The editors of the Journal of Virology are in a privileged position to observe our field grow and develop. While the rational prediction of things to come is based on extrapolation of what we know now, we cannot anticipate the surprises that result from the serendipity of research. The discoveries emanating from virology in the past 50 years have been simply astounding, and few of them could have been predicted or even imagined based on prior knowledge. It is no accident that virologists have played major roles in the biological revolutions of the last century. Viral gene products engage all the key nodes of biology, ranging from the atomic to the organismal, and thus serve as ideal tools to dissect the most intricate life processes. Our joys and challenges are to identify and understand these biological nodes and extrapolate from this information how viruses replicate, disseminate, and sometimes cause disease. We can say with certainty that virology in the 21st century will continue to prosper. We discuss several general forces driving the future of our discipline: technology development, public health, information processing, and, of course, personal curiosity. Perhaps more importantly, to ensure maximal scientific return we must continue to give imagination and serendipity a chance.
A HISTORICAL PERSPECTIVE
Viruses and viral diseases have been at the centers of science, agriculture, and medicine for millennia, and some of our greatest challenges and triumphs have involved virology. Smallpox is a prime example: humankind's greatest killer, which literally changed the course of history during the European conquest of the New World, is also the only disease ever eradicated from the globe. This remarkable achievement began with Edward Jenner's scientific demonstration in 1796 that inoculation with cowpox lesions provided protection against the far-more-virulent variola major virus. A concerted worldwide vaccination effort against smallpox led by the World Health Organization resulted in the eradication of the disease by 1979. The smallpox vaccination breakthrough was only the first in a series of important investigations and discoveries inspired by the study of viruses. Tables 1 and 2 list many of these advances and highlight the contributions of virology and virologists to our understanding of basic cellular functions and disease mechanisms.
TABLE 1.
Eras in virology
| Era | Years | Descriptiona |
|---|---|---|
| Protovirology | 1796-1885 | Before viruses were recognized |
| Auroravirology (named for the Roman goddess of dawn) | 1892-1933 | Dawn of virology |
| Meridiovirology (from Latin for midday, sequel to dawn) | 1934-1955 | From the demonstration that bacteriophages are composed of protein and nucleic acid and the crystallization of TMV to the in vitro assembly of infectious TMV from purified RNA and protein |
| Janovirology (named for the Roman god of endings and beginnings) | 1956-1975 | Spans the interval between classic virology and the beginning of the era dominated by viral sequence information; encompasses the elucidation of essential features of gene structure, expression, and regulation and the development of essential techniques, including cloning and restriction sequence mapping |
| Neovirology | 1976-present | Begins with the first complete sequencing of viral genomes and atomic resolution structures of intact viruses |
TMV, tobacco mosaic virus.
TABLE 2.
Landmarks in the study of virusesa
| Era and year | Landmark (virus or scientist) |
|---|---|
| Protovirology | |
| 1798 | Cowpox lesions used to vaccinate against smallpox (Jenner) |
| 1882 | Transmission of tobacco mosaic disease with cell-free extracts (Mayer) |
| 1885 | Development of rabies vaccine (Pasteur, Roux) |
| Auroravirology | |
| 1892 | Description of filterable infectious agent (TMV) (Ivanovsky) |
| 1898 | Development of concept of virus as contagious element (TMV) (Beijerinck) |
| Discovery of first animal virus (FMDV) (Loeffler, Frosch) | |
| 1901 | Discovery of first human virus (yellow fever virus) (Reed) |
| 1903 | Discovery of rabies virus (Remlinger, Riffat-Bay) |
| 1908 | Discovery of first leukemia-causing virus (Ellerman, Bang) |
| 1909 | Discovery of poliovirus (Landsteiner, Popper) |
| 1911 | Discovery of first solid tumor virus (RSV) (Rous) |
| Discovery of measles virus (Goldberger, Anderson) | |
| 1913 | Virus cultivation in tissue culture (VV) (Steinhardt, Lambert) |
| 1915 | Discovery of bacterial viruses (bacteriophages) (Twort, d'Hérelle) |
| 1917 | Development of the plaque assay and discovery of the particulate nature of viruses (bacteriophage) (d'Hérelle) |
| 1931 | Propagation of virus in embryonated chicken eggs (Woodruff, Goodpasture) |
| 1932 | Discovery of first mammalian tumor virus (MMTV) (Little, Bittner) |
| 1933 | Discovery of human influenza virus (Smith) |
| Discovery of rabbit papillomavirus (Shope) | |
| First description of viral mutants (TMV) (Jensen) | |
| Meridiovirology | |
| 1934 | Discovery that bacteriophages are composed of protein and nucleic acids (Schlesinger) |
| 1935 | Crystallization of TMV (Stanley) |
| 1938 | Development of yellow fever vaccine (Theiler) |
| Use of electron microscopy for viruses (TMV) (von Borries, Ruska, Ruska) | |
| 1939 | Description of one-step growth cycle (bacteriophage) (Ellis, Delbrück) |
| 1941 | Discovery of first virus-associated enzymes (influenza virus) (Hirst) |
| 1943 | Discovery of genetic origins of mutations (bacteriophage) (Luria, Delbruck) |
| 1945 | Development of influenza vaccine (Francis) |
| 1946 | Discovery of genetic recombination by bacteriophage (Delbruck) |
| Replication of poliovirus in nonneuronal cell cultures (Enders, Weller, Robbins | |
| Discovery of eclipse phase of virus infection (bacteriophage) (Doermann) | |
| 1951 | Discovery of bacteriophage λ (E. Lederberg) |
| Discovery that lysogenic phages produce diphtheria toxin (Freeman) | |
| 1952 | Plaque assay of animal virus (poliovirus) (Dulbecco) |
| Discovery that viral genome is nucleic acid (Hershey, Chase) | |
| Transduction of genetic information by bacteriophage (Zinder, J. Lederberg) | |
| 1953 | Discovery of host-controlled restriction and modification (Luria, Bertani, Weigle) |
| 1954 | Development of polio vaccines (Salk, Sabin) |
| 1955 | Culture of human cells (HeLa) (Gey) |
| Optimization of cell growth medium (Eagle) | |
| Definition of a gene (cis-trans test) (bacteriophage) (Benzer) | |
| In vitro assembly of infectious virus (TMV) (Fraenkel-Conrat, Williams) | |
| Janovirology | |
| 1956 | Discovery of mRNA in bacteriophage infection (Volkin, Astrachan, Brenner, Jacob, Meselson) |
| Discovery that virus particles are composed of identical subunits (Watson, Crick) | |
| Discovery that RNA can carry genetic information (TMV) (Schramm, Fraenkel-Conrat, Williams) | |
| 1957 | Discovery of interferon (Isaacs, Lindemann) |
| Discovery of respiratory syncytial virus (Chanock) | |
| 1958 | Discovery of bacteriophage λ regulation paradigm (Pardee, Jacob, Monod, Lwoff) |
| 1960 | Discovery of SV40 (Sweet and Hilleman) |
| Demonstration of the triplet nature of the genetic code (bacteriophage) (Crick) | |
| Elucidation of nonsense codons (bacteriophage) (Campbell, Epstein, Bernstein) | |
| 1962 | Studies of virus structure (Klug, Caspar) |
| 1964 | Demonstration of the colinearity of gene with polypeptide chain (bacteriophage) (Brenner) |
| Discovery of first human tumor virus (EBV) (Epstein, Barr, Burkitt) | |
| 1965 | Autocatalytic in vitro synthesis of bacteriophage DNA (Spiegelman) |
| 1966 | Experimental transmission of spongiform encephalopathy to primates (kuru) (Gajdusek, Gibbs, Hadlow) |
| 1967 | Discovery of hepatitis B virus (Blumberg) |
| Isolation of bacteriophage λ repressor (Ptashne) | |
| Discovery of viroids (Diener) | |
| Discovery of first virion-associated polymerase (VV) (McAuslan, Kates) | |
| 1970 | Discovery of retroviral reverse transcriptase (Temin, Baltimore) |
| 1971 | Discovery of RNA polyadenylation (Darnell, Edmonds) |
| 1972 | Development of first recombinant DNA molecules (phage λ, SV40) (Berg) |
| Proposal that reassortment of influenza virus segments is the origin of pandemic strains (Webster, Laver) | |
| 1973 | Development of first restriction map (SV40) (Nathans) |
| Discovery of major histocompatibility locus restriction of viral antigen recognition (Doherty, Zinkernagel) | |
| Discovery of human rotavirus (R. Bishop) | |
| 1974 | Development of first transgenic mouse (SV40) (Mintz) |
| 1975 | Discovery of mRNA capping (Shatkin, Moss) |
| Neovirology | |
| 1976 | First RNA virus genome sequenced (bacteriophage MS2) (Fiers) |
| Demonstration that retroviral oncogenes are derived from cells (J. M. Bishop, Varmus) | |
| 1977 | First DNA virus genomes sequenced (ΦX174, SV40) (Sanger, Fiers, Weissman) |
| Discovery of RNA splicing (adenovirus) (Roberts, Sharp) | |
| Discovery of tumor suppressor p53 (SV40) (Levine, Crawford) | |
| Description of first virus crystal structure (TBSV) (Harrison) | |
| 1978 | Development of the first infectious molecular clone of an RNA virus (Qbeta, Weissmann) |
| 1979 | Declaration of smallpox eradication by World Health Organization |
| First in vitro replication of eukaryotic viral DNA (adenovirus, SV40) (Kelly, Hurwitz, Stillman) | |
| Development of first in vitro mRNA transcription system (adenovirus) (Roeder) | |
| Discovery of first highly active, template-specific, RNA-dependent RNA polymerase from a eukaryotic source (BMV) (Hall) | |
| Discovery of tyrosine kinases (Hunter, Erikson, Eckhart) | |
| 1980 | Discovery of first human retrovirus (HTLV-1) (Gallo) |
| 1981 | Development of first infectious molecular clones of an animal RNA virus (poliovirus) (Baltimore) |
| Discovery of transcriptional enhancers (Chambon, Khoury, Schaffner) | |
| Development of hepatitis B virus vaccine | |
| Identification of mammalian transcription factors (MMTV, SV40) (Yamamoto, Tjian) | |
| Discovery of insertional activation of cellular oncogenes by retroviruses (Hayward, Astrin) | |
| Identification of polyadenylation signal (Shenk) | |
| Discovery of the Cre/lox recombination system in phage P1 (Sternberg) | |
| 1982 | Development of antiviral and other drugs (Elion, Hitchings) |
| Definition of prions (Prusiner) | |
| 1983 | High-risk human papillomaviruses identified and linked to cervical cancer (zur Hausen) |
| Discovery of AIDS virus (HIV) (Montagnier, Barre-Sinoussi, Gallo) | |
| 1984 | Discovery of nuclear localization signals (Smith, Butel) |
| Production of first infectious, multicomponent virus from cloned DNA (BMV) (Ahlquist) | |
| 1986 | Development of first recombinant viral vaccine (hepatitis B virus) |
| Generation of transgenic virus-resistant plants (TMV) (Beachy) | |
| Discovery of hammerhead ribozymes (TRSV, ASV) (Bruening, Symons) | |
| 1988 | Discovery that DNA virus oncogene products bind cellular tumor suppressor proteins (adenovirus, SV40, HPV) (Harlow, Weinberg, Livingston, Howley) |
| Development of first ribozyme with engineered specificity (Haseloff) | |
| Discovery of internal ribosome entry sites (poliovirus) (Wimmer, Sonenberg) | |
| 1989 | Discovery of hepatitis C virus (Houghton) |
| 1990 | Development of first human gene therapy with a retrovirus vector (Anderson, Blaese) |
| 1991 | Discovery of viral antiapoptotic proteins (baculovirus) (Miller) |
| 1995 | Development of HAART treatment for AIDS |
| 1998 | Discovery of gene silencing by double-stranded RNA, an antiviral response (Fire, Mello) |
| Use of plant virion for synthesis of nanoparticles (Young) | |
| Discovery that plant viruses encode suppressors of RNAi (Vance, Baulcombe) | |
| 2001 | Discovery of the caveosome (SV40) (Helenius) |
| 2002 | Worldwide outbreak and containment of SARS |
| 2005 | Reconstruction and sequencing of the 1918 influenza virus genome (Palese, Garcia-Sastre, Tumpey, Taubenberger) |
| 2006 | Development of vaccine against human papillomavirus, the first vaccine designed to prevent human cancer |
Discoveries recognized by a Nobel prize are highlighted in bold. This list of landmarks in virology is not comprehensive. We apologize to the many colleagues whose important contributions could not be included because of space and time constraints. Abbreviations: ASV, avocado sunblotch viroid; BMV, brome mosaic virus; EBV, Epstein-Barr virus; HAART, highly active antiretroviral therapy; HPV, human papillomavirus; HTLV, human T-cell leukemia virus; MMTV, mouse mammary tumor virus; RNAi, RNA interference; RSV, Rous sarcoma virus; TBSV, tomato bushy stunt virus; TMV, tobacco mosaic virus; TRSV, tobacco ringspot virus; VV, vaccinia virus.
Much of the initial attention of virologists was focused on viruses as disease-causing agents, and great progress continues to be made in this area. Many acute viral infections are prevented or controlled in much of the world through vaccination and other public health measures. As a result, viral scourges such as measles, poliomyelitis, rabies, and yellow fever are now rare in the developed world. Numerous effective antiviral drugs are also in widespread use. We now recognize that a substantial fraction of the world cancer burden is caused by viral infections, most commonly hepatitis B virus and human papillomavirus infections, and both can be prevented by vaccination. All of these advances flowed from basic studies of viral replication, transmission, and pathogenesis. However, substantial challenges remain. New viruses periodically emerge and cause great personal and societal tragedy. AIDS, caused by human immunodeficiency virus type 1 (HIV-1), remains the defining epidemic of our time, the true cost of which cannot be calculated. Although the severe acute respiratory syndrome (SARS) epidemic was brief, dengue and West Nile viruses continue to smolder, and Chikungunya virus, monkeypox virus, and Ebola and other hemorrhagic fever viruses crouch in the darkness. H5N1 avian influenza virus continues to sporadically infect humans in Southeast Asia and elsewhere. The emergence of a new influenza pandemic or a viral bioterrorism attack could have catastrophic consequences on public health, commerce, and civic discourse.
Viruses also cause serious diseases in plants and livestock. The 2001 epidemic of foot-and-mouth disease in the United Kingdom devastated its beef industry. Plum pox virus, which has decimated stone fruit trees in Europe since the early 1900s, has now spread to the United States and Canada. Viruses have been implicated in a disease that is ravaging our honeybees, threatening natural pollination cycles and thus much of agriculture.
Beyond their medical and agricultural importance, viruses are great teachers, and their lessons are not restricted to viral diseases. Viral replication is strictly dependent on cell structure, metabolism, and biochemical machinery. As a consequence, viral gene products interact with crucial regulatory nodes that control cell function, a situation that facilitates the identification and characterization of these nodes and the networks they control. Indeed, the roster of important discoveries uncovered by studies of viral replication and transformation is long: the existence of mRNA and mRNA processing, including splicing, capping, and polyadenylation; transcriptional control elements and transcription factors; gene silencing mechanisms; cellular oncogenes and tumor suppressor proteins; and signal transduction pathways and tyrosine kinases, to name just a few. The structural biology revolution, the initially outlandish idea that life processes can be understood at the chemical and eventually at the atomic level, was championed by the crystallization of tobacco mosaic virus by Wendell Stanley in the 1930s. This line of inquiry has produced high-resolution images of the structures of viral proteins and virus particles themselves, the largest biological structures known at the atomic level. Molecular biology emerged from studies of bacterial viruses. Studies of “unconventional viruses” resulted in the discovery of viroids and prions and the concept of protein-folding diseases.
Viral genomes encode gene products that modulate host defenses, including the immune response, an elaborate system that evolved in large part to protect us against invading microorganisms like viruses. Ideally, pathogens are cleared by immune defenses with minimum damage to the host. However, in the process, the immune defenses themselves can also cause damage (immunopathology). Indeed, much of viral clinical disease is immunopathological in nature, as shown in infections ranging from the common cold to AIDS. Studies of the interactions between viruses and cells have revealed many aspects of immunity, including the elucidation of histocompatibility antigen function, intrinsic cell defense mechanisms such as apoptosis, interferons, and RNA interference, and sophisticated viral countermeasures to evade or antagonize host immune responses. In fact, this discipline has been coined “anti-immunology” by some to highlight the close evolutionary relationship between the vertebrate immune system and microbial pathogens.
Many technologies employed to study cellular genes were first developed and perfected by using smaller and more easily manipulated viral genomes, including restriction enzyme mapping, molecular cloning, and genome sequencing. Indeed, the field of genetic engineering and the biotechnology industry were incubated in virology laboratories. Viruses and viral gene products have also emerged as valuable tools to study many aspects of biology and, potentially, to treat disease. These tools include reverse transcriptase for the synthesis of cDNA, viral vectors for gene delivery and protein production, transgenic animal technology, vaccination, and oncolytic therapy, which attempts to harness the capacity of some viruses to specifically infect and kill cancer cells. Studies to determine whether this approach has efficacy in the treatment of human cancers are under way.
Critical knowledge may also come from unexpected sources. Simple, highly expressed plant viruses have been developed into model systems to identify host factors involved in viral replication, translation, and other processes fundamental to all viruses. Plant viruses are also excellent tools for biotechnology and nanotechnology. For the latter, virions provide natural reaction chambers for the precise synthesis of nanoparticles, as well as digital memory components when complexed with metals.
As briefly outlined in this section, virology played a major role in 20th-century biology. The numerous Nobel prizes awarded to virologists are one measure of the impact of virology (Table 2). In this essay, we highlight some of the areas where virology will continue to address substantial challenges and provide new and important insights.
TECHNOLOGY DEVELOPMENT
Until recently, the only microbiota that we could identify in a complex community (e.g., gut flora or seawater) were those we could cultivate. Research in the 21st century will allow the identification of new families of organisms (including viruses) by high-speed sequencing of RNA and DNA. For example, the technology of “deep sequencing” of mixed populations found in respiratory secretions and gastrointestinal contents is revealing novel virus families, both pathogenic and nonpathogenic. Indeed, new polyomaviruses, marine viruses, and bacteriophages have been identified by using sequence-based techniques coupled with genomic and metagenomic analyses. Strikingly, some of the viral proteins revealed by these studies show little genetic similarity to known viruses, suggesting the existence of a universe of novel viruses awaiting study.
As technical advances drive the discovery of viral pathogens, we will obtain a greater understanding of the pathogenesis of “orphan” infectious diseases, a wider appreciation of zoonotic cycles, and an increased understanding of how viral infections directly or indirectly cause or modulate chronic diseases (e.g., autoimmune syndromes, cancers, cardiovascular disease, and neurological illnesses). This new information will highlight the role of viruses in emerging infectious diseases, the interface between viral gene products and host defense mechanisms (cell autonomous defenses as well as innate and acquired immunity), and the forces that drive patterns of acute and persistent infections in plants and animals. We may find viruses that occupy niches equivalent to those of commensal bacteria. Genomic approaches are currently guiding these new discoveries. When deep sequencing of nucleic acids in a complex sample is merged with other powerful technologies, including mass spectrometry, proteomics, optical imaging, and high-throughput screening using small molecules and short hairpin RNAs (shRNAs), we can be sure that the discovery pipeline for novel viruses and their antagonists will be full indeed.
In a similar vein, high-throughput sequencing and gene-mapping techniques, the availability of the genome sequences of humans and other organisms, and proteomics and metabolomics will provide us with the ability to study host determinants of viral virulence in ways previously unimagined. The complete sequencing of mammalian genomes and the development of RNA interference technology make it practical to systematically test the role of every cellular gene in a virus infection. The use of such screens in studies of HIV, hepatitis C virus, and West Nile virus replication has identified hundreds of cellular genes required for infection. Similarly, whole-genome association studies and other sorts of genetic analyses will identify additional genes required for infection, pathogenesis, and transmission in various hosts. The identification of host genes and pathways that confer susceptibility or resistance to infection in the context of the whole organism will lead to novel antiviral therapies and improved viral prevention strategies.
Exciting discoveries notwithstanding, the identification of new viruses brings a serious challenge. Are these viruses true pathogens or could they actually have symbiotic relationships with their host organisms? For example, perhaps infection by these agents stimulates local and systemic immune responses that protect against or suppress responses that contribute to pathogenesis by more-virulent microbes. Dissecting these complicated microbial relationships will undoubtedly yield unanticipated insights about viruses and their hosts. This work will require careful epidemiological and clinical studies aided in part by advances in technology.
A systems approach to virology.
Instead of studying one gene or gene product at a time, examining large groups of genes or gene products allows the identification of fundamental biological networks. By networks, we mean complex and interconnected intracellular processes that control, for example, gene expression, organelle biogenesis, and metabolism, as well as networks of intercellular communication at the tissue, organ, and whole-organism level. The fundamental premise is that information flows through these networks and disease arises when these networks are perturbed, causing changes in network architecture and the dynamics of information flow. Future studies of viral pathogenesis may be seen in terms of specific viral signatures of network imbalance that do not affect just one pathway but alter the fundamental homeostatic balance of a cell, organism, or population. Interactions of viral gene products with these networks will likely differ in different cell types and tissues. Technological advances in cell and organ culture will allow the in vitro study of viral infections under conditions that more precisely mimic the in vivo environment. This effort will extend our understanding of the interplay of microbial communities and host cells within an entire organism. Once such an understanding is achieved, we may be able to better identify cellular genes associated with disease risk and therefore predict which human or animal hosts should be vaccinated or prophylaxed.
The technical advances in systems biology will open doors to “systems microbiology.” Viruses will be increasingly viewed not in isolation with their cellular or organismal hosts but in the real world of a microbial ecosystem where a single host is infected with a plethora of microbes, including many viruses. An understanding of the interactions between a host and several viral or other microbial agents that simultaneously or sequentially infect it is likely to be informative in many ways. For example, some viral infections are associated with atherosclerosis and obesity. However, it is not clear whether the associated viral infections are causal, serve as essential cofactors, or are completely irrelevant. Components of the host inflammatory response to an initial infection, namely cytokines, chemokines, and cells of the innate and adaptive immune systems, can regulate the outcome of infection by a second agent, not only by acting on the infected cell but also by influencing uninfected cells of the same organism. Such interactions can be synergistic or antagonistic. The degree of susceptibility and the resultant response of a virus-infected cell to a secondary infection can be modulated by many cellular factors, such as receptors, antiviral proteins, microRNAs, and protein-trafficking machinery. A systems microbiology approach will also allow us to understand immunopathological disease to a greater extent and thereby be in a position to minimize the untoward effects of an overexuberant immune response.
We see a robust future for the field of “interactomics,” which we define as the process by which viral gene products interact with cellular gene products to affect different phenotypes. This field has flourished in studies of the oncogenic activity of DNA tumor viruses, but similar interactions surely underlie much of the cellular response to viral infection. Research on virus-host interactions and network dynamics will produce new insights into why some viruses can occasionally enter into new host species to cause new and unexpected diseases. In fact, the study of how zoonotic viruses become human pathogens has already become a major focus of 21st century virology.
One of the tenets of systems biology is that networks process information; the output can vary depending on the action at key nodes of the network. Therefore, an important use of systems biology is not simply to collect reams of data but rather to perturb the networks and predict the changed outcomes. Viral infections provide the opportunity to take a system from state A (uninfected) to state B (infected) with synchrony and technical control. This approach to generate and test hypotheses will be a powerful tool to understand homeostatic control and viral pathogenesis. Just as studies of viral gene products and their interactions with specific cellular components have yielded many fundamental insights into individual cellular functions, virology has much to contribute to our understanding of more-complex interacting systems.
New and old: coupling new technology with established procedures.
With the advent of such exciting new experimental approaches, will the methods of traditional virology continue to be necessary? Will the plaque assay go the way of the Model T? Can we turn off our incubators and freeze down our cell lines? We think not. Experiments using cell culture, classical biochemistry, animal models, clinical trials, and population-based analyses will continue to be essential components of contemporary and future virology research. The new technologies will not displace their predecessors but join and complement them. For example, the characterization of new viruses discovered through deep sequencing will require cultured cells to investigate viral replication biology and host organisms to investigate viral pathogenesis and disease outcomes. The causal association of viruses with specific disease phenotypes will require experimental infections and intervention trials. Thus, the virology toolbox will be enhanced by technological innovations rather than replaced by them.
PUBLIC NEED: RESEARCH FUNDING, CRISES AND PUBLIC PERCEPTION, AND ADVOCACY
Support and advocacy for virology research.
Our nation's economic stability is tightly interwoven with its scientific progress. The highest purpose of science is the search for a greater understanding of the world around us. In virology, this purpose translates into advances in our understanding of basic biology and improvements in the health of the flora and fauna that inhabit our plant. We cannot shy away from the need to educate the public and our national leaders about the important contributions our field can make. Engaging our lay colleagues and political leaders at all levels about our research and its impact will be essential to secure sufficient funding to support our efforts.
One example is the halt in the last decade of the 20th century to the enormous progress being made in using RNA interference to protect crops against infection by highly destructive viruses. The withdrawal of genetic modification of crops based on unsubstantiated fears has left few alternatives to deal with potentially devastating losses of crops to viral disease. With the prospect of worldwide food shortages caused by climate change, decreased investment in genetic technology has serious implications for human health. These and similar concerns provide a rich opportunity for the scientific community to advance ideas in the public domain about new technological developments and evidence-based risk assessments.
Emerging infections.
We will continue to observe new or previously unrecognized infections in humans, animals, and plants. In humans, such infections will more often be zoonotic (i.e., the transmission of a virus from wild or domesticated animals to humans with attendant disease). As we discover new viruses and relate new and well-known viruses to specific diseases, we can anticipate public pressure to rapidly develop antivirals and vaccines. Depending on the sense of urgency, the tendency will be to shift resources to the “threat” of the day. For example, at the start of the 21st century, we witnessed several examples of emerging infections followed by exaggerated (but not necessarily unwarranted) public reactions. The global concern and almost immediate response of scientists and health officials to the SARS and West Nile virus epidemics are cases in point. Soon after these events, we saw the spread of Chikungunya virus to several countries where it was hitherto unknown. What will tomorrow bring? How will we deal with these new infections? Improved surveillance, more-rapid reagent sharing and information transfer, more-effective quarantine procedures, and various public health measures will undoubtedly contribute to controlling emerging diseases, but increasing attention and resources are likely to be devoted to maintaining, as well as expanding, the roster of antivirals and vaccines.
One view is that we should accelerate the development of new antiviral strategies to protect the public from these emerging infections. To be effective, antiviral drugs must be safe and potent and must be administered soon after infection. These requirements constitute substantial impediments to drug discovery, which has limited the number of antivirals in clinical use for acute infections relative to antibiotics. Nonetheless, numerous highly effective antiviral drugs are in widespread use, particularly those against HIV. Advances in genetics, biochemistry, structural biology, and computational biology provide a strong platform for the future development of additional antiviral drugs. Although we must certainly prepare for future threats, antiviral-drug development should not ignore viruses that currently account for a substantial burden of disease.
The need for vaccines.
Vaccines are among the most cost-effective means of preventing infectious disease morbidity and mortality. However, recent progress in vaccine development has been uneven. We saw the introduction of effective human papillomavirus and rotavirus vaccines yet witnessed many unsuccessful attempts to develop an HIV vaccine. Why is it that many of our most successful vaccines were introduced 20 to 50 years ago (e.g., vaccines for hepatitis B, influenza, measles, mumps, and rubella viruses)? In addition to HIV, many globally important viruses (e.g., dengue virus, hepatitis C virus, human cytomegalovirus, and respiratory syncytial virus) still lack vaccines. There are several challenges to the development of these vaccines. Critical knowledge gaps must be filled before a “product” can be developed, and funding decisions must be tailored to these needs. First, we must understand the basic biology of viral evolution and quasispecies. Second, we need to define what constitutes a protective immune response. Third, we have to acknowledge the economics of vaccine development and the risk to the private sector, recognizing that the necessity of immunizing a healthy naïve population to prevent a disease will be unacceptable if there are significant vaccine-associated adverse events.
The challenges for the development of new generations of vaccines are substantial at the levels of basic biology and efficacy. But even greater challenges arise in introducing vaccines to the public. Many believe that children are already “overvaccinated” in infancy. In addition, there is a growing public perception that vaccines actually cause disease (e.g., autism, attention deficit disorder, or multiple sclerosis) despite substantial evidence to the contrary. Few people are well versed in the analysis of large epidemiologic studies designed to identify low-frequency associations. Moreover, disproving cause and effect in the face of well-established unscientific beliefs is a difficult task. Successful vaccine efforts will require both sound science and forceful public advocacy.
Natural and unnatural events.
Concern exists that highly pathogenic viruses, either in their wild-type state or after genetic manipulation, will be used to perpetrate acts of terrorism. Presently, the means for wide dissemination of these infectious agents are not available. Arguably the most dangerous virus, variola major virus (which causes smallpox), is not accessible to those wishing to inflict harm, but there are worries that covert stocks of variola major virus may remain undeclared. There is also much discussion about whether it is possible to develop “designer viruses” containing virulence factors from more than a single source. At the present time, it is not possible to predict the outcome of such genetic engineering approaches. However, viruses have evolved so precisely that even subtle genetic changes usually result in attenuation. Therefore, the risks of viral bioterrorism are thought to be low, but even low-probability, high-impact events can be devastating. There is no question that virologists and the scientific community in general should be vigilant to the misuse of scientific information as the field advances.
Indeed, nature is likely to be a much more dangerous terrorist, acting through zoonoses. Just as the AIDS pandemic was initiated by the transmission of HIV from nonhuman primates to humans and SARS-coronavirus was transmitted to humans from bats and civet cats, other viruses are likely to make similar leaps. Influenza virus does so on a regular basis, with many virologists predicting that it is only a matter of time before the next highly virulent and transmissible strain catches humans without preexisting immunity, resulting in a new pandemic. In 2003, a shipment of rodents from West Africa to the United States caused an outbreak of monkeypox virus in the Midwestern United States, which occurred because some of the rodents were subclinically infected with monkeypox virus. This virus, in turn, infected North American prairie dogs (not previously known to be hosts), which then transmitted the infection to about 100 people. Fortunately, nobody died during this miniepidemic, but the incident demonstrates that pathogenic viruses can move rapidly and unexpectedly into new populations and that Mother Nature is the consummate “bioterrorist.”
Balancing risks of dangerous-pathogen research.
Heightened concerns about potential viral pandemics and bioterrorism have resulted in the construction of high-containment research facilities and increased scrutiny about the safety of research on pathogens designated by the CDC or USDA as potential biological weapons (i.e., “select agents”). This designation mandates strict regulatory oversight of research that is aimed primarily at reducing the risks of misuse of these pathogens. However, select-agent designations also dramatically increase costs and slow the pace of research, discouraging some scientists from pursuing these studies. For instance, if HIV and SARS-coronavirus had been designated select agents, then responses to these outbreaks might have been much slower, perhaps with catastrophic consequences. Thus, attempts to reduce the risks of performing or misusing infectious-disease research should be balanced by consideration of the risks of hindering the research required to protect society against important pathogens. Furthermore, select agents are often endemic in some areas of the world where they infect humans by natural exposure and could be obtained by unauthorized individuals without regulatory oversight. Accordingly, future regulatory decisions about the designation and control of “select agents” should be based as much as possible on scientific factors and realistic risk assessments.
High-level containment does provide appropriate protection for the most dangerous pathogens, with the resurrection of the H1N1 influenza virus that caused the 1918 pandemic a case in point. This virus was reconstructed in biosafety level 4 containment facilities by using viral RNA sequences obtained from human autopsy specimens. The rationale for, and the advisability of, reconstruction were questioned by some, but results gathered in subsequent studies substantially enhanced our understanding of influenza pathogenesis and left us better prepared to anticipate and combat the next influenza pandemic.
Political issues impacting virology: climate change as an example.
Among many issues on the political agenda, climate change captures considerable attention. Climate science seems an unlikely subject for virologists, but it may be prudent to think seriously about this topic. Many regard changes in weather patterns and sea level as the primary effects of global warming. However, the influence of climate on the ecology of microbial systems may be as large as, or even larger than, these meteorological effects. Microbial pathogens unexpectedly gain access to new hosts when natural cycles of host-pathogen relationships are interrupted or altered. Insect and rodent vectors, weather, floods, and social interactions are affected by climate and contribute to the highly interactive cycles of host-pathogen engagement. Despite more than 100 years of studying microbes, we have minimal knowledge of the natural microbial world and how microbial communities evolve. Only recently have we become aware that the oceans are teeming with bacteriophages that modulate the aquatic bacterial population, thereby affecting the ocean's chemistry, ecology, and overall well-being.
The effect of climate on microbial communities is not understood. Indeed, the molecular and cellular biology of microbial communities is only now being examined in a rigorous way. For example, new methods of sequencing demonstrate a remarkable diversity of microorganisms in every ecosystem examined. These findings emphasize the need to predict how climate affects biological systems. If the temperature rose 1°C on average, major biological communities would change dramatically as competition for resources removed the less fit and opened new niches for competitors. We need more research on the molecular and cellular biology of populations to understand and model the evolution of interacting communities in nature. Laboratory biology must be better integrated with field biology: e.g., the techniques of deep sequencing, proteomics, and metabolomics should be brought to the field. For entrepreneurs, it is certain that innovative and lucrative technologies will emerge.
TRAINING VIROLOGISTS FOR THE FUTURE
How can virologists take advantage of all of these new opportunities for scholarship and application? To address this question, we must consider what constitutes optimum training for virologists in the 21st century. Tomorrow's advances will require a combination of small, tightly focused groups and large, multidisciplinary teams. Traditional teams of basic scientists working with clinicians will be augmented by mathematicians, physicists, and population biologists, among others. Integrating new technology advances with classical epidemiology and clinical approaches will not be a simple pedagogical exercise. Training the next generation of scientists to be capable of undertaking this research will require more-diverse course offerings, enhanced training opportunities, especially involving interdisciplinary collaboration and computational approaches, and instruction in teamwork. To facilitate professional advancement, we will need to develop new strategies to recognize and reward individual contributions to group scientific efforts as team science becomes more and more prevalent.
Systems biology approaches, large-scale genetic screens, and the metagenomics of host and viral genes from related viruses present unique challenges for training. These approaches will produce enormous amounts of information that will challenge our capacity to integrate it into useful conceptual frameworks. Imaging technology will reveal the dynamics of biological interactions at every level yet produce huge sets of data that strain current systems for information storage and retrieval. Methods to search, screen, recover, and use this information will provide new avenues for discovery and require a new generation of virologists with special expertise in computational methods and information technology.
PERSONAL CURIOSITY
The power of a single mind to identify a problem and solve it is one of the greatest joys of humanity. No executive, laboratory head, business manager, or marketing director can dictate discovery. The very meaning of the word is that we did not know what we would find when we started looking. The process of discovery follows paths that often seem simple or elegant in retrospect but almost always reflect individual intellect, personal curiosity, and luck, which cannot be defined or packaged. Although the world of science is changing in many ways, the role of the individual scientist in discovery cannot be underestimated. Another look at Table 2 emphasizes the importance of serendipity as an essential force in discovery. Someone has to see something no one else did. Many of the investigators who will make discoveries in the 21st century will do so because they want to understand how something works, not because something is trendy.
One of the motivating forces for virology research is the concept of the “model system.” Even with the current unprecedented rate of discovery, we are unlikely to know everything about every organism. A model system is a convenient organism that can be analyzed in great depth so that the knowledge obtained can be applied to other organisms that may not be amenable to study. What will constitute model systems for the 21st century? Model systems are often chosen to serve particular problems. For example, very few virologists study viruses of algae. Who knows, if we start driving cars fueled by genetically altered algae, it might be good to know more about algal viruses.
Continuing research should not only focus on conventional viruses but also enhance our understanding of new classes of subviral infectious agents such as prions. Prions are infectious misfolded host-derived proteins that can spread disease or phenotypic traits without carrying their own nucleic acid genome. A variety of prions have been identified in species ranging from fungi to mammals, but much remains to be discovered about the diversity, structure, and impact of these enigmatic agents in biology. Practical diagnostic tests and treatments must be developed for mammalian prion diseases such as bovine spongiform encephalopathy and Creutzfeldt-Jacob disease. An additional concern is the theoretical possibility that a number of common protein-misfolding diseases such as Alzheimer's disease and other amyloidoses might be transmissible under some circumstances, due to the prion-like behavior of misfolded proteins. The extent to which such possibilities are biologically relevant will be an important area for future investigation.
THE CRYSTAL BALL
These are exciting times for biology. The general trends likely to drive virology research in the next decade include systems biology of virus-host interactions, viral ecology and the virosphere, evolution of viruses, and improved vaccines and therapeutics. Many of these advances will be accelerated by technologic innovations in high-throughput sequencing, the complete synthesis of viral genomes, small-molecule and shRNA screens, and the imaging of cells and whole organisms, in concert with traditional methods used by virologists. The knowledge, techniques, new ideas, and urgency to learn more are stronger than ever. The importance of studying the basic biology of viruses, even those that today may not seem relevant to human, animal, and plant disease, cannot be overstated. As an example, studies of avian Rous sarcoma virus led to the discovery of cellular oncogenes and guided the initial studies of HIV. Nonpathogenic viruses have been widely utilized as gene transfer and vaccine vectors. Even replication-defective endogenous viruses have been informative, as they have provided a glimpse into virus-host battles fought by our distant ancestors. History has proven again and again that understanding the basic biology of viruses leads to new and often unexpected insights. We anticipate that studies of viruses will continue to yield surprising glimpses into the inner workings of their host cells. In fact, recent research on simian virus 40 (SV40) entry led to the discovery of an entirely new organelle, the caveosome.
We anticipate a rich future for viral pathogenesis research. Just as studies of viral infections of single cells have led to astonishing insights into basic biological processes, we think studies of viral infections of host organisms will continue to teach us much about physiology in health and disease. The application of new technologies will allow us to approach with increasing sophistication complex questions about how viruses invade, disseminate, target specific tissues, elicit host defenses, and cause disease. These questions cannot be addressed solely using cell culture but in addition require animal models, clinical trials, and population-based studies. In this context, we will likely explore the concept that viruses or viral components can be harnessed to protect us from other microbial or inflammatory diseases. In fact, individual “anti-host defense” proteins derived from viral genomes are currently being tested in clinical trials as novel classes of drugs to treat diseases associated with systemic inflammation, such as atherosclerosis and heart disease.
If we could point to one sea change in virology that will affect us all, it would be that we now function in a cross-disciplinary environment. Just look at the author list or acknowledgments in the latest papers and note the number of disciplines that are represented. It is likely that you will see not only authors with expertise in cell biology, immunology, and chemistry but also authors who are experts in ecology, computer and information science, mathematics, and physics, among others. Interdisciplinary research in virology is essential for future progress. However, “Biology” with a capital “B” is at the root of all this excitement. Those in other disciplines who do not master the biology of viruses are likely to provide technical expertise, but they will not share with virologists the joys of understanding fundamental biology, making discoveries, and improving the health and well-being of our planet.
Figure 1.
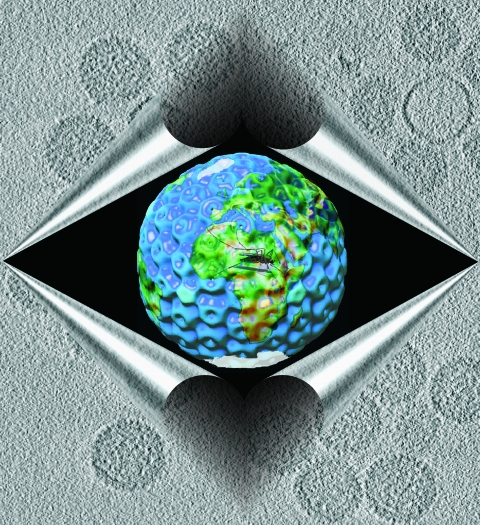
Viruses pose important global public health challenges. Concern is heightened by population growth and environmental changes, which might facilitate the transmission of animal viruses into humans. This trend may be accelerated by global warming. This figure shows the three-dimensional organization of Rift Valley fever virus (RVFV) revealed by cryoelectron tomography. RVFV (Bunyaviridae, Phlebovirus) is an emerging human and veterinary pathogen responsible for recurring epidemics throughout Africa and the Arabian Peninsula. RVFV has the potential to cause hemorrhagic fever in humans. Tomographic reconstruction of RVFV vaccine strain MP-12 revealed a capsid containing 122 capsomeres arranged in an icosahedral lattice with T=12 quasisymmetry. The virus particle is enwrapped with a map of the earth looking down at the African continent, and the mosquito represents the vector for RVFV. Frozen-hydrated RVFV MP-12 particles are shown in the foreground. (This figure first appeared on the cover of the Journal of Virology, November 2008, vol. 82, no. 21. [See related article on p. 10341.])
Figure 2.
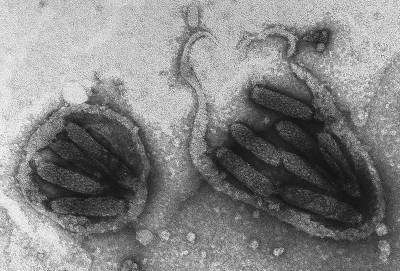
Viruses come in all shapes and sizes. Unbiased sequencing efforts have revealed an astonishing diversity of viruses. Shown here is an image of negatively stained nucleocapsids of a polydnavirus from the ichneumonid parasitoid Glypta fumiferanae. Mature virions consist of several nucleocapsids surrounded by two envelopes; the latter cannot be distinguished here because of the detergent treatment used to expose the nucleocapsids. Each nucleocapsid is believed to package more than one and possible many double-stranded circular DNA molecules, but it remains unclear whether each nucleocapsid, or virion, contains the full spectrum of more than 100 genome segments. (Micrograph by Don Stolz.) (This figure first appeared on the cover of the Journal of Virology, January 2008, vol. 82, no. 2. [See related article in June 2007, vol. 81, no. 12, p. 6491.])
Figure 3.
Whole viral genome sequences have revolutionized our ability to identify and characterize viral genes and have revealed evolutionary relationships between viruses. Nudiviruses are proposed to be a new genus of viruses isolated from different orders of insects, e.g., Orthoptera, Coleoptera, and Lepidoptera. The complete genome of a nudivirus infecting the cricket Gryllus bimaculatus suggests a common ancestor of nudiviruses and baculoviruses. Despite their differing morphology, these viruses share similar genes involved in virus structure, the infection process, and gene transcription. (The genome map was drawn using Genevision software; the contribution of G. Rossen, C. Bauser, and T. Bopp to the artwork is acknowledged.) (This figure first appeared on the cover of the Journal of Virology, December 2007, vol. 81, no. 23. [See related article in May 2007, vol. 81, no. 10, p. 5395.])
Figure 4.
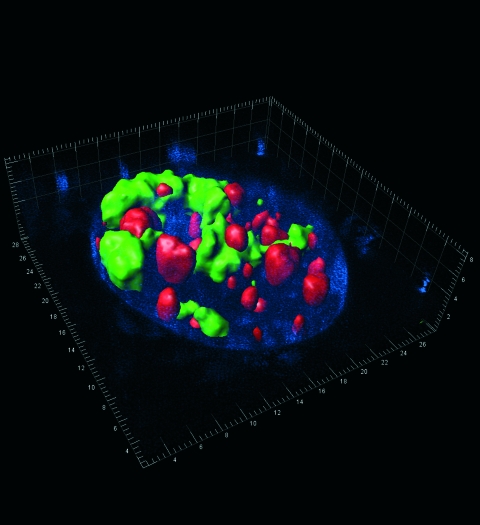
Numerous technical advances, including the ability to label and visualize viral genes and gene products, combined with sophisticated imaging techniques, have yielded unprecedented insights into the details of viral replication, including the impact of coinfection with more than one virus. Studies of interactions between viruses in coinfected hosts are likely to uncover important new strategies for viral commensalism and parasitism. Live covisualization of competing adeno-associated virus (AAV) and herpes simplex virus type 1 (HSV-1) DNA replication. Replicating AAV DNA containing lac operator sequences was visualized by binding of a red fluorescent protein fused to lac repressor protein (red), while the replication of HSV-1 DNA containing tetracycline operator sequences was visualized by binding of enhanced yellow fluorescent protein fused to the tetracycline repressor DNA binding domain (green). AAV and HSV-1 DNA replication occurred in spatially separate nuclear compartments, which were often found in juxtaposition. Blue, Hoechst stain; scale in micrometers. (This figure first appeared on the cover of the Journal of Virology, May 2007, vol. 81, no. 9. [See related article on p. 4732.])
Figure 5.
Viral-sequence information has revolutionized viral epidemiology, allowing the spread of an epidemic to be tracked and its evolution through space and time to be monitored. The transmission history of foot-and-mouth disease virus (FMDV) can be elucidated from complete genome sequence analysis of the virus, enabling epidemiological tracing of the virus between infected premises. The United Kingdom map shows the locations of premises infected during the 2001 FMDV outbreak. The structure is a representation of FMDV British field strain (serotype O), showing the alpha-carbon backbone (accession IFOD), courtesy of Nick Knowles, Institute for Animal Health, Pirbright, United Kingdom. (This figure first appeared on the cover of the Journal of Virology, January 2007, vol. 81, no. 1. [See related article in November 2006, vol. 80, no. 22, p. 11274.])
Figure 6.
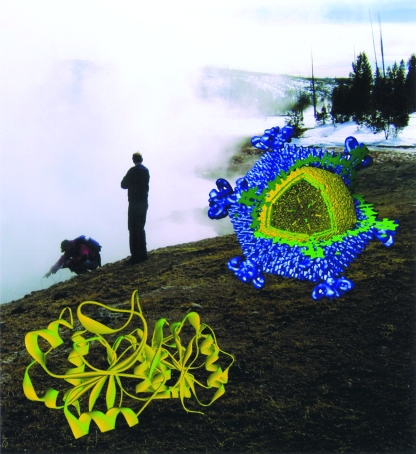
Viruses inhabit all ecological niches. The isolation and characterization of viruses from unconventional habitats are providing new views of virus diversity, evolution, and function. Wherever life is found, so are viruses. The hot and acidic waters of hot springs, such as those in Yellowstone National Park, are no exception. Species of the archaeal organism Sulfolobus thrive at high temperatures and low pH and are host to a number of virus strains, including the double-stranded DNA virus Sulfolobus turreted icosahedral virus (STIV). The characterization of STIV particles and virus-encoded proteins is leading to a better understanding of the origin and evolution of this group of viruses. (This figure first appeared on the cover of the Journal of Virology, August 2006, vol. 80, no. 16. [See related article in August 2006, vol. 80, no. 15, p. 7625.])
Figure 7.
Since the discovery of the first virus, tobacco mosaic virus, more than 100 years ago, the study of plant viruses has provided fundamental insights into numerous aspects of biology, including biochemistry, structural biology, genetics, and, as illustrated here, evolutionary biology. Shown here is a schematic diagram of the distribution of virus diversity in a single plant host, with different colors of branches and leaves illustrating the diversity of haplotypes isolated from different locations on a single, chronically infected host tree. The results demonstrate that several distinct subpopulations of Plum pox virus differentiate and evolve independently in different locations of a single tree. Closely related colors represent closely related haplotypes. (Photo provided by Michel Yvon, Chiraz Jridi, and Stéphane Blanc.) (This figure first appeared on the cover of the Journal of Virology, June 2006, vol. 80, no. 12. [See related article in March 2006, vol. 80, no. 5, p. 2349.])
Footnotes
Published ahead of print on 18 March 2009.