goat anti-rabbit IgG, and goat anti-rat IgG (Jackson Immunoresearch Laboratories). Secondary antibodies for immunofluorescence and flow cytometry coupled Alexa fluorophores were from Invitrogen and included anti-mouse Alexa 488 (A-11029), Alexa 594 (A-11032), and Alexa 405 (A-31553); anti-rabbit Alexa 488 (A-11008) and Alexa 594 (A-11037); and anti-rat Alexa 594 (A-11007). Additional reagents included DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen), propidium iodide (catalog no. PPI777; BioShop), paraformaldehyde (Electron Microscopy Sciences), and Lipofectamine 2000 reagent (Invitrogen).
Infections and DNA transfections.
For infections, cells were plated in six-well dishes and infected with viruses diluted in infection medium (0.2 mM CaCl2, 0.2 mM MgCl2, and 2% serum in phosphate-buffered saline [PBS]) for 90 min before its removal and the addition of normal growth medium. A multiplicity of infection (MOI) of 35 PFU per cell was used for viral vectors, whereas MOIs of 5 PFU/cell for wt virus time course studies and 1 PFU/cell for virus mutants were used. The final amounts of viral vectors per well were adjusted to a total MOI of 70 using the viral vector AdLacZ. For infections and cotransfections, cells were either mock infected or infected with AdLacZ at an MOI of 35 PFU/cell for 90 min; the cells were then transfected with 0.75 μg of pCDNA3-E4orf6 and/or 1.75 μg of pCDNA3-E1B55K plasmid DNAs using Lipofectamine 2000 reagent according to the manufacturer's recommendations. The final amount of DNA per well was adjusted to 2.5 μg using the vector plasmid pcDNA3.
Protein extraction.
Cells were washed with PBS and removed at different times postinfection by incubation for 5 min with gentle agitation with 0.53 mM EDTA. Cells were collected by centrifugation, and the pellets were incubated for 5 min on ice in CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} buffer (4% CHAPS [BioShop], 30 mM Tris-HCl [pH 8.5], 50 mM NaCl) plus inhibitors (protease inhibitor cocktail [Sigma], 1 mM Na3VO4, 10 mM NaPPi, 10 mM NaF). Cells were then lysed by sonication, and the protein concentrations of the lysates determined using Bio-Rad DC protein assay reagents according to manufacturer's protocol. Extracts were brought to 7 M of urea-2 M thiourea (GE Healthcare Biosciences) and clarified by centrifugation at 13,000 × g for 5 min, and the protein concentrations were recalculated according to the increase in volume.
2D-DIGE and image analysis.
H1299 cells infected with AdE1B55K and/or AdE4orf6 adenovirus vectors were harvested at 48 h postinfection (hpi), and cell extracts were prepared in CHAPS buffer (see above) to reach a protein concentration of ∼10 mg/ml. The following procedures were carried out using the CIAN two-dimensional difference gel electrophoresis (2D-DIGE) platform (McGill University). Portions (50 μg) of protein were minimally labeled with CyDye (GE Healthcare) according to the manufacturer's recommendations (400 pmol of dye/50 μg of protein). The control sample consisting of a mixture of 50 μg of each sample was labeled with Cy2. Labeled samples were combined (with 50 μg of Cy2-labeled control) in 2D-DIGE rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 0.002% bromophenol blue, 0.5% immobilized pH gradient [IPG] buffer [pH 4 to 7], and 18 mM dithiothreitol) according to the method illustrated in Fig. 1A. IPG strips (pH 4 to 7, 24 cm; GE Healthcare) were rehydrated with Cy-labeled samples in the dark at 30 V and 50 μA/strip 20°C for 15 h. First-dimension isoelectric focusing (IEF) was performed using an Ettan IPGPhor II electrophoresis system (GE Healthcare) as recommended by the supplier. After reduction and alkylation of disulfide bonds with 1% dithiothreitol and 4% iodoacetamide, respectively, in equilibration buffer (50.25 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% sodium dodecyl sulfate [SDS], 0.00125% bromophenol blue), the second-dimension separation was performed by SDS-polyacrylamide gel electrophoresis (PAGE) using 10% polyacrylamide gels in an Ettan DALT VI electrophoresis system (GE Healthcare) according to the manufacturer's recommendations. A slow entry phase at 30 V for 1 h at room temperature was followed by a 1-W/gel constant at 15°C until the front exited the gel. The 2D gels were scanned on a Typhoon 9400 imager (GE Healthcare). Excitation and emission wavelengths were chosen specifically for each dye according to the manufacturer's recommendations. Intra- and intergel matching, statistical analysis (Student t test, P < 0.05), and spot filtering were performed using DeCyder software v6.5 (GE Healthcare).
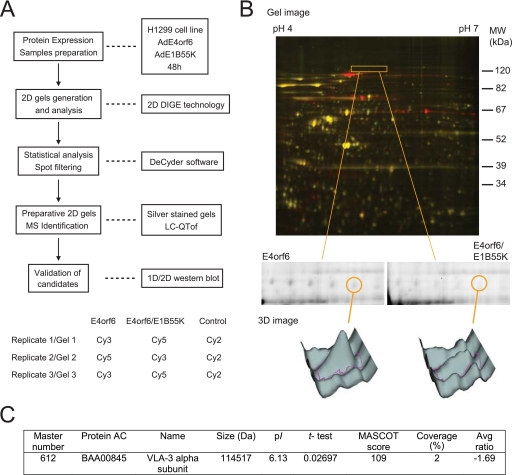
FIG. 1.
2D-DIGE proteomic screen. VLA-3a is decreased in cells expressing E4orf6/E1B55K. (A) The upper panel shows the experimental design of the 2D-DIGE identification procedure. The lower panel shows the sample labeling and mixing strategy for the three replicates to generate 2D gels. (B) The top panel shows an example of a three-channel 2D-DIGE overlay image comparing H1299 cells expressing E4orf6/E1B55K to cells expressing E4orf6 alone. The boxed region shows a region of interest containing spot 612. In the middle and bottom panels, grayscale images represent individual scans, as well as 3D images, of the spot 612 intensity for each condition. (C) Summary of the VLA-3a 2D-DIGE and mass spectrometry data.
Preparative 2D gels for protein identification were prepared as follows. Portions (500 to 1,000 μg) of unlabeled samples were rehydrated individually, as described above, and loaded onto IPG strips (pH 4 to 7, 13 cm; GE Healthcare) for the first-dimension IEF according to the manufacturer's recommendations. After reduction and alkylation of the disulfide bonds, the second-dimension SDS-PAGE was performed in Hoefer Ruby SE600 (GE Healthcare) using a slow entry phase at 30 V for 1 h at room temperature and then a 50- to 75-V constant at 4°C until the front exited the gel. The 2D gels were silver stained by using a procedure adapted from Blum et al. (6). The protein spots of interest were manually excised from the gel. In-gel trypsin digestion was performed on a MassPrep robotic workstation (Perkin-Elmer), and digested peptides were processed by using tandem mass spectrometry on an LC-QTof Micro apparatus (Waters) operated by the McGill University and Genome Quebec Innovation Center. Subsequent identification of peptides and proteins from complex mixtures was done by using Mascot (Matrix Science) with the NCBI nonredundant database as a reference.
Immunoblotting.
Equal amounts of proteins were separated by SDS-PAGE and then transferred to polyvinylidene difluoride membranes (Millipore) that had been blocked using 5% skimmed milk. Primary antibodies were added on membranes for 2 to 3 h at room temperature or overnight at 4°C. Membranes were washed with PBS containing 0.1% Tween 20, and the secondary antibody was added for 1 h at room temperature. Detection was performed using Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer).
Immunofluorescence microscopy.
Cells grown on coverslips and infected as described above were washed once in PBS and fixed with ice-cold methanol for 15 min at −20°C. Methanol was then replaced by TBS-BG blocking buffer (20 mM Tris-HCl [pH 7.4], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 0.05% Tween 20, 66 mM glycine, 5 g of bovine serum albumin/liter) for 20 min at room temperature. Cells were incubated with primary antibody diluted in PBS for 2 h at room temperature and then washed two times for 5 min each time with PBS, followed by incubation with secondary antibody diluted in PBS for 1 h at room temperature in the dark. Coverslips were washed again and incubated with DAPI diluted in PBS for 10 min at room temperature in the dark. Cells were washed twice with PBS and once with water before being mounted on slides in mounting medium (Immu-Mount; Thermo Scientific). Images were acquired at the McGill Life Sciences Complex Imaging Facility on a Axiovert 200M fully motorized confocal microscope using Zeiss LSM 5 Pascal software.
Flow cytometry.
Cells were removed from the plate using EDTA as described above and washed twice with PBS containing 2% fetal calf serum (FCS). One million cells were incubated with primary antibody (anti-integrin α3β1 M-KID2 or isotype-matched normal mouse IgG1) diluted in PBS-2% FCS for 20 min at 37°C. Cells were washed once in PBS-2% FCS and then incubated with anti-mouse antibody labeled with Alexa 488 for 20 min at 37°C in the dark. The cells were washed again with PBS-2% FCS and resuspended in 0.6 ml of PBS-2% FCS containing propidium iodide for 10 min at 37°C in the dark. Cells were washed twice as described above, fixed in 0.5 ml of 2% paraformaldehyde, and analyzed immediately by flow cytometry using a FACScan analyzer (Becton Dickinson). The data were analyzed by using FCS Express software (De NoVo Software) with dead cells excluded from the analysis using the propidium iodide gate.
Detachment assay.
Cells grown and infected in six-well plates were washed once with PBS and incubated in 1 ml of 0.13 mM EDTA with gentle agitation. Photographs of cells at the center of the well were taken at 0 min of EDTA treatment and every 2 min thereafter.
RESULTS
Establishment of a proteomic screen using E4orf6/E1B55K-expressing cells.
In previous work we studied the activity of the Ad5 E4orf6/E1B55K E3 ubiquitin ligase complex using p53-null H1299 cells transfected with plasmid DNAs expressing these Ad5 proteins in the absence of other viral products (4, 5, 41, 43). We were also able to show degradation of endogenous Mre11 using adenovirus vectors expressing these two viral proteins (data not shown). We thus utilized the adenovirus vector system to establish a proteomic screen to identify additional E4orf6/E1B55K substrates. Preliminary experiments were conducted using endogenous Mre11 protein as control, and the results indicated that optimal levels of degradation of this substrate were observed using H1299 cells harvested 48 h after infection with both the adenovirus vectors AdE4orf6 and AdE1B55K, at an MOI of 35 PFU/cell, with cells infected with AdE4orf6 alone as controls (data not shown). Thus, these conditions were used in a screen to evaluate the degradation of substrates according to the experimental procedure depicted in Fig. 1A, upper panel.
Three independent 2D-DIGE gel analyses were performed comparing the proteomes of H1299 cells expressing E4orf6 alone to those expressing both E4orf6 protein and E1B55K (Fig. 1A, bottom panel). An internal control containing protein extracts of all of the samples in the study was labeled with Cy2 and included in all gels to serve as a reference for spot normalization. Dye swapping was also performed to minimize labeling-dependent bias. Analysis of the Cy2, Cy3, and Cy5 gel images using the DeCyder software revealed the presence of a number of protein spots that differed significantly in levels, with statistical variance of the E4orf6/E1B55K versus E4orf6 spot volume ratios within the 95th confidence level (Student t test: P < 0.05). We intend to publish full details of the entire proteomic analyses elsewhere; however, Fig. 1B shows an example of a three-channel 2D-DIGE overlay image that exhibits several proteins (red, Cy5) that were significantly increased in E4orf6/E1B55K-expressing cells compared to proteins not varying in intensity (yellow, Cy5 and Cy3 combination). Because our interest was on species that decreased in response to expression of E4orf6 protein, we focused on spots (green, Cy3) showing a significant decrease in intensity in E4orf6/E1B55K-expressing cells relative to the E4orf6 alone control, with an average ratio less than or equal to −1.5. The 12 species showing the lowest average ratios and confirmed visually to decrease in at least two of three gels were excised from preparative 2D gels, and the species were identified by mass spectrometry.
Identification of the VLA-3 alpha subunit as a substrate of the E4orf6/E1B55K complex.
Figure 1B illustrates one of these 12 species (termed 612 in our analysis) that migrated as a polypeptide of ∼130 kDa and that exhibited an E4orf6/E1B55K versus E4orf6 alone average ratio of −1.69 at P < 0.027 (highlighted area in the top panel and isolated in the middle panel; the grayscale images in the lower panel represent individual scans as well as 3D images of the spot intensity for each condition). Figure 1C shows that analysis by mass spectrometry revealed that this species corresponded to the VLA-3 alpha subunit (α3 subunit). Two peptides were identified covering 2% of the whole protein sequence (Fig. 1C and data not shown). Individual ion scores for the two peptides were 62 and 47 for a total of 109, where ion scores greater than 36 indicate identity or extensive homology (P < 0.05) (Matrix Science). The peptides identified by mass spectrometry spanned the whole protein and were present both in the alpha and beta spliced forms of the α3 integrin (14; data not shown).
An observed decrease in spot intensity in this type of 2D gel analysis may be explained either by a decrease in protein level or by a change in migration position of the species due to one or more newly induced posttranslational modifications that could affect molecular mass and/or the pI. To validate that the decrease in α3 was due to a general decrease in protein levels, this ∼130-kDa species was analyzed by Western blotting with an antibody specific for the α3 subunit (clone 29A3). Figure 2A (top panel) shows that the expression of either E4orf6 or E1B55K proteins alone by viral vectors had little or no effect on α3, whereas the presence of both viral proteins dramatically decreased α3 protein levels (Fig. 2A). This decrease was not due to stress caused by higher viral vector content with E4orf6/E1B55K coexpression because cells received the same amount of total virus through infection by a control adenovirus vector expressing β-galactosidase (3). In addition to these immunoblotting experiments, the identity of the α3 species was also confirmed by immunoprecipitation and RNA interference studies using reagents specific for α3 (data not shown).
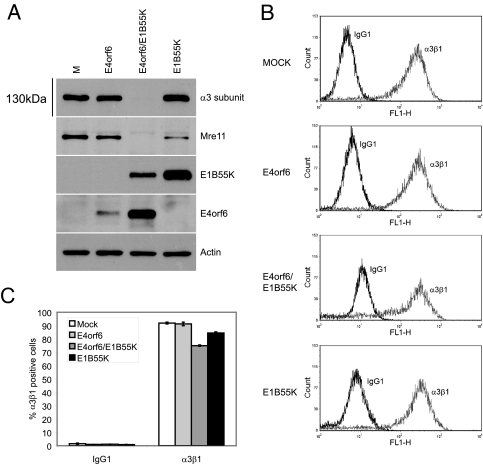
FIG. 2.
Analysis of total and surface α3β1 pools in cells expressing E4orf6 protein and E1B55K. (A) H1299 cells were infected with viral vectors expressing E4orf6, E1B55K, or with both at an MOI of 35 PFU/cell per vector. The final amounts of vectors per well were kept to an MOI of 70 using the AdLacZ control vector. Cells were harvested at 48 hpi, and cell extracts were analyzed by SDS-PAGE, followed by Western blotting with appropriate antibodies (anti-α3 clone 29A3 and anti-E1B55K 2A6). (B) Representative histograms of the surface levels of α3β1 integrin are shown. H1299 cells expressing E4orf6, E1B55K, or both proteins were gated for the removal of dead cells. Solid black lines represent the isotype control and the solid gray lines represent the anti-integrin α3β1 M-KID2. (C) Bar diagrams of median value (top panel) or percentages (bottom panel) of H1299 cells positive for surface α3β1 marker. To evaluate the percentage of cells expressing surface α3β1, a marker was set just after the isotype-matched IgG1 control median peak, and this marker was translated to the α3β1 antibody peak. The results shown are averages obtained from three independent labelings of the same pool of cells. Error bars indicate the standard errors of the means.
The α3β1 integrin is composed of the α3 and β1 subunits. Although much of α3β1 integrin at any one time is intracellular, a portion is present at the cell surface and functions as an attachment factor to the extracellular matrix and in cell-cell adhesion (reviewed in references 21, 26, and 63). To determine whether the surface pool of α3β1 is also influenced by the presence of E4orf6 and E1B55K proteins, surface levels of α3β1 were evaluated by flow cytometry using the mouse monoclonal anti-integrin α3β1 M-KID2 antibody in a triplicate labeling of the same pool of cells. Figures 2B and C show that a small but significant decrease in surface α3β1 was evident when both E4orf6 and E1B55K were expressed. A more significant decrease in α3β1 was observed when the cells were permeabilized before flow cytometry staining (data not shown), confirming the immunoblotting data addressing the status of total and/or intracellular α3 subunit pool. Thus, the data in Fig. 2A to C suggested that the total cellular pool of α3 was highly reduced in the presence E4orf6 protein and E1B55K but that decreases in the small extracellular pool of α3β1 were more modest, at least after 48 h in the presence of these two viral polypeptides.
Degradation of α3 requires infection of cells by adenovirus particles.
We had previously developed a plasmid-based degradation assay to evaluate the activity of the E4orf6-mediated E3 ligase complex (4, 41, 43) and thus, instead of using adenovirus vectors, we applied this approach to examine the degradation of α3. H1299 cells were transfected with plasmid DNAs that express the E4orf6 and E1B55K proteins, and the degradation of endogenous α3 and Mre11 was examined by Western blotting with appropriate antibodies. Figure 3 (lanes marked “Mock”) indicated that although Mre11 clearly was degraded specifically in response to E4orf6 and E1B55K, repeated studies indicated only at best a very modest reduction of α3 levels. Because these results differed so markedly from those presented in Fig. 1B and 2A in studies that used adenovirus vectors to express E4orf6 and E1B55K, we sought to determine whether the effect of E4orf6 and E1B55K on α3 required cells to be infected with adenovirus particles to promote the degradation. Thus, the same degradation assay was repeated; however, in this case prior to DNA transfection, cells were infected with a control adenovirus vector expressing the LacZ protein. Figure 3 (lanes marked “AdLacZ”) indicates that both α3 and Mre11 were efficiently degraded by E4orf6 and E1B55K. These results clearly suggested that, unlike Mre11 degradation, the degradation of α3 requires factors or processes associated with adenovirus infection to proceed. This somewhat surprising finding is discussed in more detail below.
FIG. 3.
Effect of infection by adenovirus particles on degradation of α3. H1299 cells were sequentially mock infected or infected with AdLacZ at an MOI of 35 PFU/cell and then transfected with either pcDNA3, pcDNA3-E4orf6, pcDNA3-E1B55K, or pcDNA3-E4orf6/pcDNA3-E1B55K. Cells were harvested at 48 h posttransfection, and cell extracts were analyzed by SDS-PAGE, followed by Western blotting with appropriate antibodies indicated on the right (anti-VLA-3a clone 29A3 and anti-E1B55K 2A6).
α3 is decreased in wt adenovirus infection.
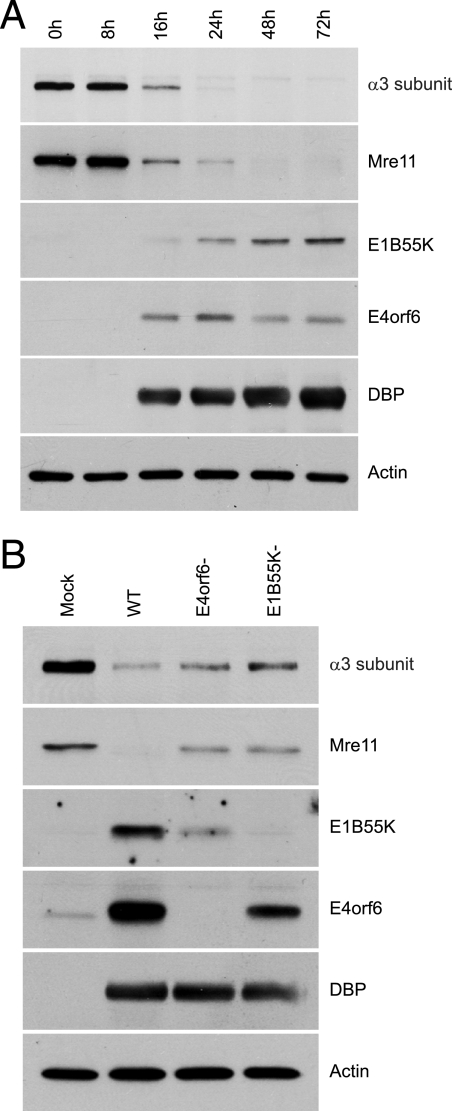
To determine whether a reduction of α3 protein levels was apparent during a wt adenovirus infection, cells were infected, and cell extracts prepared at various times after infection were analyzed by Western blotting for the presence of α3 and Mre11, as well as for several virus proteins, using appropriate antibodies. Figure 4A shows that the level of α3 started to decrease by 16 hpi, the time at which E1B55K and E4orf6 proteins were first detected, and by 48 hpi α3 was completely undetectable. Figure 4A also shows a very similar pattern of degradation of Mre11, suggesting that the E4orf6/E1B55K complex affects both proteins in a similar manner. To confirm the dependency on E4orf6 and E1B55K for the degradation of α3 and Mre11 in the virus context, H1299 cells were infected either by wt Ad5 or mutants in the same genetic background that fail to produce either E4orf6 protein or E1B55K. The E1B55K− virus contains mutations that convert the codons encoding the first four amino-terminal methionine residues to “stop” codons to prevent any translation initiation from the first or cryptic start sites. The E4orf6-null virus contains a “stop” codon after Pro66, thus yielding a severely truncated unstable E4orf6 product (5). Figure 4B shows that in cells infected with wt virus and harvested at 24 hpi most of the α3 had been degraded; however, in the absence of either E4orf6 or E1B55K, clearly higher levels of α3 were present, although in both cases these were lower than in wt-infected cells. Comparable results were also obtained with Mre11 (Fig. 4B), suggesting that in the context of virus infection, although the E4orf6/E1B55K E3 ubiquitin ligase complex seems to be implicated in the degradation of both Mre11 and α3, other processes may also affect their stability.
FIG. 4.
Analysis of α3 levels during adenovirus infection. (A) H1299 cells were infected with H5pg4100 (wt) at an MOI of 5 fluorescence-forming units/cell. Cells were harvested at several different times, and cell extracts were analyzed by SDS-PAGE, followed by Western blotting with appropriate antibodies indicated on the right (anti-VLA-3a clone 29A3, anti-E1B55K 2A6). (B) H1299 cells were infected with H5pg4100 (wt), H5pm4154 (E4orf6−), or H5pm4149 (E1B55K−) at an MOI of 1 fluorescence-forming unit/cell (MOI of 4 for H5pm4149). Cells were harvested at 24 hpi, and cell extracts were analyzed by SDS-PAGE, followed by Western blotting as indicated above.
α3 is degraded by the Cul5-mediated E3 ligase complex.
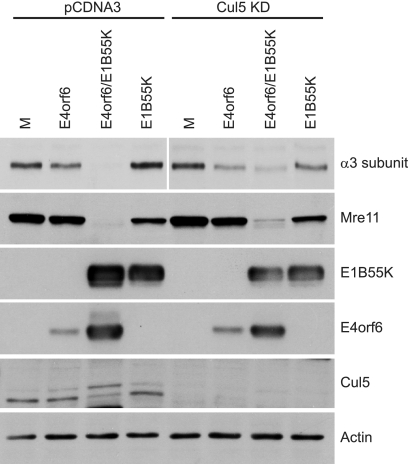
The proteins p53, Mre11, and DNA ligase IV are all substrates of the E4orf6-associated E3 ligase complex containing the cullin family member Cul5, elongins B and C, and the RING protein Rbx1 (2, 4, 18, 41, 43, 51, 56). Cul5 seems to be a key player because it is the only cullin family member found to interact with the E4orf6 complex (41). A dominant-negative mutant of Cul5 was shown to prevent the degradation of the all three known E4orf6 substrates (2, 66), and our group has shown previously that the degradation of p53 is impaired in a Cul5 knockdown cell line (11). To confirm that the E4orf6/E1B55K-dependent decrease of α3 is mediated by the E4orf6-Cul5 E3 ligase complex, our Cul5 knockdown cell line was used to examine the dependence of α3 degradation on the presence of Cul5. Figure 5 shows that in control H1299 cells (pCDNA3) significant reductions in both endogenous α3 and Mre11 levels again occurred in the presence of both E4orf6 and E1B55K. In the Cul5 KD cell line a modest relief of the E4orf6/E1B55K-mediated degradation was detected, an effect that was also apparent with the substrate Mre11. We had expected to observe a greater inhibition of α3 and Mre11 degradation in these cells; however, although the Cul5 knockdown seemed very efficient, as seen by the virtually undetectable levels of Cul5 protein indicated in Fig. 5 by Western blotting analysis, the remaining significant reductions in α3 levels may have been due to residual amounts of Cul5 that were sufficient for degradation by the E4orf6-associated E3 ligase. Nevertheless, our results suggested that significant degradation of both proteins occurs via the Cul5-based E3 ligase complex.
FIG. 5.
Analysis of protein degradation in cells expressing reduced levels of Cul5. Control (pCDNA3) or Cul5 knockdown (Cul5 KD) H1299 cell lines were infected and harvested, and cell extracts were analyzed as indicated in Fig. 2A. The level of Cul5 in these cell lines was determined by Western blotting with the anti-Cul5 antibody H-300.
E1B55K colocalizes with α3.
E1B55K is known to interact with p53, Mre11, and DNA ligase IV and has been shown to colocalize with p53 and Mre11 in perinuclear bodies (1, 2, 32). The presence of Mre11 in these bodies (called aggresomes) is believed to accelerate its degradation by proteasomes after ubiquitination by the E4orf6-mediated E3 ligase complex (32). The presence of Mre11 and p53 in these bodies is dependent on E1B55K (32) or E4orf3 (1). To determine whether E1B55K colocalizes with α3, immunofluorescence studies were conducted in H1299 cells infected with wt Ad5 and treated at 24 hpi with appropriate antibodies. Figure 6 shows that in uninfected cells (labeled as “u” in Fig. 6A), as well as in infected cells (labeled as “i” in Fig. 6A; see also cells in Fig. 6B), α3 is present in a diffuse intracellular pattern but also concentrated at the plasma membrane (this is particularly evident in Fig. 6Be and h) and in a small perinuclear bodies (indicated by arrows). These bodies may correspond to perinuclear recycling centers of membrane receptors in uninfected cells; however, in infected cells these structures are much larger with brighter fluorescence (compare the “u” and “i” in Fig. 6A), and they coincide with aggresomes since E1B55K also localizes to these structures (see merge pictures) in virtually all cells infected with wt virus (Fig. 6A and Ba to d) and cells infected with viral vectors expressing E1B55K (Fig. 6Be to h) or both E1B55K and E4orf6 (Fig. 6Bi to l). A vimentin cage is also frequently seen surrounding these structures (not shown), as is often the case around aggresomes (23). Thus, α3 and E1B55K appear to colocalize largely in aggresomes.
FIG. 6.
α3 colocalizes with E1B55K in H1299 cells. (A) Uninfected cells (u) and infected cells (i) were compared for α3 perinuclear aggregates. H1299 cells were infected with H5pg4100 (wt) at an MOI of 1 for 24 h. Cells were fixed and stained for α3 (AB1920) and E1B55K (2A6). Two equivalent fields are shown for each channel. (B) H1299 cells were infected with H5pg4100 (wt) at an MOI of 5 for 24 h (a to d), with viral vectors expressing E1B55K (e to h) or both E4orf6 and E1B55K (i to l) at 35 PFU/cell per vector for 48 h. Cells were fixed and stained for α3, E1B55K (2A6), and DAPI as indicated above.
The presence of E4orf6 protein and E1B55K promotes cell detachment from extracellular matrix.
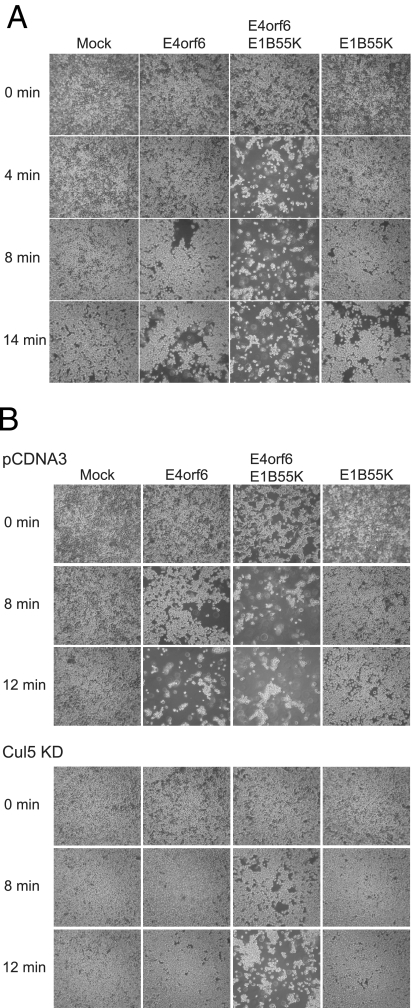
Kaabeche et al. (25) showed previously that Cbl triggers α5 integrin degradation by the proteasome, resulting in reduced osteoblast attachment via fibronectin and in caspase-dependent apoptosis. Because the surface pool of α3β1 was seen to be at least somewhat decreased in the presence of E4orf6/E1B55K, studies were conducted to determine whether this decrease affected the cell monolayer integrity and cellular attachment. E4orf6 protein and E1B55K were expressed in cells for 48 h using adenovirus vectors, and then the medium was removed and replaced with EDTA at low concentration with gentle agitation. Figure 7A shows photographs of these cells at several times after the addition of EDTA. Mock-infected cells were relatively unaffected by this treatment over 14 min. Some detachment was observed after 8 min in the presence of either E4orf6 or E1B55K alone; however, significant cell detachment was evident almost immediately after addition of EDTA when both E4orf6 and E1B55K were expressed. These results suggested that the presence of E4orf6 and E1B55K proteins reduced the attachment of cells to the matrix.
FIG. 7.
Analysis of cell adhesion in the presence of E4orf6 and E1B55K proteins. (A) H1299 cells were infected with viral vectors expressing E4orf6, E1B55K, or both proteins at an MOI of 35 PFU/cell per vector for 48 h. Cells were placed in 0.13 mM EDTA with gentle agitation, and photographs of cells were taken at the indicated times. (B) Comparison of cell detachment in control (pCDNA3) or Cul5 knockdown (Cul5 KD) H1299 cell lines in the presence of E4orf6 and E1B55K proteins.
To correlate the variation of cell attachment to the α3 protein levels, the same assay was performed in the Cul5 KD cell line in which the degradation of α3 was found to be reduced compared to the control cell line (Fig. 5). Figure 7B shows that the cells lifted quite rapidly after E4orf6/E1B55K coexpression in the control cell line in a pattern similar to that seen in Fig. 7A; however, in the Cul5 KD cell line the detachment was significantly delayed (Fig. 7B). Altogether, these results suggest that the degree of binding of cells to the matrix correlates with the α3 protein levels observed in these cell lines.
DISCUSSION
Degradation of the previously identified substrates of the E4orf6-associated E3 ligase complex appears to play a role in enhancing viral replication (2, 18, 41, 43, 56). Degradation of p53 may prevent early apoptotic death of infected cells, thus permitting the replication cycle to be completed (35, 36). Degradation of Mre11, a member of the DNA repair complex, and DNA ligase IV, a central component of the nonhomologous end-joining DNA repair system, may protect from viral genome concatenation that could interfere with viral DNA packaging into virions (2, 56). We and others have demonstrated a role for the E4orf6-associated ligase in the regulation of viral and cellular mRNA export, although the identity of the substrate(s) involved remains unknown (5, 66). With this growing list of roles of the E3 ligase complex, one might suppose that additional functions regulated by degradation of as yet unknown substrates may be identified. Analysis by 2D electrophoresis of proteins provides a reasonable approach to identifying new substrates since it allows separation and visualization of most cellular proteins to evaluate variations in protein intensity. Using this proteomic approach, we have identified the integrin α3 subunit as a substrate of the E4orf6/E1B55K complex. The identity of the ∼130-kDa species as the α3 subunit seen to be reduced in the screen and identified by mass spectroscopy was confirmed by Western blotting analysis with a α3 subunit-specific antibody. Another integrin known to be a target of an E3 ligase complex is the integrin α5 subunit, which is ubiquitinated and degraded by Cbl E3 ubiquitin ligase complex after FGFR2 activation in osteoblasts (25).
A quite unexpected observation was that infection by adenovirus virions appeared to greatly enhance E4orf6-dependent degradation of α3 since E4orf6 and E1B55K expressed from plasmid DNAs were not capable of inducing significant degradation of α3. Such was not the case with the substrate Mre11. In addition, immunofluorescence data indicated that the perinuclear α3 aggregates present in infected cells tended to be larger than those present in uninfected cells and that they colocalize with E1B55K (and probably E4orf3), which are known to localize in aggresomes. One possibility to explain these observations is that adenovirus infection may promote internalization of the secondary receptor and cause a stress signal which decreases the rate of trafficking to the cell surface. This process may result in an increased accumulation of the internalized and intracellular pool of the integrin at the perinuclear recycling centers, which coincides with the aggresomes. In these structures E1B55K could trap the integrin, thus preventing its recycling to the plasma membrane and enhancing its degradation by the E4orf6/E1B55K complex. Clearly, additional studies will be required to explain this phenomenon further.
A rather striking observation was the difference in degradation of α3β1 at the cell surface relative to the total cellular α3 pool. The data shown in Fig. 2C indicated that the surface pool of α3 appeared to be only modestly reduced by expression of E4orf6 and E1B55K proteins. It could be that in H1299 cells the levels of α3 at the cell surface represent only a minor fraction of the total in the cell, as suggested by Fig. 6B. This distribution pattern of α3 could explain why in spite of only a modest decrease in levels of α3β1 present at the cell surface in the presence of E4orf6 and E1B55K, a dramatic reduction in total cell α3 took place. Additionally and despite the fact that α3β1 has been shown to be a secondary receptor for adenovirus, it may be utilized far less than the well-known αVβ5 secondary receptor, and thus the internalization and further degradation of the surface pool may be proportional to the degree of receptor utilization. As mentioned previously, the degradation of the internal pool may also simply be triggered by an adenovirus infection but independently of the type of receptor utilized. An alternative explanation for this observed difference of degradation between the surface and intracellular pools could be that degradation of α3 within the cell may induce a vastly increased rate of transport of the remaining intracellular α3 to the cell surface, thus maintaining reasonably high levels on the surface of cells.
In the context of infection with viral vectors (Fig. 2A) or in DNA transfection experiments, with prior infection with the LacZ viral vector, the degradation of α3 appears to be completely dependent on the presence of both E4orf6 and E1B55K proteins; however, in the context of a full virus infection, only a partial rescue of degradation was observed by eliminating either E4orf6 or E1B55K. A similar effect was also observed with Mre11, suggesting that with these two substrates another pathway of degradation likely is also active during infection. Interestingly, both of these substrates are at least partially localized at the aggresomes (1, 32) (Fig. 6), suggesting that localization at this structure may be part of the alternate mechanism of degradation, and perhaps also involving E4orf3, which also localizes at the aggresomes (1, 32). It is possible that the formation of the aggresomes is sufficient to induce degradation of α3 and Mre11, which both accumulate there, since aggresomes may be able to attract ubiquitin-conjugating machinery as well as proteasomes (15, 28).
In our simple studies there appeared to be a good correlation between the levels of α3 and the tendency of the cells to detach from the extracellular matrix. Although only one integrin has been identified as a substrate of the E4orf6-mediated E3 ligase complex, other adhesion molecules may also be affected by E4orf6/E1B55K, and a well-planned effort seems to be warranted to examine the stability of this class of proteins. Additional targets of this type may explain why such a striking detachment phenotype was observed with such modest decreases in surface α3β1. The degradation of integrins followed by cell detachment may cause some form of apoptosis, a phenomenon observed with integrin α5 subunit (39). This detachment-mediated apoptosis, termed anoikis (12, 16, 46), has been observed in other types of virus infections (45, 52, 54). Adenoviruses may use this effect as an alternative virus release mechanism to the action of the E3 adenovirus death protein, which stimulates cell lysis by apoptosis to allow virus exit and spreading from the cells (57, 61, 62, 67, 68). Another mechanism that improves virus spreading has been demonstrated by Walters et al. (64). The excess fiber proteins not encapsulated or present on defective viruses compete with the CAR receptor on adjacent cells to interrupt cell-cell adhesion mediated by CAR-CAR interactions. This process allows the virus to cross tissue barriers and spread to distal sites for further infection and prevents the virus binding to cells that have already been infected, thus spreading of progeny away from the initial site of infection. The decrease of α3 may affect cell-cell interactions and improve virus spreading in a similar fashion.
Adenoviruses very typically utilize several alternative but related mechanisms to promote replication and virus spread. Thus, cell surface molecules may represent important new targets for degradation by the E4orf6-E1B55K E3 ubiquitin ligase, and further studies are under way to examine more carefully this class of cell proteins.
Acknowledgments
We thank the CIAN facility and Imaging facility (McGill University Life Sciences Complex) for their help with the 2D-DIGE technology and confocal microscopy, respectively.
This study was supported by grants from the Canadian Institutes of Health Research, from the Canadian Cancer Society through the National Cancer Institute of Canada and the Fonds de la Recherche en Santé du Québec (FRSQ) (P.E.B.), and from the Deutsche Forschungsgemeinschaft (T.D.). P.B. had an FRSQ postdoctoral fellowship during part of this work.
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 7911382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 817034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bett, A. J., L. Prevec, and F. L. Graham. 1993. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 675911-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 249619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette, P., K. Kindsmuller, P. Groitl, F. Dallaire, T. Speiseder, P. E. Branton, and T. Dobner. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 822642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 893-99. [Google Scholar]
- 7.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 731245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braithwaite, A., C. Nelson, A. Skulimowski, J. McGovern, D. Pigott, and J. Jenkins. 1990. Transactivation of the p53 oncogene by E1a gene products. Virology 177595-605. [DOI] [PubMed] [Google Scholar]
- 9.Caswell, P. T., and J. C. Norman. 2006. Integrin trafficking and the control of cell migration. Traffic 714-21. [DOI] [PubMed] [Google Scholar]
- 10.Chattopadhyay, N., Z. Wang, L. K. Ashman, S. M. Brady-Kalnay, and J. A. Kreidberg. 2003. α3β1 integrin-CD151, a component of the cadherin-catenin complex, regulates PTPmu expression and cell-cell adhesion. J. Cell Biol. 1631351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, C. Y., P. Blanchette, and P. E. Branton. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 36436-44. [DOI] [PubMed] [Google Scholar]
- 12.Chiarugi, P., and E. Giannoni. 2008. Anoikis: a necessary death program for anchorage-dependent cells. Biochem. Pharmacol. 761352-1364. [DOI] [PubMed] [Google Scholar]
- 13.DiPersio, C. M., S. Shah, and R. O. Hynes. 1995. α3Aβ1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J. Cell Sci. 108(Pt. 6)2321-2336. [DOI] [PubMed] [Google Scholar]
- 14.DiPersio, C. M., J. E. Trevithick, and R. O. Hynes. 2001. Functional comparison of the α3A and α3B cytoplasmic domain variants of the chicken α3 integrin subunit. Exp. Cell Res. 26845-60. [DOI] [PubMed] [Google Scholar]
- 15.Fabunmi, R. P., W. C. Wigley, P. J. Thomas, and G. N. DeMartino. 2000. Activity and regulation of the centrosome-associated proteasome. J. Biol. Chem. 275409-413. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore, A. P. 2005. Anoikis. Cell Death Differ. 12(Suppl. 2)1473-1477. [DOI] [PubMed] [Google Scholar]
- 17.Groitl, P., and T. Dobner. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 13029-39. [DOI] [PubMed] [Google Scholar]
- 18.Harada, N., Y. Maniwa, M. Yoshimura, M. Nagata, H. Hamada, K. Yokono, and Y. Okita. 2005. E1B-deleted adenovirus replicates in p53-deficient lung cancer cells due to the absence of apoptosis. Oncol. Rep. 141155-1163. [PubMed] [Google Scholar]
- 19.Hartl, B., T. Zeller, P. Blanchette, E. Kremmer, and T. Dobner. 2008. Adenovirus type 5 early region 1B 55-kDa oncoprotein can promote cell transformation by a mechanism independent from blocking p53-activated transcription. Oncogene 273673-3684. [DOI] [PubMed] [Google Scholar]
- 20.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 642732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemler, M. E. 1990. VLA proteins in the integrin family: structures, functions, and their role on leukocytes. Annu. Rev. Immunol. 8365-400. [DOI] [PubMed] [Google Scholar]
- 22.Huang, S., R. I. Endo, and G. R. Nemerow. 1995. Upregulation of integrins αvβ3 and αvβ5 on human monocytes and T lymphocytes facilitates adenovirus-mediated gene delivery. J. Virol. 692257-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1431883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, M. C., P. T. Caswell, and J. C. Norman. 2006. Endocytic recycling pathways: emerging regulators of cell migration. Curr. Opin. Cell Biol. 18549-557. [DOI] [PubMed] [Google Scholar]
- 25.Kaabeche, K., H. Guenou, D. Bouvard, N. Didelot, A. Listrat, and P. J. Marie. 2005. Cbl-mediated ubiquitination of α5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J. Cell Sci. 1181223-1232. [DOI] [PubMed] [Google Scholar]
- 26.Kreidberg, J. A. 2000. Functions of α3β1 integrin. Curr. Opin. Cell Biol. 12548-553. [DOI] [PubMed] [Google Scholar]
- 27.Lampe, P. D., B. P. Nguyen, S. Gil, M. Usui, J. Olerud, Y. Takada, and W. G. Carter. 1998. Cellular interaction of integrin α3β1 with laminin 5 promotes gap junctional communication. J. Cell Biol. 1431735-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, H. J., S. Y. Shin, C. Choi, Y. H. Lee, and S. J. Lee. 2002. Formation and removal of α-synuclein aggregates in cells exposed to mitochondrial inhibitors. J. Biol. Chem. 2775411-5417. [DOI] [PubMed] [Google Scholar]
- 29.Leopold, P. L., and R. G. Crystal. 2007. Intracellular trafficking of adenovirus: many means to many ends. Adv. Drug Deliv. Rev. 59810-821. [DOI] [PubMed] [Google Scholar]
- 30.Li, E., S. L. Brown, D. G. Stupack, X. S. Puente, D. A. Cheresh, and G. R. Nemerow. 2001. Integrin αvβ1 is an adenovirus coreceptor. J. Virol. 755405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, J., B. A. Ballif, A. M. Powelka, J. Dai, S. P. Gygi, and V. W. Hsu. 2005. Phosphorylation of ACAP1 by Akt regulates the stimulation-dependent recycling of integrin beta1 to control cell migration. Dev. Cell 9663-673. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y., A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7535-545. [DOI] [PubMed] [Google Scholar]
- 34.Marcellus, R. C., J. G. Teodoro, R. Charbonneau, G. C. Shore, and P. E. Branton. 1996. Expression of p53 in Saos-2 osteosarcoma cells induces apoptosis which can be inhibited by Bcl-2 or the adenovirus E1B-55 kDa protein. Cell Growth Differ. 71643-1650. [PubMed] [Google Scholar]
- 35.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 9311295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 1997. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc. Natl. Acad. Sci. USA 941206-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 731591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nevels, M., B. Tauber, T. Spruss, H. Wolf, and T. Dobner. 2001. “Hit-and-run” transformation by adenovirus oncogenes. J. Virol. 753089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oguey, D., P. W. George, and C. Ruegg. 2000. Disruption of integrin-dependent adhesion and survival of endothelial cells by recombinant adenovirus expressing isolated beta integrin cytoplasmic domains. Gene Ther. 71292-1303. [DOI] [PubMed] [Google Scholar]
- 40.Pellinen, T., and J. Ivaska. 2006. Integrin traffic. J. Cell Sci. 1193723-3731. [DOI] [PubMed] [Google Scholar]
- 41.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Querido, E., M. R. Morrison, H. Chu-Pham-Dang, S. W. Thirlwell, D. Boivin, and P. E. Branton. 2001. Identification of three functions of the adenovirus E4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsay, A. G., J. F. Marshall, and I. R. Hart. 2007. Integrin trafficking and its role in cancer metastasis. Cancer Metastasis Rev. 26567-578. [DOI] [PubMed] [Google Scholar]
- 45.Ray, R. B., A. Basu, R. Steele, A. Beyene, J. McHowat, K. Meyer, A. K. Ghosh, and R. Ray. 2004. Ebola virus glycoprotein-mediated anoikis of primary human cardiac microvascular endothelial cells. Virology 321181-188. [DOI] [PubMed] [Google Scholar]
- 46.Reddig, P. J., and R. L. Juliano. 2005. Clinging to life: cell to matrix adhesion and cell survival. Cancer Metastasis Rev. 24425-439. [DOI] [PubMed] [Google Scholar]
- 47.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128480-484. [DOI] [PubMed] [Google Scholar]
- 48.Roth, J., C. Konig, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 728510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salone, B., Y. Martina, S. Piersanti, E. Cundari, G. Cherubini, L. Franqueville, C. M. Failla, P. Boulanger, and I. Saggio. 2003. Integrin α3β1 is an alternative cellular receptor for adenovirus serotype 5. J. Virol. 7713448-13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120510-517. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz, R. A., S. S. Lakdawala, H. D. Eshleman, M. R. Russell, C. T. Carson, and M. D. Weitzman. 2008. Distinct requirements of the adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 829043-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shahgasempour, S., S. B. Woodroffe, G. Sullivan-Tailyour, and H. M. Garnett. 1997. Alteration in the expression of endothelial cell integrin receptors α5β1 and α2β1 and α6β1 after in vitro infection with a clinical isolate of human cytomegalovirus. Arch. Virol. 142125-138. [DOI] [PubMed] [Google Scholar]
- 53.Shigeta, M., N. Sanzen, M. Ozawa, J. Gu, H. Hasegawa, and K. Sekiguchi. 2003. CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J. Cell Biol. 163165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 762518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sriramarao, P., P. Steffner, and K. R. Gehlsen. 1993. Biochemical evidence for a homophilic interaction of the α3β1 integrin. J. Biol. Chem. 26822036-22041. [PubMed] [Google Scholar]
- 56.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 57.Subramanian, T., S. Vijayalingam, and G. Chinnadurai. 2006. Genetic identification of adenovirus type 5 genes that influence viral spread. J. Virol. 802000-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Symington, B. E., Y. Takada, and W. G. Carter. 1993. Interaction of integrins α3β1 and α2β1: potential role in keratinocyte intercellular adhesion. J. Cell Biol. 120523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takada, Y., E. Murphy, P. Pil, C. Chen, M. H. Ginsberg, and M. E. Hemler. 1991. Molecular cloning and expression of the cDNA for α3 subunit of human α3β1 (VLA-3), an integrin receptor for fibronectin, laminin, and collagen. J. Cell Biol. 115257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takada, Y., J. L. Strominger, and M. E. Hemler. 1987. The very late antigen family of heterodimers is part of a superfamily of molecules involved in adhesion and embryogenesis. Proc. Natl. Acad. Sci. USA 843239-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tollefson, A. E., A. Scaria, T. W. Hermiston, J. S. Ryerse, L. J. Wold, and W. S. Wold. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 702296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toth, K., H. Djeha, B. Ying, A. E. Tollefson, M. Kuppuswamy, K. Doronin, P. Krajcsi, K. Lipinski, C. J. Wrighton, and W. S. Wold. 2004. An oncolytic adenovirus vector combining enhanced cell-to-cell spreading, mediated by the ADP cytolytic protein, with selective replication in cancer cells with deregulated wnt signaling. Cancer Res. 643638-3644. [DOI] [PubMed] [Google Scholar]
- 63.Tsuji, T. 2004. Physiological and pathological roles of alpha3beta1 integrin. J. Membr. Biol. 200115-132. [DOI] [PubMed] [Google Scholar]
- 64.Walters, R. W., P. Freimuth, T. O. Moninger, I. Ganske, J. Zabner, and M. J. Welsh. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110789-799. [DOI] [PubMed] [Google Scholar]
- 65.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73309-319. [DOI] [PubMed] [Google Scholar]
- 66.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yun, C. O., E. Kim, T. Koo, H. Kim, Y. S. Lee, and J. H. Kim. 2005. ADP-overexpressing adenovirus elicits enhanced cytopathic effect by induction of apoptosis. Cancer Gene Ther. 1261-71. [DOI] [PubMed] [Google Scholar]
- 68.Zou, A., I. Atencio, W. M. Huang, M. Horn, and M. Ramachandra. 2004. Overexpression of adenovirus E3-11.6K protein induces cell killing by both caspase-dependent and caspase-independent mechanisms. Virology 326240-249. [DOI] [PubMed] [Google Scholar]