Abstract
In this study, we have identified an internal ribosome entry site (IRES) from the highly infectious herpesvirus Marek's disease virus (MDV). The IRES was mapped to the intercistronic region (ICR) of a bicistronic mRNA that we cloned from the MDV-transformed CD4+ T-cell line MSB-1. The transcript is a member of a family of mRNAs expressed as immediate-early genes with two open reading frames (ORF). The first ORF encodes a 14-kDa polypeptide with two N-terminal splice variants, whereas the second ORF is contained entirely within a single exon and encodes a 12-kDa protein also known as RLORF9. We have shown that the ICR that separates the two ORFs functions as an IRES that controls the translation of RLORF9 when cap-dependent translation is inhibited. Deletion analysis revealed that there are two potential IRES elements within the ICR. Reverse genetic experiments with the oncogenic strain of MDV type 1 indicated that deletion of IRES-controlled RLORF9 does not significantly affect viral replication or MDV-induced mortality.
Translation is a fundamental process for the expression of genetic material. Translation initiation of capped eukaryotic mRNA is postulated to involve scanning by the ribosome from the 5′ end to the initiator AUG codon (27). In contrast, studies on the translation of uncapped RNAs from picornaviruses revealed an alternative mode of 40S recruitment to the mRNA. This cap-independent mechanism is directed by an RNA sequence within the message known as an internal ribosome entry site (IRES) (24, 34). Subsequently, many IRESs were identified in both RNA and DNA viral genomes (7, 13, 14, 16, 23, 38, 41, 43). To date, at least 85 cellular IRESs have been described (5), although the experimental grounds on which proof of some of them rests have been the subject of dispute (6, 28).
It appears that although many RNA and DNA viruses use cap-dependent translation initiation, they have also evolved IRES-mediated translation. So what are the benefits of having two alternative mechanisms of translation initiation? There is ample evidence to suggest that IRES elements have important functions in the viral life cycle, mostly to ensure efficient viral translation when components of the host translation machinery are limited (37) due to virus-induced modification or host-induced antiviral response, such as phosphorylation of eIF2 (17). It must be remembered that viruses rarely create entirely unique biochemical mechanisms. Instead, they mimic or capitalize on existing cellular mechanisms. Thus, cellular and viral IRESs must have evolved to provide a physiological advantage. Indeed, many cellular IRESs have been shown to be active when cap-dependent protein synthesis is greatly reduced (for a review, see references 20 and 25). IRES-mediated translation is therefore viewed as a cellular backup plan for survival under various stress conditions. It is even argued that initiation of translation in early eukaryotes was IRES driven and cap independent and was then superseded by a cap-dependent mechanism (18).
Marek's disease (MD) is a commercially important, rapidly progressive lymphomatous disease of chickens caused by the highly infectious herpesvirus Marek's disease virus (MDV-1). MD is a valuable model for human lymphomas (11), including Hodgkin's and non-Hodgkin's lymphomas (8). In this study, an IRES element from MDV-1 was revealed. The IRES was mapped to the intercistronic region (ICR) of a bicistronic viral mRNA that we cloned from MSB-1, an MDV-transformed CD4+ T-cell line (1). Interestingly, a similar cDNA was previously identified in MDV-1-infected primary cells and was found to be a member of a family of RNAs expressed as immediate-early genes with two open reading frames (ORF) (21, 35). The first ORF encodes a 14-kDa polypeptide with two N-terminal splice variants (21, 22), whereas the second ORF is contained entirely within a single exon and encodes a 12-kDa protein also known as RLORF9 (9, 31). Using bicistronic reporter assays, we show that the ICR functions as an IRES that controls the translation of RLORF9, with no apparent promoter activity or cryptic splicing. Deletion analysis revealed two potential IRES elements within the ICR. Reverse genetic experiments with the oncogenic strain of MDV-1 indicated that the deletion of IRES-controlled RLORF9 did not significantly affect viral replication or MD-induced mortality.
MATERIALS AND METHODS
cDNA library from MSB-1 cells.
Total RNA was extracted from the MDV-transformed T-cell line MSB-1 by use of Trizol (Invitrogen). Poly(A)+ mRNA was purified using an LNA enhanced oligo-T20 capture probe (Exiqon). A specific subset of poly(A)+ mRNA was enriched according to a protocol that will be described elsewhere (A. Tahiri-Alaoui et al., unpublished results). The resulting mRNA was converted into cDNA and cloned into lambda ZAP-CMV-XR vector following the protocol provided by the manufacturer (Stratagene). Mass excision of an aliquot from the library was performed according to the manufacturer's instructions (Stratagene), followed by bacterial plating. Insert-positive clones were identified by PCR screening using T3 and T7 universal primers. Selected clones were sequenced in both directions using T3 and T7 primers and dye terminator cycle sequencing with a quick start kit from Beckman Coulter. The sequencing reaction mixtures were run on a CEQ8000 DNA sequencer (Beckman Coulter). The resulting sequences were subjected to a BLAST search to determine the identity of the cloned poly(A)+ RNAs.
Bicistronic vectors.
In order to assess IRES activity, we constructed a bicistronic vector that was based on the psiCHECK-2 vector (Promega). The psiCHECK-2 vector was digested with NotI and ApaI to remove the herpes simplex virus thymidine kinase promoter and the synthetic poly(A) RNA. This digestion removed the first 10 amino acid residues from the firefly ORF. These were restored by cloning two annealed oligonucleotides (oligo 1, 5′-GGCCGCGGACTAGTCATGGCCGATGCTAAGAACATTAAGAAGGGCC-3′; and oligo 2, 5′-GCGGACTAGTCATGGCCGATGCTAAGAACATTAAGAAG-3′) and at the same time allowed the addition of a SpeI site upstream of the firefly ORF. The resulting construct was named psiRF Vector. The ICR spanning the region from nucleotides 131117 to 131566 or the selected deletions (genomic coordinates of MDV-1 strain Md5 [GenBank accession number AF243438]) was PCR amplified using the viral cDNA and cloned into a bicistronic psi RF vector between PmeI and SpeI sites. The resulting construct was called psiRF-ICR. The simian virus 40 (SV40) promoter was removed from the psi-ICR construct by double digestion with BglII and NheI, blunt ended with T4 DNA, and religated. The promoterless construct was named psiRF-ICR/pLess. All clones were verified and confirmed by sequencing. For fluorescence microscopy studies, the pIRES2-AcGFP1 vector (Clontech) was modified to allow cloning of the ICR from MDV-1. The encephalomyocarditis virus (EMCV) IRES sequence was removed from pIRES2-AcGFP1 by restriction digestion with BstXI, blunt ended with T4 DNA, and then digested with BamHI before ligation of the PCR-amplified ICR with compatible ends. Red fluorescent protein (dsRed) was cloned upstream of the EMCV IRES or MDV-1 ICR as an XbaI-EcoRI fragment, and the resulting construct was designated pRG-ICR/EMCV or pRG-ICR/MDV-1, respectively.
Transient expression assays.
Primary chicken embryo fibroblast (CEF) cells were prepared from 10-day-old, specific-pathogen-free embryos (obtained from flocks maintained at the Institute for Animal Health, Compton, United Kingdom) and seeded the day before transfection at a density of 1.5 × 105 cells/well in a 24-well plate. The DF-1 cell line, derived from line zero CEF (19), was seeded as before, at a density of 0.5 × 106 cells/well. The human embryonic kidney (HEK) 293T cell line (ATCC) was seeded at a density of 1 × 105 cells/well the day before transfections. Transfection with bicistronic constructs for analysis of IRES activity in DF-1 and 293T cells was carried out using Lipofectamine 2000 (Invitrogen). Transfection of CEF was performed using Lipofectamine (Invitrogen) following the manufacturer's instructions. CEF and DF-1 cells were incubated at 38.5°C and 5% CO2. HEK 293T cells were incubated at 37°C and 5% CO2. Firefly and Renilla luciferase levels were measured at 24 h posttransfection, using a Dual-Glo luciferase assay system (Promega) and an Anthos Lucy1 microplate luminometer (Anthos Labtec Instruments, Austria).
RNA transfection of DF-1 cells was performed as described before (40). The cells were incubated for 6 h after transfection, harvested, and assayed for luciferase activity. Capped RNA used for transfection was produced by in vitro transcription of linearized plasmid as described below, using a T7 mMessage mMachine kit (Ambion). The RNA was also polyadenylated using a poly(A) tailing kit from Ambion following the manufacturer's protocol.
In vitro transcription and translation.
The psiRF-Vector constructs were linearized with BamHI. Capped transcripts were synthesized by using a T7 mMessage mMachine kit (Ambion), and uncapped transcripts were synthesized by using Megascript T7 (Ambion), both according to the manufacturer's instructions. These transcripts were translated using Retic Lysat IVT (Ambion) according to the manufacturer's instructions. We used a ratio of 2:3 (low-salt to high-salt mix) to obtain a 100 mM potassium acetate concentration in the translation mixes. Assays supplemented with m7GpppG were carried out as previously described (2). Firefly and Renilla luciferase activities were measured from in vitro translation reaction mixes by using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's instructions. Polypeptides from [35S]methionine-labeled translations were analyzed by using 4 to 15% NuPAGE Novex Bis-Tris gels (Invitrogen) followed by autoradiography.
shRNA.
Small interfering RNA (siRNA) sequences were predicted using a target-specific siRNA online design site (http://genomics.jp/sidirect). The siRNA sequence (5′-GTCGCTGTTTGCACATTATCA-3′) targeted the ICR within the MDV-1 genome at nucleotides 131150 to 131170 (GenBank accession no. AF243438). As a negative control, we used a siRNA that was designed against the EMCV IRES (5′-GTAACATGGCGTAGTAGAAAC-3′). All siRNAs were cloned as short hairpin RNAs (shRNAs) by a previously described method (29). All constructs were verified by sequencing.
Viral bacterial artificial chromosome (BAC) mutagenesis.
We used recombination-mediated genetic engineering (30, 42) to examine the requirement of the viral protein RLORF9 (sequence from nucleotides 131569 to 131890 [GenBank accession no. AF243438]) for in vitro replication and tumorigenesis of pRB-1B5 (36). This allowed us to generate a series of RLORF9 deletion mutants of infectious pRB-1B5. Briefly, to knock down the first copy of RLORF9, a kanamycin resistance gene (Kanr) cassette flanked by frt sites was amplified from the plasmid pKD13 (32), using the forward primer 5′-AGATGTTGTAGGGTTCGAGAGGGGTGAGACCTAAACATGCAGTCGCATGCCGTGTAGGCTGGAGCTGCTTC-3′ and the reverse primer 5′-CCGGTCATACATTCTATGTAAACAAGGAAGTTATCCCCTTTGCTTCCGTACATTCCGGGGATCCGTCGAC-3′). The underlined sequences are complementary to pKD13, whereas the nucleotides in bold are complementary to regions immediately flanking the RLORF9 exon. Agarose gel-purified PCR products were electroporated into Escherichia coli EL250 cells (kindly provided by N. Copeland, NCI, Frederick, MD), and recombinant chloramphenicol- and kanamycin-resistant colonies were selected. The Kanr cassette was subsequently excised by the induction of FLPe recombinase, using 0.2% arabinose. This resulted in a single-copy RLORF9 deletion in pRB-1B5, which was designated pRB-1B5-SCΔRLORF9. To knock out the second copy of RLORF9, we used the same strategy as that described above, but using a different reverse primer (5′-TTATCGATAATCGGCTCCGATCCCGATTCCGCGGACCAAGCATTGGCTGCCATTCCGGGGATCCGTCGAC-3′). The nucleotides in bold are complementary to a region within the RLORF9 exon (nucleotides 131844 to 131893 [GenBank accession no. AF243438]), and the underlined sequence is complementary to pKD13. The double-copy RLORF9 deletion of pRB-1B5 was designated pRB-1B5-ΔRLORF9. To make a single-copy RLORF9 revertant, we used the same strategy to put a single copy of the RLORF9 gene back into pRB-1B5-ΔRLORF9. First, we amplified a genomic fragment from pRB-1B5, using the forward primer 5′-CGCGGGGACGAGCAAAGCGTGCGGTGCGGGCAG-3′ and the reverse primer 5′-AGGGCATAGCGCCCGGCTCTGGCTCCTGAGAC-3′. This amplified a genomic region spanning nucleotides 127808 to 134901 (GenBank accession no. AF243438). The resulting PCR product was cloned into the pGEMT vector, resulting in pGEM-RLORF9. Second, a spectinomycin resistance (Specr) gene cassette flanked by frt sites was amplified from plasmid pL451 (32), using the forward primer 5′-ATCGATAAGCTTGATATCGAATTCCGAAGTTCC-3′ and the reverse primer 5′-TATTATGTACCTGACTGATGAAGTTCCTATAC-3′, and the resulting product was cloned into XbaI-digested pGEM-RLORF9. This resulted in the vector pGEM-RLORF9-Spec. This was used as a template to amplify the cassette that contained the RLORF9 gene and the Specr marker with the forward primer 5′-CGCGGGGACGAGCAAAGCGTGCGGTGCGGGCAG-3′ and the reverse primer 5′-TATTATGTACCTGACTGATGAAGTTCCTATAC-3′. Finally, the resulting PCR product was gel purified and electroporated into competent pRB-1B5-ΔRLORF9, and selection was carried out as described before to yield the single-copy RLORF9 revertant, designated pRB-1B5-SCrevRLORF9.
Growth curve and virus assay.
Each virus (pRB-1B5 wild type, pRB-1B5-ΔRLORF9 double-copy deletion, pRB-1B5-SCΔRLORF9 single-copy deletion, and pRB-1B5-SCrevRLORF9 single-copy revertant) was plated on CEF to study its in vitro growth characteristics. Briefly, 100 PFU was inoculated onto freshly seeded CEF (2 × 106 cells) in 60-mm-diameter dishes. After 0, 24, 48, 72, 96, and 120 h, the infected cultures were trypsinized, diluted, and seeded onto fresh CEF in triplicate. MDV plaques were counted after visualization by immunohistochemistry, using an MDV-specific monoclonal antibody directed against glycoprotein B (HB3) and a horseradish peroxidase-conjugated secondary antibody, followed by colorimetric detection.
Challenge experiment with mutagenized and control pRB-1B5.
We used 36-day-old, specific-pathogen-free P-line (B19/B19) chickens. For each group, 12 birds were infected via the intraperitoneal route with 1,000 PFU of the pRB-1B5 wild type, pRB-1B5-ΔRLORF9 double-copy deletion, or pRB-1B5-SCrevRLORF9 single-copy revertant. At 0, 4, 7, 14, 21, 28, and 35 days postinfection, 150 μl peripheral blood was taken from the wing vein into 3% sodium citrate. Peripheral blood lymphocytes (PBLs) were prepared by centrifugation over Histopaque as previously described (3). All experiments and sampling procedures were carried out according to the United Kingdom Home Office guidelines, including the culling of birds showing symptoms of MD.
DNA preparation and qPCR.
DNA from PBLs was prepared using a Qiagen DNeasy 96 blood and tissue kit following the manufacturer's instructions. Absolute quantitation of MDV-1 genomes was performed using quantitative real-time duplex PCR (qPCR), using an ABI7500 instrument, essentially as previously described (3, 4). Sequences of primers and probes for the reference gene (chicken ovotransferrin gene) and the MDV-1 meq gene have been reported (3). For each assay run, standard curves were prepared using 10-fold serial dilutions of DNAs from MDV-1-infected cells (for MDV-1 meq reaction) and noninfected cells (for ovotransferrin gene reaction), which had been calibrated accurately against plasmid constructs of known target gene copy number. Viral genomes were quantified per 104 PBLs.
RESULTS AND DISCUSSION
Identification of a bicistronic transcript incorporating p14 lytic protein and RLORF9.
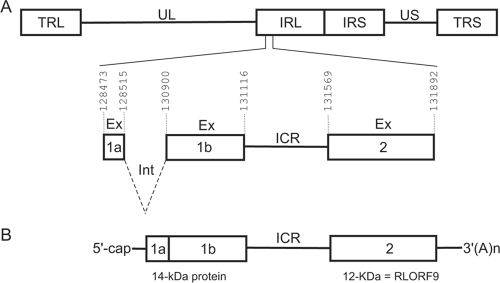
We have identified a viral transcript from MSB-1, an MDV-transformed CD4+ T-cell line derived from a spleen lymphoma (1) (Fig. 1). The cDNA was the product of an immediate-early gene and belonged to a family of transcripts known as 1.8-kb RNA (26). Previous studies showed that it resulted from alternative splicing and that it was expressed in chicken cells lytically infected with oncogenic and attenuated strains of MDV-1 (21, 26, 35). Analysis of the cDNA revealed that it was bicistronic. The transcript that we identified corresponded to the full-length cDNA (GenBank accession no. L26394) previously reported by Hong and Coussens (21). The availability of the complete genomic sequences of the attenuated (GenBank accession no. DQ530348) and oncogenic (GenBank accession no. AF243438) strains of MDV-1 allowed us to precisely map the boundaries of exons within the viral genome (Fig. 1A). The first ORF encoded a protein with a predicted molecular mass of 10.2 kDa, but when the protein was detected by Western blotting (21, 22), it had the apparent mass of the 14-kDa protein (p14). The p14 protein existed as two N-terminal splice variants, whereas the second ORF was contained entirely within a single exon and encoded a 12-kDa protein (RLORF9) (Fig. 1B). Protein products from both ORFs were also identified in the proteomes of MDV-transformed cell lines (9, 21, 31).
FIG. 1.
Genomic structure of MDV and positions of ORFs on the bicistronic transcript from the rightward transcriptional unit within the BamHI-H region. (A) Schematic representation of MDV genomic structure, consisting of unique long (UL) and unique short (US) regions, each bounded by a set of inverted repeats (TRL, IRL, IRS, and TRS). Intron (Int) and exon (Ex) sequences are shown, as well as the ICR between exon 1b and exon 2. (B) Schematic representation of the bicistronic transcript that we and others (21) cloned as cDNA. All genomic coordinates are according to the MDV-1 Md5 strain (GenBank accession number AF243438).
The ICR between the p14 lytic protein and RLORF9 coding regions has IRES activity.
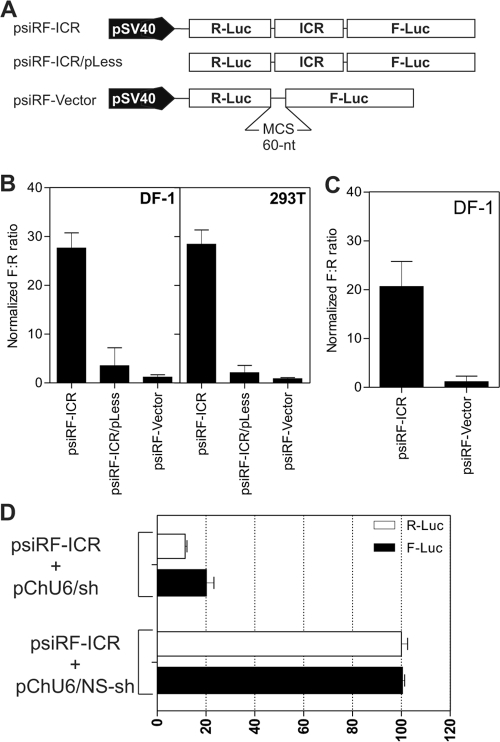
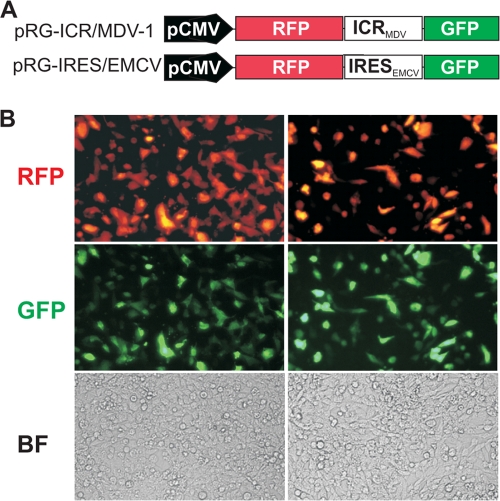
We have made the observation that the ICR of the naturally occurring bicistronic transcript has sequence and predicted structural features reminiscent of an IRES, such as predicted multiple stem-loop structures, multiple AUG codons, and a pyrimidine-rich tract (39). Indeed, using a genetic algorithm to predict the secondary structure (15) of various fragments from the ICR, we found several stable stem-loops that seemed to fold independently (data not shown). To test the ICR for IRES activity, we used the psiRF vector (Fig. 2A). This plasmid encodes a bicistronic mRNA with the Renilla luciferase gene as the first cistron, followed by a 60-nucleotide multicloning site (MCS) and the gene for the firefly (Photinus) luciferase as the second cistron. In this configuration, the transcription of both cistrons in the psiRF vector is driven by the SV40 promoter. The translation of the first cistron (R-Luc) is cap dependent, whereas translation of the second cistron (F-Luc) would require the presence of an IRES. We used the MCS spacer as a negative control. A PCR fragment from the ICR spanning the region from nucleotides 131117 to 131566 (genomic coordinates of MDV-1 strain Md5 [GenBank accession number AF243438]) was cloned into the MCS of the psiRF vector. It was essential to show that the ICR from MDV-1 has no inherent promoter activity that would result in transcription initiation at this internal site, resulting in a transcript containing only the second cistron (28). This would produce the F-Luc protein, resulting in false prediction of an IRES. The constructs shown in Fig. 2A were used to transfect avian and mammalian cells. After 24 h, the cells were harvested and assayed for luciferase activity. The ratio of the firefly luciferase activity to Renilla luciferase activity (F:R ratio) was calculated following a luciferase assay of the cell lysate. The ratio obtained for the negative control was set to 1, and the ratios from the other constructs were normalized to that value. The results show clearly that there was an approximately 30-fold increase in the F:R ratio for the psiRF-ICR construct compared with that for the negative control (Fig. 2B). A <10-fold increase in the F:R ratio was observed with the construct lacking the SV40 promoter, which may indicate the presence of very weak cryptic promoter activity within the MDV-1 ICR, spanning the region from nucleotides 131117 to 131566 (Fig. 2B), but there is the possibility that the weak cryptic promoter activity may originate from the backbone vector. To more conclusively demonstrate IRES activity within the ICR, we transfected DF-1 cells directly with RNA that was in vitro transcribed from DNA constructs. This approach should reveal IRES activity exhibited by mRNA without subjecting it to nuclear processing. Indeed, the F:R ratio was >20-fold higher than that for the control mRNA which lacked the MDV-1 ICR (Fig. 2C). These results indicate that even though there may be a weak cryptic promoter activity within the ICR, this does not preclude its ability to internally initiate translation. Similar observations have been made for other IRES sequences (10, 40). We also wanted to rule out other mechanisms inherent to the use of bicistronic vectors and that could generate monocistronic mRNA and therefore lead to false prediction of IRES activity. Such a mechanism could be due to splicing events. Accordingly, we designed shRNA that targets the MDV-1 ICR. We reasoned that equivalent knockdowns of both Renilla and firefly luciferase activities would happen only if the two cistrons are on the same bicistronic mRNA. We cotransfected DF-1 cells with a vector encoding MDV-1 ICR shRNA and with the bicistronic construct psiRF-ICR. As expected for an authentic bicistronic mRNA, the luciferase assay revealed equivalent knockdowns of about 80% for both Renilla and firefly luciferases (Fig. 2D). Additional experiments, including reverse transcription-PCR with primers corresponding to the 5′ end of the mRNA and to the coding region of the firefly luciferase gene, were performed and confirmed the presence of the full-length bicistronic transcripts, therefore ruling out the possibility of cryptic splicing (see Fig. S1 in the supplemental material). We also used a different bicistronic construct to demonstrate the IRES activity of the MDV-1 ICR. The construct was based on the pIRES2AcGFP backbone. The presence of two fluorescent proteins, green fluorescent protein (GFP) and red fluorescent protein (RFP), allowed us to use live-cell imaging and fluorescence microscopy to visualize translational activity. In this construct configuration, the GFP was controlled by either the EMCV IRES or the IRES from the MDV-1 ICR, whereas the RFP was under the control of cap-dependent translation (Fig. 3A). In both constructs, the transcription of the bicistronic mRNA was driven by a cytomegalovirus promoter. Transfection of DF-1 cells revealed that all cells that were positive for GFP were also positive for RFP (Fig. 3B). The ensemble of these results strongly indicated that the ICR from the MDV-1 immediate-early transcript has intrinsic IRES activity.
FIG. 2.
The ICR from the MDV-1 bicistronic transcript has IRES activity in a dual-luciferase reporter assay. (A) Bicistronic luciferase constructs used for transfection. The sequence (ICR) to be tested for IRES activity was inserted between the Renilla (R-Luc) and firefly (F-Luc) luciferase genes in the MCS spacer. In the psiRF-ICR/pLess construct, the SV40 promoter was removed. (B) Results of luciferase assay using DNA transfection of DF-1 and HEK 293T cells. The F:R ratio for each DNA construct was normalized to that obtained with the psiRF vector containing the MCS spacer as a negative control, whose F:R was set to 1. (C) Same as panel B, but the transfection was done with RNAs produced by in vitro transcription. (D) shRNA knockdown of the ICR. The data are results of cotransfection experiments with DF-1 cells. The firefly and Renilla luciferase activities were determined and expressed as percentages of the activity with control nonsilencing shRNA (pChU6/NS-shRNA). pChU6/sh refers to the plasmid encoding the siRNA that targets the ICR from nucleotides 131150 to 131170 (GenBank accession number AF243438).
FIG. 3.
The ICR in the MDV-1 bicistronic transcript has IRES activity by fluorescence microscopy. (A) List of bicistronic constructs used for transfection of DF-1 cells. The constructs are based on the pIRES2-AcGFP vector. The sequence to be tested for IRES activity (ICR) was cloned between the RFP and GFP genes. The EMCV IRES was used as a positive control. (B) Results of transfection with the pRG-ICR/MDV DNA construct (left) and the pRG-IRES/EMCV DNA construct (right). Live cells were examined under a fluorescence microscope with the appropriate filters to reveal fluorescent cells or under a bright field (BF) to visualize all cells.
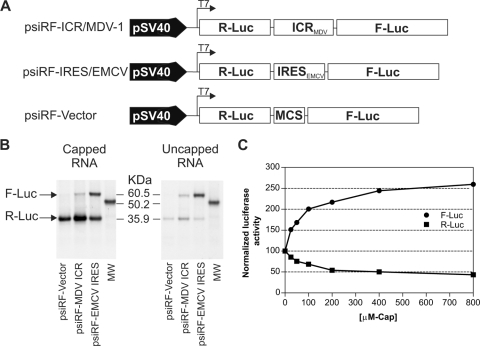
To further demonstrate that the ICR from MDV-1 has the ability to initiate cap-independent translation, in vitro-transcribed RNAs from the bicistronic constructs (Fig. 4A) were translated in rabbit reticulocyte lysate. In this study, we compared the efficiencies of translation of capped versus uncapped RNA transcripts with the ICR and used the EMCV IRES as a positive control and the multicloning spacer as a negative control. The [35S]methionine-labeled proteins resulting from in vitro translation were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and revealed by autoradiography (Fig. 4B). The results showed efficient translation of the Renilla luciferase cistron compared with the firefly luciferase cistron in all three capped transcripts. There was virtually no detection of the firefly luciferase cistron from the psiRF vector, which further demonstrated the requirement of an IRES for efficient translation of the second cistron. However, when uncapped transcripts were used for in vitro translation, the Renilla luciferase cistron was only weakly translated. In contrast, the firefly luciferase cistron, which was controlled by the ICR IRES from MDV-1 or the EMCV IRES, was still efficiently translated. It was also clear from these results that the EMCV IRES had strong activity compared with the ICR IRES from MDV-1. These results indicated that the ICR IRES had the ability to internally initiate translation in a cap-independent manner. To examine in more detail the competitive interaction between the cap-dependent and ICR IRES-driven translation, we carried out in vitro translation of the capped bicistronic ICR transcript in the presence of increasing concentrations of the free m7GpppG cap analogue. The rationale behind this was that the free m7GpppG cap analogue would bind to the cap-binding pocket of the eIF4E component of eIF4F (2). This binding would sequester eIF4E and prevent the competitive influence of translation of capped transcripts, which should result in redirection of all translational resources to the IRES. Indeed, as predicted, increasing the concentration of the free cap analogue in the translation reaction mix resulted in an increase in the activity of the ICR IRES and a concomitant decrease in the amount of cap-dependent translation (Fig. 4C). These results suggest that under conditions where cap-dependent translation might be inhibited due to virus-induced stress, the ICR IRES could provide a selective advantage for efficient translation of a subset of viral genes (37, 43).
FIG. 4.
The ICR IRES from the MDV-1 bicistronic transcript is capable of initiating translation when cap-dependent translation is inhibited. (A) List of DNA constructs used as templates to generate capped and uncapped RNA transcripts in vitro. (B) RNA transcripts were used for in vitro translation using rabbit reticulocyte lysate, and the resulting [35S]methionine-labeled proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and revealed by autoradiography. The positions of the firefly (F-Luc) and Renilla (R-Luc) proteins are indicated by arrows relative to the positions of the molecular size marker, obtained from the mRNA reference provide with a Retic-Lysat kit from Ambion. (C) Capped bicistronic RNA from the psiRF-ICR/MDV construct was translated in vitro in the presence of increasing amounts of free cap analogue. Firefly and Renilla luciferase activities were determined and expressed as percentages of the control reaction in the absence of cap analogue, whose value was set to 100%.
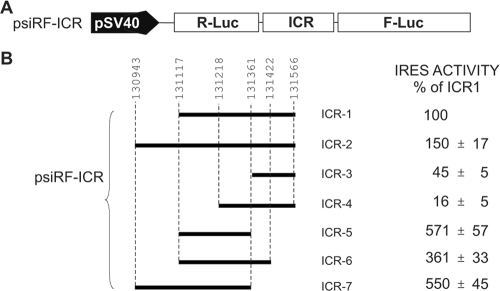
Deletion mapping suggests the presence of two IRES elements within the ICR.
We next used deletion analysis of the region spanning nucleotides 131117 to 131566 to better define the boundaries of the ICR IRES. These experiments were done using the bicistronic psiRF vector (Fig. 5). Deletion and extended fragments were generated by PCR, using MDV-1-specific primers that flanked the region of interest. The IRES activity of each insert is shown relative to the activity of the ICR IRES spanning the region from nucleotides 131117 to 131566, which was set at 100% (Fig. 5B). The 3′-end and 5′-end deletions were chosen based on the predicted secondary structure obtained with the genetic algorithm from the STAR program (15). Particularly, we noticed that the 3′ end of the ICR IRES (nucleotides 131361 to 131566), corresponding to ICR3, adopted an independent fold as a Y-shaped structure in isolation as well as within the full-length ICR (data not shown). The Y-shaped structure was consistently predicted using either of two independent RNA folding programs (15, 33). When ICR3 was tested for IRES activity, it was found to have about 45% of the activity of the full-length element. Interestingly, extending the 5′ end of ICR3, giving ICR4, caused a dramatic reduction in IRES activity, to the background level. Furthermore, there was a 50% increase in IRES activity when the 5′ end of ICR1 was extended to give ICR2, which included part of exon 1b (Fig. 1A). This suggested that part of the IRES activity was contributed from the first ORF. Surprisingly, the highest IRES activity was found with ICR5, which was the result of deleting ICR3 from ICR1 and extending the 5′ end of ICR1. The resulting ICR5 had IRES activity that was fivefold higher than that of ICR1. Extending the 3′ end of ICR5, which gave ICR6, resulted in a decrease of the IRES activity. Strikingly, when the domain containing ICR3 was removed from ICR2 to give ICR7, the IRES activity was restored to the level for ICR6. The results of the deletion analysis seemed to indicate that the ICR has a modular composition. It suggested the existence of at least two functional IRESs within the ICR. Whether these modular IRESs are all functional in cells remain to be determined. However, this observation is not unique for MDV-1 intercistronic IRESs. Other cellular and viral IRESs were reported to have multiple functional modules (for example, see references 40 and 43). We speculate that the presence of modular IRESs may play an important role in the translational control of RLORF9. This can be achieved through the existence of a dynamic equilibrium such that there is only one functional structure at any one time. Alternatively, regulation of the functional IRES module might be achieved by the presence of IRES trans-activating factors of viral or cellular origin. We have preliminary data showing that the activity of the ICR IRES is modulated by small noncoding RNAs of viral origin (unpublished data).
FIG. 5.
Deletion analysis revealed the existence of two potential IRESs within the ICR of the MDV-1 bicistronic transcript. (A) Bicistronic construct used to clone different fragments from the ICR. (B) Activities of all ICR fragments that were analyzed. The activities are shown as percentages of the full-length ICR1 activity.
The protein encoded by RLORF9 and controlled by IRES does not appear to be essential for tumor formation in MDV-infected chickens.
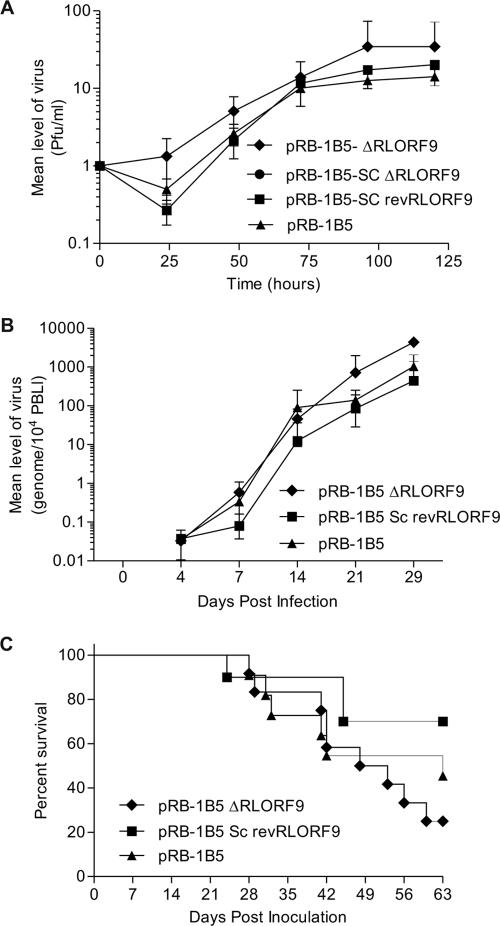
Nothing is known about the physiological function of the MDV-1 p12 protein encoded by RLORF9. The protein is highly conserved between different strains of MDV-1 and is expressed in both oncogenic and attenuated strains of the virus (8, 21). We have shown in the present study that the translation of RLORF9 is cap independent and IRES mediated. In order to gain insights into the physiological function of this viral protein, we took advantage of the availability of an infectious BAC clone of the highly oncogenic RB-1B strain (36) and used recombination-mediated genetic engineering (30, 42) to delete the IRES-controlled RLORF9. Various molecular tests were performed to assess the authenticity of the mutant and revertant viruses (see Fig. S2 and S3 in the supplemental material). We started by evaluating the ability of recombinant viruses to replicate and grow in CEF (Fig. 6A). Deletion of both copies of RLORF9 seemed to allow the virus to replicate and grow more rapidly in these cells (Fig. 6A). This apparent beneficial effect was more visible at the early stages of viral replication. There was no measurable difference in growth properties between wild-type pRB-1B5, the single-copy deletion mutant pRB-1B5-SCΔRLORF9, and the single-copy revertant pRB-1B5-SCrevRLORF9. These results indicate that RLORF9 is not essential for the in vitro growth of the virus. In fact, it appeared that the virus replicated slightly better in the absence of RLORF9. We then assessed the ability of the pRB-1B5 recombinants to replicate and induce disease in chickens. In the bird challenge experiment, we excluded the single-copy deletion mutant pRB-1B5-SCΔRLORF9, as this showed similar in vitro growth properties to the single-copy revertant pRB-1B5-SCrevRLORF9. We used qPCR on PBL DNA to estimate the viral copy number (3) and found that all three recombinants of MDV-1 replicated in the blood with comparable kinetics (Fig. 6B). These results indicated that the protein encoded by RLORF9 was not directly involved in viral replication. Postmortem analysis of inoculated chickens either at the time of showing symptoms of persistent neurological disease (12) or at the end of the trial revealed typical MD pathology, such as the presence of tumors in the spleen, kidney, and gonad. Overall, there was no significant difference in the tumor incidence between all three MDV recombinants (see Fig. S4 in the supplemental material), indicating that the RLORF9 protein was not directly involved in MDV pathogenesis, including tumor formation.
FIG. 6.
Challenge experiment revealing that the ICR IRES-controlled RLORF9 is not essential for viral replication or tumor formation. (A) Growth curves for wild-type pRB-1B5, the double-copy deletion mutant pRB-1B5-ΔRLORF9, the single-copy deletion mutant pRB-1B5-SCΔRLORF9, and the single-copy revertant pRB-1B5-SCrevRLORF9. Fresh CEF were infected with the indicated viruses. After 0, 24, 48, 72, 96, and 120 h, the infected cultures were trypsinized and plated on fresh CEF in triplicate. MDV-1 plaques were counted after visualization by immunohistochemistry. Each point represents the average for two plates. (B) Time course analysis of MDV-1 genome copy number in the blood of chickens infected with wild-type pRB-1B5, the double-copy deletion mutant pRB-1B5-ΔRLORF9, and the single-copy revertant pRB-1B5-SCrevRLORF9. The viral genome copy number was determined by qPCR, using DNAs extracted from PBLs and primers specific to the MDV-1 genome. (C) Survival curves for chickens inoculated with the indicated recombinant viruses. A log rank (Mantel-Cox) test revealed no statistical difference between pRB-1B5 and pRB-1B5-ΔRLORF9 (P = 0.4627) or between pRB-1B5 and pRB-1B5-SCrevRLORF9 (P = 0.2419).
In conclusion, we have identified an intercistronic IRES within a transcript that is known to be the product of an immediate-early gene from MDV. This is the first report of an IRES in MDV, which is a major avian oncogenic herpesvirus. We showed that the ICR IRES was active under conditions that inhibited cap-dependent translation. Furthermore, through deletion analysis, we revealed that the ICR IRES was modular, in that it seemed to harbor at least two functional domains. The presence of modular IRESs may play an important role in the translational control of RLORF9, as indicated by the competitive nature of the functional domains within the ICR. However, the physiological relevance of these functional domains remains to be determined. The IRES activity of the full-length ICR seemed to be moderate in comparison with that of the prototypical EMCV IRES. The moderate activity of the ICR IRES may reflect the need for limited expression of the protein encoded by RLORF9 and/or to keep it under tight control. Indeed, our preliminary data showed that constitutive expression of RLORF9 under the control of a strong cytomegalovirus promoter induced cell death. Nothing is known about the physiological role of RLORF9 in MDV pathogenesis. Our data from reverse genetic analysis showed that RLORF9 was not directly involved in viral replication and tumor formation. It would be interesting to evaluate the effect of overexpression of the protein encoded by RLORF9 on viral replication and MDV pathogenesis.
Supplementary Material
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC), United Kingdom.
Footnotes
Published ahead of print on 18 March 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akiyama, Y., and S. Kato. 1974. Two cell lines from lymphomas of Marek's disease. Biken J. 17105-116. [PubMed] [Google Scholar]
- 2.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 757854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baigent, S. J., L. J. Petherbridge, K. Howes, L. P. Smith, R. J. Currie, and V. K. Nair. 2005. Absolute quantitation of Marek's disease virus genome copy number in chicken feather and lymphocyte samples using real-time PCR. J. Virol. Methods 12353-64. [DOI] [PubMed] [Google Scholar]
- 4.Baigent, S. J., L. P. Smith, R. J. Currie, and V. K. Nair. 2005. Replication kinetics of Marek's disease vaccine virus in feathers and lymphoid tissues using PCR and virus isolation. J. Gen. Virol. 862989-2998. [DOI] [PubMed] [Google Scholar]
- 5.Baird, S. D., M. Turcotte, R. G. Korneluk, and M. Holcik. 2006. Searching for IRES. RNA 121755-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baranick, B. T., N. A. Lemp, J. Nagashima, K. Hiraoka, N. Kasahara, and C. R. Logg. 2008. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl. Acad. Sci. USA 1054733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 751864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess, S. C., J. R. Young, B. J. Baaten, L. Hunt, L. N. Ross, M. S. Parcells, P. M. Kumar, C. A. Tregaskes, L. F. Lee, and T. F. Davison. 2004. Marek's disease is a natural model for lymphomas overexpressing Hodgkin's disease antigen (CD30). Proc. Natl. Acad. Sci. USA 10113879-13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buza, J. J., and S. C. Burgess. 2007. Modeling the proteome of a Marek's disease transformed cell line: a natural animal model for CD30 overexpressing lymphomas. Proteomics 71316-1326. [DOI] [PubMed] [Google Scholar]
- 10.Dumas, E., C. Staedel, M. Colombat, S. Reigadas, S. Chabas, T. Astier-Gin, A. Cahour, S. Litvak, and M. Ventura. 2003. A promoter activity is present in the DNA sequence corresponding to the hepatitis C virus 5′ UTR. Nucleic Acids Res. 311275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein, M. A. 2001. Historical background. Philos. Trans. R. Soc. Lond. B 356413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gimeno, I. M., R. L. Witter, and W. M. Reed. 1999. Four distinct neurologic syndromes in Marek's disease: effect of viral strain and pathotype. Avian Dis. 43721-737. [PubMed] [Google Scholar]
- 13.Griffiths, A., and D. M. Coen. 2005. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 1029667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 751857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gultyaev, A. P., F. H. van Batenburg, and C. W. Pleij. 1995. The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol. 25037-51. [DOI] [PubMed] [Google Scholar]
- 16.Han, F., and X. Zhang. 2006. Internal initiation of mRNA translation in insect cell mediated by an internal ribosome entry site (IRES) from shrimp white spot syndrome virus (WSSV). Biochem. Biophys. Res. Commun. 344893-899. [DOI] [PubMed] [Google Scholar]
- 17.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 151593-1612. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez, G. 2008. Was the initiation of translation in early eukaryotes IRES-driven? Trends Biochem. Sci. 3358-64. [DOI] [PubMed] [Google Scholar]
- 19.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248295-304. [DOI] [PubMed] [Google Scholar]
- 20.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6318-327. [DOI] [PubMed] [Google Scholar]
- 21.Hong, Y., and P. M. Coussens. 1994. Identification of an immediate-early gene in the Marek's disease virus long internal repeat region which encodes a unique 14-kilodalton polypeptide. J. Virol. 683593-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong, Y., M. Frame, and P. M. Coussens. 1995. A 14-kDa immediate-early phosphoprotein is specifically expressed in cells infected with oncogenic Marek's disease virus strains and their attenuated derivatives. Virology 206695-700. [DOI] [PubMed] [Google Scholar]
- 23.Isaksson, A., M. Berggren, and A. Ricksten. 2003. Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene 22572-581. [DOI] [PubMed] [Google Scholar]
- 24.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 622636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komar, A. A., and M. Hatzoglou. 2005. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 28023425-23428. [DOI] [PubMed] [Google Scholar]
- 26.Kopacek, J., L. J. Ross, V. Zelnik, and J. Pastorek. 1993. The 132 bp repeats are present in RNA transcripts from 1.8 kb gene family of Marek disease virus-transformed cells. Acta Virol. 37191-195. [PubMed] [Google Scholar]
- 27.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak, M. 2005. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 336593-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambeth, L. S., Y. Zhao, L. P. Smith, L. Kgosana, and V. Nair. 2008. Targeting Marek's disease virus by RNA interference delivered from a herpesvirus vaccine. Vaccine 27298-306. [DOI] [PubMed] [Google Scholar]
- 30.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 31.Liu, H. C., E. J. Soderblom, and M. B. Goshe. 2006. A mass spectrometry-based proteomic approach to study Marek's disease virus gene expression. J. Virol. Methods 13566-75. [DOI] [PubMed] [Google Scholar]
- 32.Liu, P., N. A. Jenkins, and N. G. Copeland. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13476-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 34.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334320-325. [DOI] [PubMed] [Google Scholar]
- 35.Peng, F., G. Bradley, A. Tanaka, G. Lancz, and M. Nonoyama. 1992. Isolation and characterization of cDNAs from BamHI-H gene family RNAs associated with the tumorigenicity of Marek's disease virus. J. Virol. 667389-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petherbridge, L., A. C. Brown, S. J. Baigent, K. Howes, M. A. Sacco, N. Osterrieder, and V. K. Nair. 2004. Oncogenicity of virulent Marek's disease virus cloned as bacterial artificial chromosomes. J. Virol. 7813376-13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarnow, P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 772801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarnow, P., R. C. Cevallos, and E. Jan. 2005. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 331479-1482. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28130-136. [DOI] [PubMed] [Google Scholar]
- 40.Timmerman, S. L., J. S. Pfingsten, J. S. Kieft, and L. A. Krushel. 2008. The 5′ leader of the mRNA encoding the mouse neurotrophin receptor TrkB contains two internal ribosomal entry sites that are differentially regulated. PLoS ONE 3e3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 661476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 975978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, Y., and J. C. Alwine. 2006. 19S late mRNAs of simian virus 40 have an internal ribosome entry site upstream of the virion structural protein 3 coding sequence. J. Virol. 806553-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.