Abstract
The cell-to-cell transport of plant viruses depends on one or more virus-encoded movement proteins (MPs). Some MPs are integral membrane proteins that interact with the membrane of the endoplasmic reticulum, but a detailed understanding of the interaction between MPs and biological membranes has been lacking. The cell-to-cell movement of the Prunus necrotic ringspot virus (PNRSV) is facilitated by a single MP of the 30K superfamily. Here, using a myriad of biochemical and biophysical approaches, we show that the PNRSV MP contains only one hydrophobic region (HR) that interacts with the membrane interface, as opposed to being a transmembrane protein. We also show that a proline residue located in the middle of the HR constrains the structural conformation of this region at the membrane interface, and its replacement precludes virus movement.
Plant viruses encode movement proteins (MPs) that mediate the intra- and intercellular spread of the viral genome via plasmodesmata, membranous channels that traverse the walls of plant cells and enable intercellular transport and communication. There is a range of diversity in the number and type of viral proteins required for viral movement (21). Research on tobacco mosaic virus (TMV) has played a leading role in understanding MP activity (2). The genome of TMV encodes a single 30-kDa multidomain protein, the namesake of the 30K superfamily (7). Viral RNA is associated with the membrane of the endoplasmic reticulum (ER) and microtubules in the presence of this MP (23, 30).
A large number of plant viruses have 30K MPs, which share common abilities, including binding nucleic acids, localizing and increasing the size exclusion limit of plasmodesmata, and interacting with the ER membrane. A topological model has been proposed in which the TMV MP has two putative transmembrane (TM) helices, both the N and C termini oriented toward the cytoplasm, and a short loop exposed in the ER lumen (4). There is less experimental information for other 30K MPs, but they are likely to have some membrane interaction.
Direct experimental evidence of the integration of MPs into the membrane has been obtained only for small hydrophobic MPs that do not belong to the 30K superfamily. There are two TM segments in the p9 protein of carnation mottle virus (41), whereas the p6 protein of beet yellow virus (29) and the p7B protein of melon necrotic spot virus (22) have a single TM segment. In viruses with genomes that include three partially overlapping open reading frames, termed the triple-gene block (TGB), all three TGB proteins are required for movement where the two smaller proteins, TGBp2 and TGBp3, are also TM proteins (24). Furthermore, cross-linking experiments with carnation mottle virus p9 protein demonstrated that its membrane insertion occurs cotranslationally in a signal recognition particle-dependent manner and throughout the cellular membrane integration components, the translocon (33, 34).
Prunus necrotic ringspot virus (PNRSV) is a tripartite, positive-strand RNA virus in the genus Ilarvirus of the family Bromoviridae. RNAs 1 and 2 encode the polymerase proteins P1 and P2, respectively. RNA 3 is translated into a single 30K-type MP. The coat protein is translated from a subgenomic RNA 4 produced during virus replication.
The present study tackled the association of the PNRSV MP with biological membranes. The in vitro translation of model integral membrane protein constructs in the presence of microsomal membranes demonstrated that the hydrophobic region (HR) of the PNRSV MP did not span the membranes. Different biochemical and biophysical experiments suggested that the protein is tightly associated with, but does not traverse, the membrane, leaving both its N- and C-terminal hydrophilic regions facing the cytosol. Finally, a mutational analysis of the HR revealed that both the helicity and hydrophobicity of the region are essential for viral cell-to-cell movement.
MATERIALS AND METHODS
Enzymes and chemicals.
All enzymes, as well as plasmid pGEM1, the RiboMAX SP6 RNA polymerase system, rabbit reticulocyte lysate, and dog pancreas microsomes, were from Promega (Madison, WI). [35S]Met and 14C-methylated markers were from GE Healthcare. Restriction enzymes and proteinase K were from Roche Molecular Biochemicals. The DNA plasmid, RNA cleanup, and PCR purification kits were from Qiagen (Hilden, Germany). The oligonucleotides were from Isogen (Maarseen, The Netherlands) or Thermo (Ulm, Germany).
Computer-assisted analysis of TM helices.
The prediction of TM helices for the PNRSV MP sequence was performed using two of the most commonly used prediction methods available on the Internet: TMHMM (17) (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and SOSUI (15; http://bp.nuap.nagoya-u.ac.jp/sosui/). The prediction of free-energy differences for TM helix insertion was performed using the ΔG Prediction Server v1.0 available on the Internet (http://www.cbr.su.se/DGpred/; 13, 14). All user-adjustable parameters were left at their default values.
DNA manipulation.
Plasmid pGEM-p32 was created by subcloning the PNRSV MP (p32) sequence into NcoI/NdeI restriction sites in the pGEM-Lep vector (26). The HR from p32 was introduced into Lep (leader peptidase) by replacing the H2 DNA sequence, as described previously (41). Construction of chimeric pGFP/MP was previously described (12, 32). The engineered glycosylation sites, the replacement of Pro 96 by Ala and Gln 99 by Leu, and deletion of the p32 HR in all the plasmids were done using the QuikChange mutagenesis kit from Stratagene (La Jolla, CA). All DNA manipulations were confirmed by sequencing of plasmid DNAs.
Protein expression in vitro.
Lep-derived constructs and the full-length PNRSV MP were transcribed and translated in the presence of [35S]methionine as previously described (28, 41). Full-length p32 DNA was amplified from the pGEM-p32 plasmid using a reverse primer with a stop codon at the end of the PNRSV MP sequence (p32-derived expression). Transcription and translation in the presence of reticulocyte lysate, [35S]Met, and dog pancreas microsomes were done as described previously (28). Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were visualized on a Fuji FLA3000 phosphorimager using ImageGauge software. The extent of glycosylation of a given mutant was calculated as the quotient of the intensity of the glycosylated band divided by the summed intensities of the glycosylated and nonglycosylated bands for each analyzed lane (35). For the proteinase K protection assay, after translation, the mixture was subjected alternatively to digestion by 200 or 400 μg/ml proteinase K for 40 min on ice. The reaction was stopped by adding 2 mM phenylmethylsulfonyl fluoride. The membrane fraction was then collected by centrifugation and analyzed by SDS-PAGE.
Cysteine modification.
After in vitro translation in the presence or absence of microsomal membranes, samples were supplemented with 20 mM 4-acetamido-4′-maleimidostilbene-2,2′-disulfonate (AMS) (Molecular Probes) in 50 mM sodium phosphate (pH 7) and incubated for 1 h at room temperature. The membranes were collected by ultracentrifugation and analyzed by SDS-PAGE.
Membrane sedimentation.
The translation mixture was diluted in 8 volumes of buffer A (35 mM Tris-HCl, pH 7.4, and 140 mM NaCl) for the membrane sedimentation or 4 volumes of buffer A supplemented with 100 mM Na2CO3 (pH 11.5) for the alkaline wash. The samples were incubated on ice for 30 min and clarified by centrifugation (10,000 × g; 20 min). The membranes were collected by ultracentrifugation (100,000 × g; 20 min; 4°C) of the supernatant onto a 50-μl sucrose cushion. The pellets and supernatants from the ultracentrifugation were analyzed by SDS-PAGE.
Membrane flotation.
We examined the membrane association of the full-length PNRSV MP using vesicle flotation (8). The translation mixture was adjusted with 0.2 M Na2CO3 (pH 11.5) to a final volume of 250 μl and incubated for 1 hour on ice. Samples were then mixed with 4 M sucrose, 0.1 M Na2CO3 to obtain a 1.8 M sucrose solution. The sucrose mixture was transferred to centrifugation tubes and overlaid with 275 μl of 1.25 M sucrose, 0.1 M Na2CO3 and with 250 μl of 0.25 M sucrose, 0.1 M Na2CO3. After centrifugation for 4 h in a Beckman TLS-55 rotor, four fractions (numbered from the top) of approximately 200 μl each were withdrawn from the top and analyzed by SDS-PAGE. The pellet was dissolved with the last 200-μl fraction. The results were always processed in parallel with the sample distribution of the model membrane protein Lep (the leader peptidase from Escherichia coli).
Expression of PNRSV MP and Lep proteins in planta, membrane sedimentations, and Western blot assay.
The PNRSV MP and Lep genes were first amplified by PCR using specific antisense primers containing the hemagglutinin (HA) sequence and then introduced into the pKS+ vector under the control of the 35S promoter from cauliflower mosaic virus and the potato proteinase inhibitor terminator (PoPit), using the NcoI-NheI restriction enzymes. The resultant 35S-PNRSVMP:HA-PoPit and 35S-Lep:HA-PoPit cassettes were introduced into the XhoI-digested pMOG800 binary vector. Cultures of Arabidopsis tumefaciens strain C58C1 transformed with the different binary pMOG800 plasmids were used to infiltrate Nicotiana benthamiana plants as described previously (12). Three days postinfiltration, the leaves were processed to obtain enriched membranous fractions as described previously (29). The resultant membranous enriched pellet was resuspended in buffer A (20 mM HEPES, pH 6.8, 150 mM potassium acetate, 250 mM mannitol, 1 mM MgCl2, 2.5 ml of protease inhibitor cocktail for plant cell and tissue extracts; Sigma) and divided into three aliquots for the untreated, alkaline wash, or urea treatments as described previously. The membranes were collected by ultracentrifugation (100,000 × g; 20 min; 4°C). For Western analysis, pellets and supernatants from the ultracentrifugation were electrophoresed through 12% SDS-polyacrylamide gels, electrotransferred to polyvinylidene difluoride membranes following the manufacturer's instructions (Amersham), and probed with anti-HA (Sigma) following the manufacturer's instructions (Amersham). Antibody binding was detected by immunoreaction with the appropriate immunoglobulin G coupled to horseradish peroxidase and subsequent chemobioluminescence detection as recommended by the manufacturer (Amersham).
Peptide synthesis.
Synthetic peptides, including the PNRVS MP HR (RVFLVYVPIIQATTSGLITLK-NH2) and the mutant P96A/Q99L (RVFLVYVAIILATTSGLITLK-NH2), were synthesized using a 433A automatic synthesizer (Applied Biosystems) as previously described (27).
Circular dichroism.
All measurements were carried out on a Jasco J-810 CD spectropolarimeter with a Neslab RTE 110 water bath as a thermostat. The CD spectra were the average of a series of 10 scans taken at 0.2-nm intervals. The peptide concentration was 30 μM in all cases.
Interfacial adsorption of peptides.
The interfacial adsorption of synthetic peptides derived from the PNRSV MP HR sequence was assayed using a specially designed surface balance, as described previously (38). The microbalance was filled with 1.5 ml of buffer (5 mM Tris, 150 mM NaCl, pH 7), and after 5 min of equilibration, different amounts of peptide, dissolved in methanol, were injected into the subphase. Changes in surface pressure (π) were then monitored over time. The subphase was continuously stirred and maintained at 25°C with a thermostat. Subphases were prepared with double-distilled water. Injections of equivalent volumes of pure methanol did not produce any detectable change in surface pressure.
Insertion of peptides into preformed lipid monolayers.
The insertion of peptides into phospholipid monolayers preformed at different initial surface pressures (πi) was monitored from the changes in surface pressure over time (Δπ) after peptide injection into the subphase. Phospholipid monolayers were preformed by spreading concentrated solutions of lipids (palmitoyloleoylphosphatidylcholine [POPC] or palmitoyloleoylphosphatidylglycerol [POPG]) in chlorophorm/methanol (3:1 [vol/vol]) on top of the aqueous surface. After 10 min of stabilization, 5 μg of peptide in methanol was injected into the subphase. Δπ was plotted against the initial pressure, allowing calculation of the critical insertion pressure (πc), defined as the maximum π value at which the peptide could still insert into the lipid monolayer. The subphase was steadily stirred and kept at 25°C.
Inoculation of P12 protoplasts and plants.
pGFP/MP/CP plasmids containing either the wild type (WT) or the different mutants of the PNRSV MP were linearized with PstI and transcribed with T7 RNA polymerase. Protoplasts were extracted from P12 transgenic Nicotiana tabacum plants, and 2.5 × 105 protoplasts were inoculated by the polyethylene glycol method (20) with 6 μl of the transcription mixture. P12 plants were grown and inoculated with RNA transcripts, as previously described (40). The green fluorescent protein (GFP) expression in protoplast and plants was analyzed with a Leica TCS SL confocal laser scanning microscope (Leica), with excitation at 488 nm and emission at 500 to 535 nm.
RESULTS
Sequence analysis of the PNRSV MP.
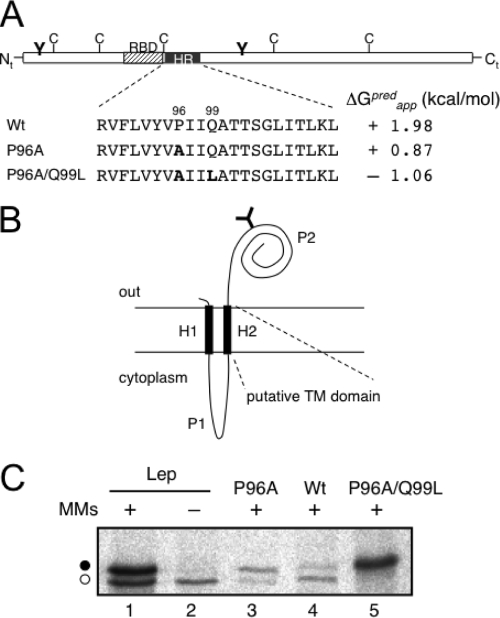
A highly conserved HR was previously identified within the MPs of all ilarviruses (31). Nevertheless, a sequence analysis of the PNRSV MP, utilizing several algorithms (see Materials and Methods), failed to identify any membrane-spanning domain. The more-hydrophobic region (residues 89 to 110) found using the ΔG Prediction Server v1.0 (http://www.cbr.su.se/DGpred/), in which the prediction of TM helices comes from the apparent free-energy difference (ΔGapp) from insertion into ER membranes (13, 14), is shown in Fig. 1A. The positive value of ΔGapp predicted that the HR is not TM. A closer analysis of the output data highlighted Pro96 and Gln99 as precluding the TM orientation. Hence, replacing Pro96 with alanine reduced the predicted ΔGapp by more than 1 kcal/mol, whereas the double mutant Pro96Ala/Gln99Leu was predicted to favor insertion due to the negative value of ΔGapp (−1.06 kcal/mol) (Fig. 1A).
FIG. 1.
Membrane association of the PNRSV MP HR. (A) Schematic representation of the PNRSV MP highlighting the HR (black box) and mutations in that region. Predicted (pred) ΔGapp values are shown. The striped box denotes the RNA-binding domain (RBD). The positions of natural cysteines are indicated, as well as the engineered glycosylation sites (Y-shaped symbol). (B) Membrane topology of the model protein Lep. The TM segments H1 and H2 are connected by a charged cytoplasmic loop (P1). The large periplasmic domain P2 translocates across the ER membrane. Engineered sites in the Lep coding region allow the exchange of H2 for the HR from PNRSV MP. (C) In vitro translation in the presence (+) or absence (−) of microsomal membranes (MMs). Bands of nonglycosylated protein are indicated by the white dot, and glycosylated proteins are indicated by the black dot.
Insertion of the HR into biological membranes.
In order to examine the propensity of the HR of the PNRSV MP to form a TM helix in biological membranes, an in vitro transcription/translation assay in the presence of microsomal membranes and [35S]methionine was performed. The assay relied on the use of a model integral membrane protein derived from the leader peptidase of E. coli (Lep) with an N-terminal TM segment (H1), followed by a positively charged loop (P1), a second TM segment (H2), and a large C-terminal domain (P2) containing an engineered glycosylation acceptor site (Fig. 1B). The presence of H1 ensured proper targeting and insertion of the protein in chimeras with the N terminus oriented toward the lumenal side of the microsome (10, 26). The glycosylation of a protein translated in vitro in the presence of microsomes indicates the exposure of the nascent chain to the lumenal side of the ER membrane. Therefore, the protein will be glycosylated only when H2 inserts into the membrane, translocating the P2 domain (Fig. 1B). In such a system, the potential for a sequence to function as a TM segment can be tested by replacing H2 with a putative TM domain, performing in vitro transcription/translation (using radioactively labeled amino acids) of the corresponding chimera in the presence of ER microsomes, and determining possible glycosylation through the observation of an approximately 2.5-kDa increase in the expected molecular mass by SDS-PAGE. Based on the predictions made by the ΔG Prediction Server, residues 89 to 110 of the PNRSV MP were selected for analysis using Lep chimera membrane insertion. The efficiency of Lep glycosylation (∼73%) is illustrated in Fig. 1C, where a new radioactive band corresponding to an increased molecular mass of the glycosylated protein (compare lanes 1 and 2) was observed in the presence of microsomal membranes. When we assayed the HR of the PNRSV MP, the glycosylation efficiency decreased to approximately 23% (Fig. 1C, lane 4), which is indicative of a poor membrane insertion capacity. However, P96A increased the glycosylation efficiency to approximately 80% (Fig. 1C, lane 3). Furthermore, the P96A/Q99L double mutant gave rise to full glycosylation (>95%) (Fig. 1C, lane 5). This behavior suggests that the HR of the PNRSV MP inserts poorly into biological membranes, even in the context of the integral membrane protein Lep, which agrees with the predicted positive ΔGapp value.
PNRSV MP is not a TM protein.
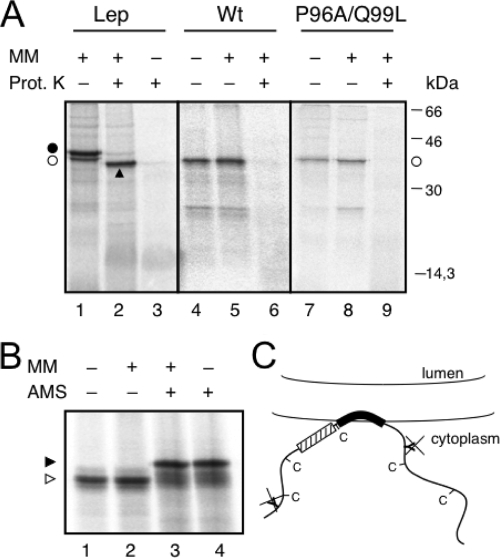
Next, we analyzed the insertion into the membrane of full-length MP. In order to determine the membrane translocation of the N- and C-terminal hydrophilic domains, we engineered two glycosylation sites on both sides of the HR (Fig. 1A). The translocation of one hydrophilic domain would result in a monoglycosylated population, whereas translocation of the whole molecule into the microsome lumen would generate double-glycosylated proteins. The addition of proteinase K should digest cytoplasmically exposed protein regions, and translocated fragments would be protected. We found that translation in the presence of microsomes yielded nonglycosylated molecules (Fig. 2A, lane 5) sensitive to proteinase K (Fig. 2A, lane 6). As a control, the translation of Lep in the presence of membranes yielded a mainly glycosylated population (Fig. 2A, lane 1) and a protected, glycosylated H2-P2 fragment upon proteinase K treatment (Fig. 2A, lane 2). Interestingly, the double mutant P96A/Q99L was not glycosylated (Fig. 2A, lane 8) and, accordingly, was digested by proteinase K (Fig. 2A, lane 9). To corroborate the results of glycosylation mapping, we assessed the membrane disposition of the PNRSV MP by labeling the cysteine residues with a membrane-impermeable thiol-reactive reagent (AMS). The AMS labeling was similar when the PNRSV MP was translated in vitro in the absence or presence of microsomal membranes (Fig. 2B, compare lanes 3 and 4). The electrophoretic mobility shift of the labeled relative to the unlabeled molecules (Fig. 2B, lanes 1 and 2) was correlated with the modification of the five cysteines, strongly suggesting a cytosolic exposure of the five cysteines present in the MP sequence. Notably, the similar levels of protein translation in the absence and presence of microsomes (Fig. 2A and B) suggests that the protein does not require the cell machinery for membrane targeting and association. The P96A/Q99L mutant was cysteine labeled to a similar extent (see Fig. S1 in the supplemental material). Taken together, these results suggest that, rather than being an integral membrane protein, the PNRSV MP is a peripherally associated membrane protein including one HR with both the N- and C-terminal hydrophilic domains oriented toward the cytoplasm (Fig. 2C).
FIG. 2.
Full-length protein expression in vitro. (A) Proteinase K (Prot. K) treatment of microsomes carrying in vitro-translated WT Lep (lanes 1 to 3), the full-length PNRSV MP WT sequence (lanes 4 to 6), and mutant P96A/Q99L (lanes 7 to 9). Nonglycosylated and glycosylated molecules are indicated by the white dots and a black dot, respectively. An arrowhead indicates protease-protected fragments. MM, microsomal membranes. (B) The AMS derivatization of PNRSV MP. After translation, proteins were incubated with 20 mM AMS (+) or mock treated with dimethyl sulfoxide (−) in the presence (+) or absence (−) of microsomal membranes. (C) Model for the association of PNRSV MP with membranes; the engineered glycosylation sites and natural cysteines are cytoplasmically exposed.
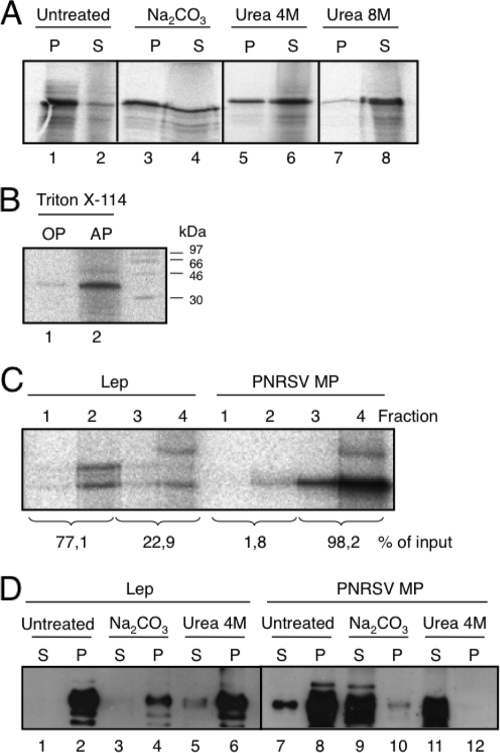
Alkaline extraction, urea, and Triton X-114 treatments have been used to distinguish between peripheral and integral membrane proteins (36). The PNRSV MP was recovered from the 100,000 × g pellet fraction (Fig. 3A, untreated lanes) upon centrifugation of the microsome-containing translation reaction mixture, indicating that it could be either a membrane-associated (peripheral or integral) protein or a lumenally translocated protein. To differentiate between these possibilities, the translation reaction mixtures were first washed with sodium carbonate (pH 11.5), which renders microsomes into membranous sheets, releasing the soluble luminal proteins (25, 29). As shown in Fig. 3A, the PNRSV MP appeared to be mainly associated (approximately 66%; lanes 3 and 4) with the membranous pellet fraction, suggesting a tight association with membranes. Further treatment with 4 M urea demonstrated that approximately 68% of the protein was in the supernatant fraction (Fig. 3A, lanes 5 and 6). More than 95% of the protein was extracted in the supernatant fraction by 8 M urea (Fig. 3A, lanes 7 and 8), suggesting that PNRSV MP is not a TM protein. The translation reaction mixtures were also treated with Triton X-114, a nonionic detergent that forms a separate organic phase to which the membrane lipids and hydrophobic proteins are segregated from the aqueous phase, which contains non-integral membrane proteins (3). The PNRSV MP was detected in the aqueous, but not the organic, phase (Fig. 3B). Again, these results indicate that the PNRSV MP is not an integral membrane protein. In addition, we examined the membrane association of the full-length PNRSV MP using vesicle flotation. As shown in Fig. 3C, when the PNRSV MP translated in the presence of microsomes was subjected to flotation gradient centrifugation, more than 98% of the protein was recovered from the bottom fractions of the gradient (fractions 3 and 4), confirming that the PNRSV MP is not an integral membrane protein. Parallel control experiments using Lep demonstrated the presence of the membrane protein in the upper fractions (fractions 1 and 2) of the gradient.
FIG. 3.
Association of the PNRSV MP with microsomal (A to C) or plant (D) membranes. (A) Segregation of [35S]Met-labeled PNRSV MP into membranous and soluble fractions (untreated) and after alkaline wash (sodium carbonate buffer) or urea treatments. P and S denote pellet and supernatant, respectively. (B) Triton X-114 partitioning. OP and AP refer to organic and aqueous phases, respectively. (C) Flotation gradient centrifugation of Lep (left) and PNRSV MP (right) translated in vitro in the presence of microsomes. For calculating the percentages of membrane binding, the signals present in all four fractions (numbered from the top of the centrifuge tubes) were summed and set to 100%. (D) Segregation of PNRSV MP and Lep proteins expressed in planta in membranous and soluble fractions. PNRSV MP and Lep proteins carrying the HA tag were expressed in N. benthamiana plants by agroinfiltration. Comparable P and S fractions obtained from untreated membranous fractions and after alkaline wash or 4 M urea treatments were analyzed by Western analysis using an anti-HA antibody.
Finally, leaf tissues infected with an HA-tagged PNRSV MP or Lep protein were used to analyze the nature of the membrane association of the MP in planta. Three days after infection, the PNRSV MP was mainly recovered from the 100,000 × g pellet fraction (Fig. 3D, lane 8). Sodium carbonate and 4 M urea treatments released the protein to the supernatant fractions (Fig. 3D, lanes 9 and 11), indicative of a peripheral association. Control immunodetection analyses of HA-tagged Lep showed that, as expected, Lep accumulates in pellet fractions under these treatments (Fig. 3D, lanes 1 to 6).
These results suggest that the PNRSV MP associates tightly with biological membranes, but its HR, though probably responsible for this association, is not a TM segment either in microsomal membranes or in planta. Peptides corresponding to the membrane-interacting portions of several viral glycoproteins have been functionally and structurally characterized (reviewed in reference 42). Many of these HRs share a common feature: the presence of a critical proline at or near the center (9, 11). This proline likely induces these regions to sit in the membrane as kinked structures. Because the HR of PNRSV MP also contains a central proline residue (Fig. 1A), which together with Glu99 precludes a TM insertion for this region (Fig. 1C), we studied the secondary structures of synthetic peptides designed from the HR sequence and characterized the association of these peptides with membrane-like interfaces.
Effects of Pro96Ala/Gln99Leu mutations on the structure of the HR.
As already shown, the PNRSV MP does not insert cotranslationally into the ER membrane (Fig. 2A). This means that the protein is likely synthesized in the cytosol of the host cell and should associate with the membrane posttranslationally (1). The calculated free energy of transfer from water to the membrane interface (ΔGwif) for the folded PNRSV MP HR sequence is −11.5 kcal/mol and is clearly larger than the value for the same unfolded sequence (ΔGwif = −0.98 kcal/mol) or the free energy of inserting the same sequence in an α-helical conformation into the bilayer hydrophobic core (ΔGwoct = −3.8 kcal/mol) (18, 43). The energy values are consistent with a preference of the HR to remain associated with the membrane interface region rather than adopting a TM configuration.
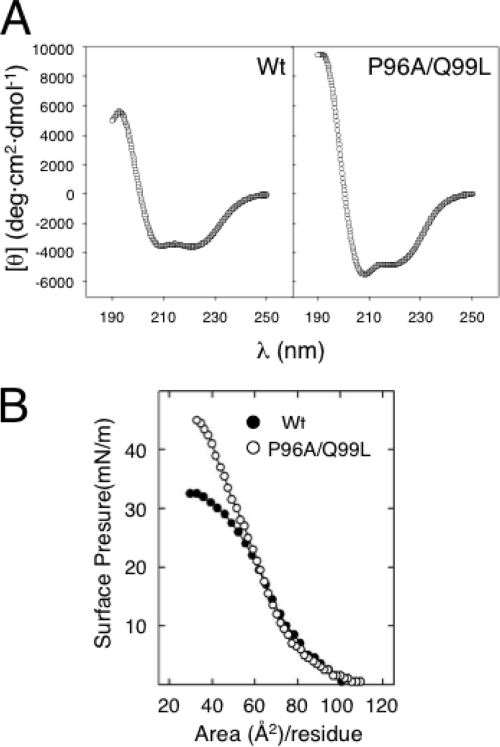
Sequence analysis of the HR suggests that Pro96 and Gln99, located roughly in the middle of the HR, display a high helical-turn potential (see Fig. S2 in the supplemental material). These residues may structurally constrain the HR sequence into a kinked α-helical conformation. To unravel the structure of this HR out of the MP context, we synthesized two peptides corresponding to residues 89 to 109 of the WT sequence and a variant bearing the P96A/Q99L mutation. Far-UV circular dichroism spectra in environments of decreasing polarity demonstrated the existence of conformational differences between the WT and P96A/Q99L peptides. Spectra in trifluorethanol (Fig. 4A) indicated mainly α-helical conformations for both peptides, with a characteristic positive band at 190 nm and two negative bands at 208 and 222 nm. The P96A/Q99L peptide had higher values, suggesting a more helical conformation for the peptide.
FIG. 4.
Structure of the HR-derived peptides. (A) Far-UV circular dichroism spectra of HR-derived synthetic peptides dissolved in trifluoroethanol at room temperature. (B) Compression π-A isotherms of monomolecular films of HR-derived peptides spread at an air-water interface. θ, molar ellipticity.
We also compared the conformational propensities of the WT and P96A/Q99L peptides at an air/water interface. The surface pressure-area (π-A) isotherms of interfacial peptide films have been widely used to obtain information on the minimal molecular areas occupied by different sequences (37). The π-A isotherms of the two synthetic peptides deposited at the interface are compared in Fig. 4B. The two sequences had similar maximal molecular areas of approximately 85 Å2/residue, corresponding to a rather extended disposition of the sequences in expanded films in which peptide molecules just touched each other. However, the two sequences had very different minimal molecular areas (Amin) at maximal compression. The P96A/Q99L variant could be compressed up to 40 Å2/residue, whereas the WT peptide had an Amin of ∼55 Å2/residue. This indicates that the WT HR peptide occupies more space at the interface at maximal packing, presumably due to the kinked conformation, than the P96A/Q99L variant.
Effects of Pro96Ala/Gln99Leu mutations on the insertion of the PNRSV MP HR into phospholipid interfaces.
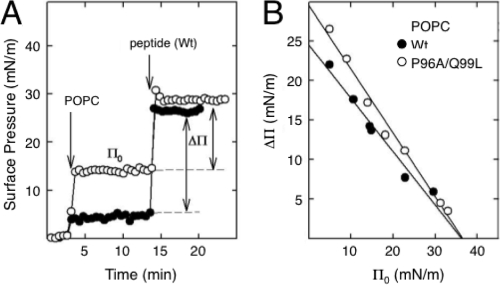
To compare the intrinsic ability of the WT and P96A/Q99L HR sequences to insert into phospholipid interfaces, we used a monolayer technique. A representative experiment with the insertion of the WT peptide into preformed POPC monolayers is shown in Fig. 5A. The association and subsequent insertion of the peptide into POPC films was detected by the rapid increase in surface pressure that followed the injection of a small volume of peptide solution under the interface. A plot of the increase in surface pressure (Δπ) versus the initial surface pressure (π0) of the preformed film allowed the calculation of the critical pressure (πc) permitting the insertion of the peptide. Values of πc at or above 30 mN/m are considered indicative of the propensity of a given peptide or protein to insert into membranes (5). The πc determination for the insertion of the WT and P96A/Q99L peptides in POPC films is shown in Fig. 5B. The two peptides had critical insertion pressures above 30 mN/m in the zwitterionic phospholipid, but also in films of the anionic POPG phospholipid (see Fig. S3 in the supplemental material), confirming their propensities to associate with and penetrate into phospholipid membranes. In POPC films, the two peptides had critical-pressure plots with different slopes, which can be interpreted as a consequence of the differences in the orientations of the peptides at the interface (39). This would support the idea that the WT and P96A/Q99L sequences associate with lipid surfaces through different conformations. No differences were observed either in πc or in the Δπ/π0 slope for the two peptides in POPG monolayers (see Fig. S3 in the supplemental material). The lipid's negative charge probably anchors both the C- and N-terminal charged ends of the two peptides to the surface, abrogating the conformational differences they exhibit in zwitterionic films, something that probably does not occur in bilayers.
FIG. 5.
Insertion of the HR-derived peptides into preformed phospholipid monolayers. (A) Two illustrative experiments are shown in which the insertion of WT peptide into POPC films formed at an initial surface pressure of either 3 (closed circles) or 13 (open circles) mN/m was followed by a concomitant increase in surface pressure (Δπ). (B) Determination of the critical insertion pressure (πc) of WT (closed symbols) or P96A/Q99L (open symbols) peptides in POPC monolayers. The πc values were determined from the abscissa intercepts.
The relative stabilities of the WT and P96A/Q99L sequences incorporated into phospholipid interfaces were also compared based on the π-A compression isotherms of dipalmitoylphosphatidylcholine films in the absence or presence of 0.75, 1.5, or 2.3% of either peptide (see Fig. S4 in the supplemental material). The P96A/Q99L peptide broadened, the liquid-expanded to liquid-condensed phase transition of dipalmitoylphosphatidylcholine to a somewhat greater extent than the WT peptide, probably because it perturbed a larger number of phospholipid molecules per peptide molecule. Furthermore, films containing the WT peptide produced isotherms exhibiting marked exclusion plateaus at 47 to 48 mN/m, whereas the P96A/Q99L peptide remained in the films at substantially higher pressures. This feature suggests that the WT sequence is less stable than the mutant in packed phospholipid layers, probably as a consequence of a lower hydrophobicity and the kinked structure.
Requirement for the PNRSV MP HR in cell-to-cell movement.
To study the potential of the PNRSV MP HR to promote cell-to-cell movement in vivo, a previous chimeric construct of Alfalfa mosaic virus (AMV) RNA 3-GFP was used (12). The construct expresses the full-length PNRSV MP fused to the C-terminal 44 amino acids of the AMV MP, a chimeric protein previously shown to be functional for the cell-to-cell transport of AMV RNA 3 (32). Using the GFP reporter, the ability of the construct to move from infected to adjacent cells in N. tabacum constitutively expressing the AMV polymerase proteins (P12 plants) was analyzed. The P12 leaves and protoplasts were inoculated with transcripts of the construct, and the fluorescent signals were monitored by confocal laser scanning microscopy (Fig. 6A). Single fluorescent cells at 2 days postinfection (p.i.) were observed to become foci at 4 days p.i., when the full-length PNRSV MP gene was expressed. Fluorescence was also detected on P12 protoplasts 18 h p.i. These results demonstrate that the chimeric construct was functionally active in the movement of chimeric AMV RNA 3.
FIG. 6.
MP expression in vivo and HR of ilarviruses. (A) Detection of green fluorescence in P12 plants and protoplasts inoculated with transcripts carrying different PNRSV MP mutants. The P12 plants and protoplasts were inoculated with RNA 3 transcripts from the following plasmids: pGFP/MP-Wt/CP, pGFP/MP-ΔHR/CP, pGFP/MP-P96A/CP, pGFP/MP-Q99L/CP, and pGFP/MP-P96A/Q99L/CP. A schematic representation of the pGFP/MP-Wt/CP construct is shown (top left). The HR of the PNRSV MP is indicated by a black box, and the amino acid sequence is shown, with the flanking residues in italics. Deleted residues are shown in gray, and replacement mutations in boldface. Fluorescence was monitored at 2 and 4 days p.i. (dpi) in P12 plants or 18 hours posttransfection in protoplasts using a confocal laser scanning microscope set with excitation at 488 nm and emission at 500 to 535 nm. The scale bars represent 200 μm (leaves) or 20 μm (protoplasts). (B) Multiple-sequence alignment of ilarvirus MPs described in the VIDE database (http://image.fs.uidaho.edu/vide/refs.htm; 6) using the CLUSTAL W2 web server at EMBL-EBI (http://www.ebi.ac.uk/Tools/clustalw2/index.html) (19). Only the HR is shown. All parameters were left at the default values. APLPV, American plum line pattern virus; ApMV, apple mosaic virus; CVV, citrus variegation virus; CiLRV, citrus leaf rugose virus; HdMV, hydrangea mosaic virus; LRMV, lilac ring mottle virus; PDV, prune dwarf virus; EMoV, elm mottle ilarvirus; TSV, tobacco streak virus; SpLV, spinach latent virus; TAMV, tulare apple mosaic virus; PMoV, parietaria mottle virus; HJV, humulus japonicus virus.
The P12 plants inoculated with transcripts from a construct in which the PNRSV MP HR was deleted rendered a hybrid virus unable to move from cell to cell, showing single infected cells at 2 days p.i. but no infection foci at 4 days p.i. (Fig. 6A). However, the construct was fully expressed in protoplasts. Remarkably, similar results were observed with the P96A mutant and transcripts bearing the P96A/Q99L mutation. Such virus transport blockage was not observed when only Q99L was present. Apparently, the absence of the proline residue is sufficient to block virus transport, indicating that the proline residue within the HR is critical for MP function.
DISCUSSION
The plant cell wall creates a sturdy barrier preventing the direct entry of viruses, which cannot infect plant cells by any independent ability to cross membranes. Plant viruses exploit small pores between adjacent cells, plasmodesmata, because they provide cytoplasm and ER membrane continuity for cell-to-cell transport. Among plant viruses, cell-to-cell transport depends on one or more virus-encoded MPs, and many of these interact with membranes. In those viruses encoding more than one small MP, there is experimental evidence demonstrating that at least one of the encoded MPs is an integral membrane protein (22, 29, 41). For the virus encoding only one large MP (30K superfamily), a topological model for TMV MP with two HRs spanning the membrane has been proposed (4). Since then, it has been assumed that a similar association with membranes exists for other MPs in this group. The PNRSV MP belongs to this group, but it bears a single HR in its sequence, which creates a topological problem. In the TMV MP model, both the N and C termini are exposed to the cytoplasm, which may facilitate RNA binding. With only one TM region, the PNRSV MP would leave only one of the two ends exposed to the cytoplasm, either the N terminus, where the RNA binding domain is located, or the large C-terminal domain.
Overall, our results show that (i) the HR of the PNRSV MP does not insert with a TM orientation when grafted onto a model membrane protein; (ii) the full-length PNRSV MP associates tightly in vitro and in planta with membranes, though not TM; (iii) the presence of a proline and a glutamine residue in the HR constrains the TM disposition, the secondary structure, and its interaction with lipid interfaces; and (iv) the replacement of the helix-bending proline residue precludes viral cell-to-cell transport in vivo. There is a striking similarity between these results and the effect exerted by proline residues present in animal virus fusion peptides, in which the replacement of proline has been shown to abolish infectivity and to interfere with the ability to induce bilayer destabilization (11, 16).
The results of the current study are consistent with the abundant data referring to an intimate association of 30K MPs, mainly TMV MP, with membranes. The full-length PNRSV MP remains in the membrane fraction unless strong denaturation conditions are applied, and a peptide harboring the HR more likely inserts into the lipid interface with a kinked α-helical conformation. This membrane disposition would allow the N and C termini of the MP to bind the viral genome and interact with cytoplasmic components of the host cell.
Given the helix-bending capacity of proline residues found in HRs and the complexity of the biological membranes, the detailed molecular mechanism behind viral cell-to-cell transport remains unclear. Interestingly, a sequence alignment of the HRs of ilarviruses demonstrated that the central proline residue is conserved in all viruses characterized so far (Fig. 6B). We also found that the alanine replacement at Pro96 abrogates viral cell-to-cell transport in vivo, whereas the presence of the glutamine residue within the HR is not critical for virus transport, as would be expected from the sequence alignment (Fig. 6B). We concluded that the presence of a proline in the middle of the HR region of the PNRSV MP is directly related to its capacity to insert into the lipid interface with a kinked α-helical conformation. We also hypothesize that Pro96Ala may unleash the interfacial kinked structure, which would be sufficient to block virus transport. Membrane interaction studies using different 30K MPs are under way with the aim of unveiling whether this tight interfacial association observed for the PNRSV MP HR, instead of the TM disposition that has been shown for plant viruses with several small MPs, is a unique case or a common theme in this important group of plant viruses.
Supplementary Material
Acknowledgments
This work was supported by grants BMC2006-08542 (to I.M.), BIO2005-07331 (to V.P.), BIO2006-03130 (to J.P.-G), and CSD2007-00010 (to J.P.-G) from the Spanish MEC and GV04B-183 from the Generalitat Valenciana (to I.M.).
José Luis Nieva (University of the Basque Country) provided valuable discussions and suggestions.
Footnotes
Published ahead of print on 25 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alder, N. N., and A. E. Johnson. 2004. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 27922787-22790. [DOI] [PubMed] [Google Scholar]
- 2.Beachy, R. N., and M. Heinlein. 2000. Role of P30 in replication and spread of TMV. Traffic 1540-544. [DOI] [PubMed] [Google Scholar]
- 3.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 2561604-1607. [PubMed] [Google Scholar]
- 4.Brill, L. M., R. S. Nunn, T. W. Kahn, M. Yeager, and R. N. Beachy. 2000. Recombinant tobacco mosaic virus movement protein is an RNA-binding, alpha-helical membrane protein. Proc. Natl. Acad. Sci. USA 977112-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockman, H. 1999. Lipid monolayers: why use half a membrane to characterize protein-membrane interactions? Curr. Opin. Struct. Biol. 9438-443. [DOI] [PubMed] [Google Scholar]
- 6.Brunt, A. A., K. Crabtree, M. J. Dallwitz, A. J. Gibbs, L. Watson, and E. J. Zurcher. 1996-2009. Plant viruses online: descriptions and lists from the VIDE database. http://image.fs.uidaho.edu/vide/. Accessed 20 August 1996.
- 7.Citovsky, V., and P. Zambryski. 1991. How do plant virus nucleic acids move through intercellular connections? Bioessays 13373-379. [DOI] [PubMed] [Google Scholar]
- 8.Deitermann, S., G. S. Sprie, and H. G. Koch. 2005. A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J. Biol. Chem. 28039077-39085. [DOI] [PubMed] [Google Scholar]
- 9.Delos, S. E., J. M. Gilbert, and J. M. White. 2000. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 741686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gafvelin, G., M. Sakaguchi, H. Andersson, and G. von Heijne. 1997. Topological rules for membrane protein assembly in eukaryotic cells. J. Biol. Chem. 2726119-6127. [DOI] [PubMed] [Google Scholar]
- 11.Gomara, M. J., P. Mora, I. Mingarro, and J. L. Nieva. 2004. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 569261-266. [DOI] [PubMed] [Google Scholar]
- 12.Herranz, M. C., J. A. Sanchez-Navarro, A. Sauri, I. Mingarro, and V. Pallas. 2005. Mutational analysis of the RNA-binding domain of the Prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell-to-cell movement. Virology 33931-41. [DOI] [PubMed] [Google Scholar]
- 13.Hessa, T., H. Kim, K. Bihlmaier, C. Lundin, J. Boekel, H. Andersson, I. Nilsson, S. H. White, and G. von Heijne. 2005. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433377-381. [DOI] [PubMed] [Google Scholar]
- 14.Hessa, T., N. M. Meindl-Beinker, A. Bernsel, H. Kim, Y. Sato, M. Lerch-Bader, I. Nilsson, S. H. White, and G. von Heijne. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 4501026-1030. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14378-379. [DOI] [PubMed] [Google Scholar]
- 16.Ito, H., S. Watanabe, A. Sanchez, M. A. Whitt, and Y. Kawaoka. 1999. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J. Virol. 738907-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 18.Ladokhin, A. S., and S. H. White. 1999. Folding of amphipathic alpha-helices on membranes: energetics of helix formation by melittin. J. Mol. Biol. 2851363-1369. [DOI] [PubMed] [Google Scholar]
- 19.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 20.Loesch-Fries, L. S., E. L. Halk, S. E. Nelson, and K. J. Krahn. 1985. Human leukocyte interferon does not inhibit alfalfa mosaic virus in protoplasts or tobacco tissue. Virology 143626-629. [DOI] [PubMed] [Google Scholar]
- 21.Lucas, W. J. 2006. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344169-184. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Gil, L., A. Sauri, M. Vilar, V. Pallas, and I. Mingarro. 2007. Membrane insertion and topology of the p7B movement protein of Melon Necrotic Spot Virus (MNSV). Virology 367348-357. [DOI] [PubMed] [Google Scholar]
- 23.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morozov, S. Y., and A. G. Solovyev. 2003. Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 841351-1366. [DOI] [PubMed] [Google Scholar]
- 25.Navarro, J. A., A. Genoves, J. Climent, A. Sauri, L. Martinez-Gil, I. Mingarro, and V. Pallas. 2006. RNA-binding properties and membrane insertion of Melon necrotic spot virus (MNSV) double gene block movement proteins. Virology 35657-67. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson, I., and G. von Heijne. 1993. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 2685798-5801. [PubMed] [Google Scholar]
- 27.Orzaez, M., E. Perez-Paya, and I. Mingarro. 2000. Influence of the C-terminus of the glycophorin A transmembrane fragment on the dimerization process. Protein Sci. 91246-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orzaez, M., J. Salgado, A. Gimenez-Giner, E. Perez-Paya, and I. Mingarro. 2004. Influence of proline residues in transmembrane helix packing. J. Mol. Biol. 335631-640. [DOI] [PubMed] [Google Scholar]
- 29.Peremyslov, V. V., Y. W. Pan, and V. V. Dolja. 2004. Movement protein of a closterovirus is a type III integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 783704-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichel, C., and R. N. Beachy. 1998. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 9511169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Navarro, J. A., and V. Pallas. 1997. Evolutionary relationships in the ilarviruses: nucleotide sequence of prunus necrotic ringspot virus RNA 3. Arch. Virol. 142749-763. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez-Navarro, J. A., C. B. Reusken, J. F. Bol, and V. Pallas. 1997. Replication of alfalfa mosaic virus RNA 3 with movement and coat protein genes replaced by corresponding genes of Prunus necrotic ringspot ilarvirus. J. Gen. Virol. 783171-3176. [DOI] [PubMed] [Google Scholar]
- 33.Sauri, A., P. J. McCormick, A. E. Johnson, and I. Mingarro. 2007. Sec61alpha and TRAM are sequentially adjacent to a nascent viral membrane protein during its ER integration. J. Mol. Biol. 366366-374. [DOI] [PubMed] [Google Scholar]
- 34.Sauri, A., S. Saksena, J. Salgado, A. E. Johnson, and I. Mingarro. 2005. Double-spanning plant viral movement protein integration into the endoplasmic reticulum membrane is signal recognition particle-dependent, translocon-mediated, and concerted. J. Biol. Chem. 28025907-25912. [DOI] [PubMed] [Google Scholar]
- 35.Sauri, A., S. Tamborero, L. Martinez-Gil, A. E. Johnson, and I. Mingarro. 2009. Viral membrane protein topology is dictated by multiple determinants in its sequence. J. Mol. Biol. 387113-128. [DOI] [PubMed] [Google Scholar]
- 36.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 164049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano, A. G., E. J. Cabre, and J. Perez-Gil. 2007. Identification of a segment in the precursor of pulmonary surfactant protein SP-B, potentially involved in pH-dependent membrane assembly of the protein. Biochim. Biophys. Acta 17681059-1069. [DOI] [PubMed] [Google Scholar]
- 38.Serrano, A. G., A. Cruz, K. Rodriguez-Capote, F. Possmayer, and J. Perez-Gil. 2005. Intrinsic structural and functional determinants within the amino acid sequence of mature pulmonary surfactant protein SP-B. Biochemistry 44417-430. [DOI] [PubMed] [Google Scholar]
- 39.Serrano, A. G., M. Ryan, T. E. Weaver, and J. Perez-Gil. 2006. Critical structure-function determinants within the N-terminal region of pulmonary surfactant protein SP-B. Biophys. J. 90238-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taschner, P. E., A. C. van der Kuyl, L. Neeleman, and J. F. Bol. 1991. Replication of an incomplete alfalfa mosaic virus genome in plants transformed with viral replicase genes. Virology 181445-450. [DOI] [PubMed] [Google Scholar]
- 41.Vilar, M., A. Sauri, M. Monne, J. F. Marcos, G. von Heijne, E. Perez-Paya, and I. Mingarro. 2002. Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J. Biol. Chem. 27723447-23452. [DOI] [PubMed] [Google Scholar]
- 42.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wimley, W. C., and S. H. White. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3842-848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.