Abstract
The mammalian reovirus (MRV) genome comprises 10 double-stranded RNA (dsRNA) segments, packaged along with transcriptase complexes inside each core particle. Effects of four small molecules on transcription by MRV cores were studied for this report, chosen for their known capacities to alter RNA duplex stability. Spermidine and spermine, which enhance duplex stability, inhibited transcription, whereas dimethyl sulfoxide and trimethylglycine, which attenuate duplex stability, stimulated transcription. Different mechanisms were identified for inhibition or activation by these molecules. With spermidine, one round of transcription occurred normally, but subsequent rounds were inhibited. Thus, inhibition occurred at the transition between the end of elongation in one round and initiation in the next round of transcription. Dimethyl sulfoxide or trimethylglycine, on the other hand, had no effect on transcription by a constitutively active fraction of cores in each preparation but activated transcription in another fraction that was otherwise silent for the production of elongated transcripts. Activation of this other fraction occurred at the transition between transcript initiation and elongation, i.e., at promoter escape. These results suggest that the relative stability of RNA duplexes is most important for certain steps in the particle-associated transcription cycles of dsRNA viruses and that small molecules are useful tools for probing these and probably other steps.
Mammalian reovirus (MRV) is the prototype of genus Orthoreovirus in family Reoviridae, a diverse family of multisegmented double-stranded RNA (dsRNA) viruses to which important pathogens of humans and other vertebrates (e.g., rotavirus and bluetongue virus) also belong. The MRV genome comprises 10 distinct, linear segments of dsRNA, each ranging in size from ∼1,200 to ∼3,900 bp (48). Within infectious virions, these segments are enclosed by two icosahedrally symmetric layers of proteins: the inner, or core, capsid, and the outer capsid (11). During cell entry, the outer capsid is largely shed, and the remaining core particle (∼52 MDa, including the genome) enters the cytoplasm (25). There, it commences to use the 10 genome segments as templates for transcribing the 10 viral mRNAs, each of which is a full-length copy of the respective genomic plus strand. Each core particle is believed to contain 10 to 12 copies of the 142-kDa viral RNA-dependent RNA polymerase (RdRp), λ3, which are anchored to the inner capsid near the 12 icosahedral fivefold axes (10, 40, 53). These RdRp molecules support simultaneous synthesis and release of the 10 mRNAs, while both genomic strands are retained in the core interior (diameter, ∼50 nm) (2, 30, 42). Moreover, the core is organized in such a way that transcription is reiterative, allowing initiation, promoter escape, elongation, and termination through successive rounds from each of the 10 templates. Lastly, as they exit the core, the MRV mRNAs are 5′-capped by other virally encoded, core-associated enzymes (17, 40).
The strategy of keeping dsRNA templates inside the core interior throughout the MRV transcription cycle has clear advantages for the virus, such as protecting the dsRNAs from cellular innate-immunity factors, but entails substantial drawbacks as well. Forced to occupy limited space, the template RNAs must overcome steric and energetic problems during transcription that would not arise to the same degree were the templates free in solution. For example, since the dsRNA helices packed in the core interior are separated by distances of only 26 to 28 Å on average (11, 19, 32, 40), one can expect that during transcription these template RNAs frequently bump into each other and also the capsid walls. This increases the effective viscosity inside the core, which in turn results in additional energy costs that MRV must pay for transcription.
In addition to the preceding phenomena, specific details of MRV transcription suggested by structural studies of its RdRp (10, 45, 53) imply other complications. From those studies, especially the λ3 crystal structure (45), the current model suggests that several different large- and small-scale movements and topological changes of dsRNA must occur both during and between each round of transcription from each template. (i) Unwinding is one of these requisite actions. The template-entry channel leading to the central, catalytic cavity of λ3 is large enough to accommodate only one RNA strand. Thus, each duplex template must be progressively unwound in order to pass only the minus strand through this channel in a 3′-to-5′ direction, while the plus strand is passed around the outside surface of λ3 in a 5′-to-3′ direction. (ii) Rewinding is another of these necessary dsRNA movements and topological changes of dsRNA. As each successive portion of the minus strand is threaded past and then beyond the catalytic site, it must next be passed out through the template-exit channel of λ3 so that it can rewind with the complementary portion of plus strand to regenerate the genomic duplex. (iii) Looping also must take place. Because the capped 5′ end of the template plus strand is thought to be continually held by the cap-binding site on the surface of λ3 near the template-entry channel, as rewinding of the duplex proceeds, the rewound regions are thought to form an expanding loop, bending away from the template-exit channel. (iv) Finally, repositioning for initiation must occur as well. At the end of each transcription cycle, the 3′ end of template minus strand must be reinserted into the template-entry channel, which likely involves some of the initial steps in unwinding described above. Since each of these steps entails unwinding, rewinding, bending, and/or other translocations of both genomic strands, all within the crowded core interior, they suggest that MRV transcription may be especially sensitive to the physical state of the genome (e.g., to duplex stability and packing density). In addition, recent studies suggest that the overall structural organization of the MRV RdRp is not unique, but rather typical of several others, including rotavirus and influenza virus (20, 31), and thus these other viruses may have similar problems with template movements and reorganizations during transcription.
Given the preceding evidence and suggestions, we undertook studies to investigate the effects of RNA duplex stability on MRV transcription in vitro. We chose the following four small molecules to promote RNA conformational changes: the polyamines spermidine and spermine to stabilize RNA duplexes and dimethyl sulfoxide (DMSO) and the prototypical betaine trimethylglycine (TMG) to destabilize them. Polyamines are components of every living cell (44), with the capacity both to stabilize nucleic acid duplexes and to compact them (7, 22, 47). The choice of DMSO and TMG for the present study was dictated by their well-known destabilizing properties for nucleic-acid duplexes in vitro (14, 21, 23, 39). In addition, both DMSO and TMG are widely used in vitro: DMSO is an essential component of reaction medium for T4 RNA ligase (13) and sometimes for PCR (4, 37), whereas TMG is a key component of PCR enhancer mix (4, 21, 37). These examples also indicate the low toxicity of DMSO and TMG for components of in vitro systems at the range of concentrations used in the present study.
The results detailed below indicate that these two pairs of small molecules affect MRV transcription in opposite directions and through different mechanisms of action. Duplex-stabilizing agents spermidine and spermine inhibited MRV transcription, largely at the transition between the end of elongation in one cycle and initiation in the next cycle. On the other hand, the duplex-destabilizing agents DMSO and TMG stimulated MRV transcription, largely at the promoter-escape step, by activating transcriptase complexes that were otherwise silent for the production of elongated transcripts. Studies with small molecules thus appear useful for probing the transcription mechanisms of MRV and other dsRNA viruses.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Murine L929 cells were maintained in Joklik's modified Eagle minimal essential medium (Irvine) supplemented with 2% fetal and 2% calf bovine sera (HyClone), 2 mM l-glutamine (Mediatech), and 100 U of penicillin and 100 μg of streptomycin (Irvine)/ml. Stocks of MRV strains type 1 Lang (T1L), type 2 Jones, and type 3 Dearing were derived from ones obtained from the late B. N. Fields (Harvard Medical School). Double-layered particles of bovine rotavirus strain UK were obtained from A. R. Bellamy (University of Auckland) by way of S. D. Trask, S. T. Aoki, and S. C. Harrison (Harvard Medical School). Viruses were amplified in L929 cells and purified by CsCl gradient centrifugation (8). Cores were isolated by protease digestion of purified MRV virions, followed by CsCl gradient centrifugation (16). Purified virions and cores were stored at 4°C in virion buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris [pH 7.5]). MRV virion and core concentrations were determined from the A260 as in previous studies (15). Spermidine, spermine, and DMSO were purchased from Sigma. TMG was obtained from E. V. Makeyev (Harvard University).
Transcription reactions. (i) Standard reactions.
Transcriptions were carried out by incubation of ∼1010 MRV cores at 45°C in 10 μl of transcription buffer (100 mM HEPES-KOH [pH 8.1], 10 mM MgCl2, 0.5 mM EDTA) that also contained 4 mM GTP and 1 mM each of ATP, CTP, and UTP (all nucleoside triphosphates [NTPs] were obtained from GE Healthcare). In experiments where [α-32P]CTP was present, the concentration of nonradiolabeled CTP was lowered to 0.2 mM. When indicated, spermidine, spermine, DMSO, or TMG was added to the reactions. Regular concentrations of the additions included the following: polyamines, 2 to 10 mM; DMSO, 6%; and TMG, 1 M. For analysis of incorporated radiolabel by liquid scintillation counting, 10- to 20-μl aliquots of transcription reactions were loaded onto a 2-by-2-cm pieces of blotting paper (VWR) impregnated with 10% trichloroacetic acid, washed three times with 5% trichloroacetic acid for 5 min, washed once with acetone, dried, and counted. For electrophoretic analysis, unless indicated otherwise, 10-μl samples were mixed with 10 μl of loading buffer (8 M urea, 50 mM EDTA [pH 8.0], bromophenol blue), heated at 100°C for 4 min, and loaded onto a 1% agarose gel. For the experiment shown in Fig. 4C, an acidic (pH 3) urea agarose gel was used (29). After electrophoresis, gels were soaked in 30% methanol and 10% acetic acid for 30 min, washed with methanol, dried, exposed to a phosphorimaging screen, and visualized with a Typhoon 9400 phosphorimager (Amersham). RNA bands were quantitated using ImageQuant (Molecular Dynamics).
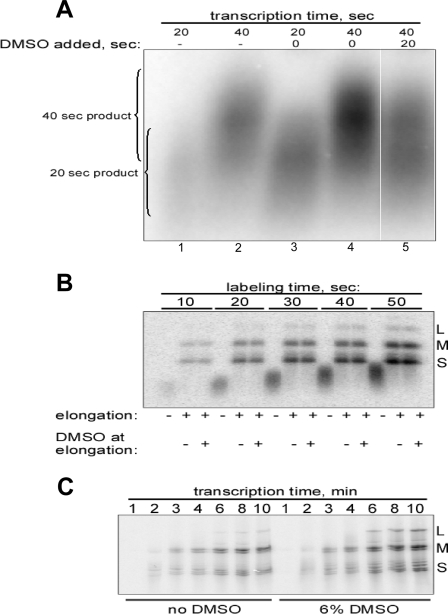
FIG. 4.
DMSO activates promoter escape in D-fraction cores. All experiments were performed with cores of MRV strain T1L. When present, DMSO concentration was 6%. Representative results are shown from a total of two or three experiments in each case. (A) Does DMSO activate elongation? Cores were subjected to very short (20- or 40-s) transcription reactions in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of DMSO. In another sample (lane 5), cores were transcribed first for 20 s in the absence of DMSO and then for another 20 s in the presence of DMSO. Transcripts were analyzed by agarose gel electrophoresis. In other gels in which DNA markers were included, the 20-s products concentrated in the 300- to 500-bp range, whereas the 40-s products concentrated in the 600- to 900-bp range. (B) Can D-fraction cores complete elongation in the absence of DMSO? Cores were allowed to transcribe for very short periods (10 to 50 s) in the presence of [α-32P]CTP and DMSO. The reactions were then stopped, and the media were exchanged for a fresh transcription mix containing no radiolabeled NTP. The reactions were next each divided into thirds, one of which was immediately prepared for electrophoresis (no elongation) and the other two of which were allowed to transcribe (elongate) for 15 min either in the absence or in the presence of DMSO as indicated. Transcripts were analyzed by agarose gel electrophoresis. (C) Do C and D fractions have similar elongation rates? Cores were allowed to transcribe for 1 to 10 min as indicated, in the absence (left, C fraction) or presence (right, C plus D fractions) of DMSO. Transcripts were analyzed by acidic urea agarose gel electrophoresis in an effort to differentiate full-length transcripts within each size class (L, M, or S).
(ii) Two-step reactions with media change.
To change the reaction medium during transcription, reactions were stopped by mixing sample with an equal volume of ice-cold transcription buffer containing 50 mM EDTA. After microfuge centrifugation at 13,000 rpm for 30 min at 4°C, the supernatant was removed, and the pellet was washed with 50 to 100 μl of ice-cold buffer (100 mM HEPES-KOH [pH 8.1], 40 mM NaCl). To ensure complete removal of radiolabel and other components of the original transcription mix, the centrifugation and washing steps were repeated. Cores were then resuspended in fresh transcription mix as indicated for each experiment, and transcription was allowed to continue at 45°C. The reproducibility of core recovery after these steps was indicated by the consistency of transcription product yields between individually pelleted samples in multiple experiments (see, for example, Fig. 2D).
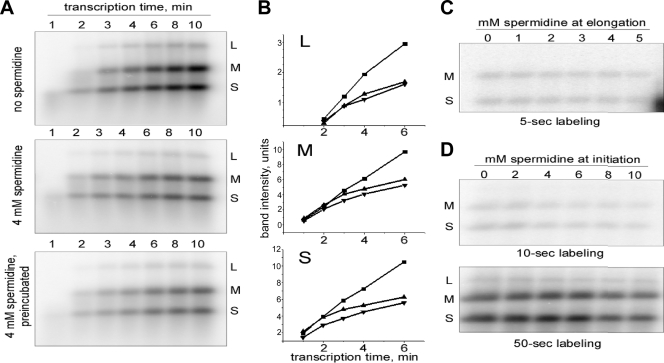
FIG. 2.
Spermidine inhibits not the first, but subsequent rounds of MRV transcription. Radiolabeled transcripts were generated by cores of MRV strain T1L at 45°C and analyzed by agarose gel electrophoresis. Representative results are shown from a total of three or four experiments in each case. (A) Time course of transcription in the absence (top) or presence (middle, bottom) of 4 mM spermidine. For the middle panel, NTPs and spermidine were added simultaneously. For the bottom panel, cores were preincubated with spermidine for 5 min at 45°C before transcription was started by the addition of NTPs. (B) Quantitative display of 1- to 6-min data from panel A. RNA bands were quantified by phosphorimaging. Each plot represents the behavior of a specific size-class of transcripts (L, M, or S as labeled in panel A, top). Symbols: ▪, data for no spermidine (A, top); ▴, data for 4 mM spermidine (A, middle); ▾, 4 mM spermidine with 5-min preincubation (A, bottom). As reflected by the unit values, the scale of the y axis has been adjusted to normalize the curves in the three graphs. (C) Effect of spermidine on elongation during the first round of transcription. Cores were first allowed to transcribe for 5 s in the presence of [α-32P]CTP in the absence of spermidine. The reaction was then stopped and, after the reaction medium was changed, the samples were allowed to elongate with nonradiolabeled NTPs for 15 min in the presence of indicated concentrations of spermidine. (D) Effect of spermidine on promoter escape during the first round of transcription. This experiment is similar to the experiment in panel C, but a labeling reaction was done for 10 s (top) or 50 s (bottom) in the presence of the indicated concentrations of spermidine. There was no spermidine present during the completion of elongation.
Abortive transcripts.
Analysis of abortive RNA synthesis was done as described previously (15) with minor modifications. Cores were subjected to 1-h transcription at 45°C in the absence or presence of 6% DMSO or 10 mM spermidine. The reaction was then stopped by the addition of EDTA to 10 mM, followed by 2 min at 100°C. After the samples cooled to room temperature, EDTA was titrated by the addition of MgCl2 to 12.5 mM. Calf intestinal phosphatase (1 U/10 μl) was then added, and the samples were incubated for 30 min at 37°C. Products were analyzed by electrophoresis on a 10 or 20% sequencing polyacrylamide gel and visualized by phosphorimaging. On a 10% gel, the abortive transcript GC migrated closely below the 20-bp DNA marker, while GCU migrated about halfway between the 10-bp DNA marker and free NTPs; on a 20% gel, the abortive transcript GC migrated closely below the 10-bp DNA marker, while GCU migrated closely above free NTPs (data not shown).
Heat inactivation.
Cores of MRV strain T1L were diluted in transcription buffer and incubated at 65°C for 1 to 20 min. After chilling on ice, 1 μl of a mixture of radiolabeled and nonradiolabeled NTPs as described above was added to 10 μl of heated sample to allow transcription, and incubation was continued at 45°C for 1 h. Products were analyzed as described in Results.
Isolation of C-fraction cores.
Approximately 1011 core particles of MRV strain T1L were transcribed for 2 min at 45°C in 100-μl transcription reactions containing 100 μM UTP in the absence or presence of 2 mM 5-bromouridine 5′-triphosphate (BrUTP) (Sigma). The reactions were then stopped and unincorporated BrUTP was washed out as described above for changing the reaction medium. Cores were resuspended in 400 μl of transcription buffer. In parallel, 15 μl of magnetic beads coated with protein G (Dynabeads Protein G, Invitrogen) were washed two times with 1 ml of buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 10% glycerol) and resuspended in 300 μl of binding buffer (50 HEPES-KOH [pH 8.1], 100 mM NaCl, 0.5 mM EDTA). Then, 5-μl portions of anti-bromodeoxyuridine (anti-BrdU) monoclonal antibodies (Sigma) were added to the beads, followed by 1-h of incubation at room temperature. Unbound antibodies were removed by two washes with binding buffer, and the beads were then resuspended in 300 μl of the same buffer. Next, 400 μl of core suspension and 300 μl of beads/antibody conjugates were mixed together, followed by incubation at 4°C overnight with shaking. After two washes and resuspension in transcription buffer, a mixture of nonradiolabeled NTPs as described above was added, and the samples (final volume, 50 μl) were incubated for 1 h at 45°C to release cores from the beads. The supernatant was collected and used directly for analysis.
RESULTS
Agents that alter RNA duplex stability affect MRV transcription.
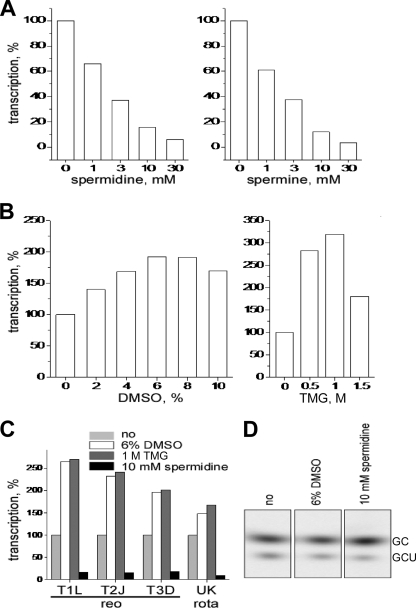
Polyamines spermidine and spermine, known to stabilize and compact RNA duplexes, substantially decreased transcription yields from MRV cores. The reduction was concentration dependent, with 50% inhibition occurring near 2 mM polyamine (Fig. 1A). In contrast, DMSO and TMG, known to destabilize RNA duplexes, substantially increased transcription yields from MRV cores. Peak levels of increase approximated two- to threefold and occurred at a concentration of 6 to 8% for DMSO and near 1 M for TMG (Fig. 1B). Transcription yields from cores of different MRV strains were similarly affected by each respective agent (Fig. 1C). Moreover, transcription yields from rotavirus double-layered particles (46) were also similarly affected (Fig. 1C). Although some virus-specific differences in the extent of activation by DMSO or TMG may be evident in Fig. 1C, other findings suggest that these largely reflect preparation-to-preparation rather than strain-to-strain differences. For example, MRV cores of the same strain but prepared on different days showed somewhat different levels of activation by DMSO (data not shown). The maximum activation observed with any preparation of MRV cores was close to 4.5-fold (strain type 2 Jones, with 1 M TMG). MRV virions and infectious subvirion particles, which are inactive for production of elongated transcripts (15, 49), were also tested for activation by DMSO and TMG. No activation was seen (data not shown), suggesting that the activation observed with cores is not due to completion of uncoating in a partially uncoated subset of the core preparation and also that the inactivation of promoter clearance provided by the MRV outer capsid (15) is not overridden by the DMSO or TMG effect.
FIG. 1.
Reagents known to stabilize or destabilize RNA duplexes affect MRV transcription. The results are for 1-h transcription reactions with cores of MRV strain type 2 Jones, or other particles as indicated, in the presence of the indicated concentrations of inhibitor or activator. Representative results are shown from a total of two to five experiments in each case. (A to C) Samples were analyzed by liquid scintillation counting of NTP-derived radiolabel incorporated into acid-insoluble material (transcripts longer than ∼50 nucleotides). Transcription yields are presented as a percentage of the yields with 0 mM concentrations of each examined reagent: spermidine (A and C), spermine (A), DMSO (B and C), or TMG (B and C) as indicated. (D) Samples were analyzed on a 10% sequencing polyacrylamide gel to detect abortive transcripts. The identities of the predominant abortives as labeled, GC and GCU, were determined in previous studies (15, 49) and confirmed in the present study by synthesis in the presence of NTP subsets (data not shown). The gel positions of the abortives relative to the DNA markers are described in Materials and Methods.
Spermidine or DMSO does not affect synthesis of abortive transcripts by MRV cores.
The current model for MRV transcription suggests that in cores before the onset of transcription, each genomic template resides in a “preinitiation” complex, with a 3′-terminal portion of minus strand unwound from the complementary 5′-terminal portion of plus strand and already threaded through the template-entry channel into the RdRp active site, ready to begin transcription as soon as nucleotides are provided (45, 53). In the absence of further conformational changes in the transcriptase complex, the products of initiation are short, abortive transcripts, mostly representing the first two to four bases [5′-GC(U)(A)] encoded by the conserved 3′-terminal bases in each template minus strand (5, 15, 49, 52). Occasionally, however, promoter escape occurs, and elongation proceeds. We therefore investigated whether any of the preceding small molecules affect the synthesis of abortive transcripts by MRV cores. In fact, effects of each of the four molecules on synthesis of abortive transcripts were limited or absent (Fig. 1D): the same abortive transcripts (predominantly GC and GCU) were synthesized to similar levels even when these molecules were present at concentrations that substantially affected synthesis of longer transcripts (compare with the results in Fig. 1A and B). These findings suggest that binding of the template RNAs to the RdRps in preinitiation complexes in MRV cores is minimally affected by these agents and is thus stronger than any effects of these agents on RNA duplex stability. The findings also suggest that the step(s) in transcription affected by each of these small molecules is subsequent to initiation events yielding abortive transcripts by newly transcribing cores.
Polyamines are known not only to stabilize RNA duplexes but also to compact them, shrinking their occupied volumes (7, 47). We thus recognized the other possibility, albeit unlikely, that the MRV dsRNAs might be compacted by polyamines into too small a volume inside the core, separating them from the RdRps and thereby reducing the number of initiation events, as well as transcription in general. The results in Fig. 1D also argue against this mechanism of polyamine action in showing that initiation events yielding abortive transcripts are little affected by spermidine.
Spermidine inhibits not the first, but subsequent rounds of MRV transcription. (i) Multicycle experiments.
We next performed further experiments in an effort to pinpoint the step(s) in transcription by MRV cores at which spermidine has its effect(s). To address whether spermidine action may vary from one transcription cycle to another, we analyzed RNA synthesized by cores at different time points over the first 10 min of transcription. Given that the elongation rate of MRV cores is reported as 7 to 12 nucleotides/s (2, 3, 42), one would expect to see the products of several rounds of transcription during a 10-min reaction, especially from the small (S) genome segments. Comparison of such early transcription kinetics by cores in the absence (Fig. 2A, top) or presence (Fig. 2A, middle) of 4 mM spermidine revealed that spermidine inhibition is clearly seen at later time points (e.g., at 8 and 10 min) but not at earlier time points (e.g., at 2 min).
One trivial explanation for the greater effect of spermidine at later times would be that it requires more time to act, for example, due to slow penetration into MRV cores. In the preceding experiment, all components except ATP and UTP were mixed at 45°C, and the reaction was then quickly started by addition of the missing NTPs. Since spermidine was added just prior to starting the reaction, therefore, it may indeed not have had enough time to permeate the cores. To rule out this possibility, we performed another experiment in which cores were preincubated with spermidine for 5 min before starting the reaction. This time of preincubation was chosen because spermidine inhibition was already evident at the 5-min time point in the preceding experiment (see Fig. 2A, middle). The results presented in Fig. 2A (bottom) are essentially identical to those obtained with no preincubation (Fig. 2A, middle), indicating that slow action by spermidine is unlikely to explain its greater effect at later time points.
Plots of quantitative data from these experiments (Fig. 2B) clearly demonstrate the biphasic nature of the spermidine effect. For the first few minutes (depending on the template size, around 2 min for the S and medium [M] segments and around 3 min for the large [L] segments), the plot lines are parallel for samples in the absence or presence of spermidine but later diverge. Accumulations of each size of transcripts in the no-spermidine samples continue at about the same rate over the full time course, while those in the spermidine-containing samples substantially slow at later times. Also, again, the samples with spermidine behave almost identically with or without preincubation.
(ii) Single-cycle experiments.
The 2- to 3-min delay in spermidine action observed in the preceding experiments might be attributed to only one transcription cycle. In other words, the delayed effect of spermidine might be explained by spermidine inhibiting not the first but subsequent rounds of transcription. The transcription cycle in other well-studied systems, most of which involve a double-stranded DNA template and a DNA-dependent RNA polymerase, is generally divided into four main phases: initiation, promoter escape, elongation, and termination (43). In MRV cores, the additional step of repositioning the 3′ region of template minus strand, after termination and before forming the initiation complex for the next round of transcription (45, 53), seems important to consider, too, as a step that might be especially sensitive to RNA duplex stability and to inhibition by small molecules that affect RNA structure.
To test whether particular steps in the first round of MRV transcription are affected by spermidine, we devised and performed single-cycle experiments to follow either elongation alone (Fig. 2C), initiation and promoter escape (Fig. 2D, top), or all three steps together (Fig. 2D, bottom). In these experiments, cores were first allowed to begin transcribing in the presence of [α-32P]CTP for 5, 10, or 50 s (labeling reaction). This reaction was then stopped by adding EDTA. Our preliminary experiments (data not shown) demonstrated that at this point the transcriptase complexes are frozen in the elongation state and upon restoration of an appropriate reaction medium are able to continue elongation of the uncompleted transcripts. After stopping the labeling reaction, the medium was changed by centrifuging, washing, and resuspending cores in full transcription mix containing no labeled NTP, followed by incubation for 15 min at 45°C to allow the transcriptases to complete elongation of the previously initiated, 5′-labeled RNAs (elongation reaction). Only these previously initiated transcripts were visible on the subsequent autoradiograph due to the absence of label in the elongation reaction. Depending on the step of interest, spermidine was present at the indicated concentrations during either the elongation reaction (Fig. 2C) or the labeling (initiation and promoter escape) reaction (Fig. 2D). Little or no inhibition was observed in either case, indicating that none of these stages in the first transcription cycle is disturbed by spermidine. These findings are consistent with those in Fig. 2A and B, suggesting that only subsequent rounds of MRV transcription are affected by spermidine, and further suggest that the transition between termination in the first round of transcription and reinitiation in the second round of transcription is the primary target of spermidine.
Initial evidence for constitutive and DMSO-activated fractions in MRV core preparations.
At least two general mechanisms might explain the activation of MRV transcription provided by DMSO. First, the same core particles or individual transcriptase complexes might be active both in the absence and in the presence of DMSO, in which case activation might be achieved by having more efficient promoter escape, faster elongation, or more efficient termination and reinitiation from the same cores or transcriptase complexes. Alternatively, not all cores or individual transcriptase complexes, but only a fraction of them, might be transcriptionally active in the absence of DMSO (“constitutive” [C] fraction), and DMSO might then activate another, physically different, fraction (“DMSO-activated” [D] fraction).
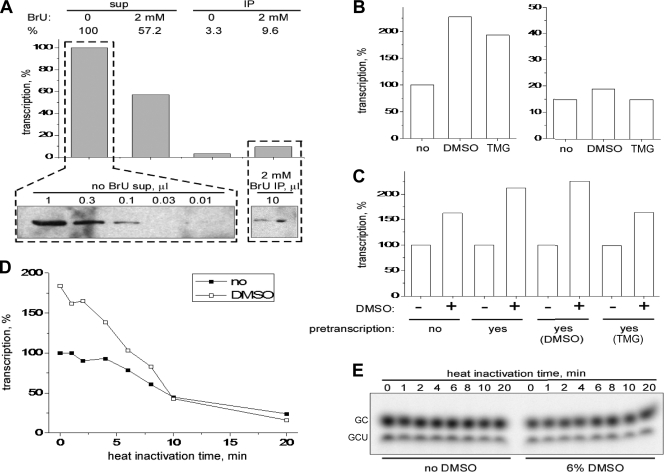
If only a fraction of MRV cores can effectively transcribe and others are inactive in the absence of DMSO, it should be possible to separate the fractions based on the presence or absence of RNA transcripts extending from the core surface. Cores were allowed to synthesize RNA in the presence of BrUTP for a short period (2 min), and transcription was then stopped by addition of EDTA. As a result of this procedure, transcriptionally active cores differ from inactive ones by the presence of protruding, BrUTP-labeled RNA. Transcriptionally active cores were then separated on magnetic beads loaded with anti-BrdU antibodies. Lastly, cores were allowed to transcribe for 1 h in the absence of BrUTP to elute them from the beads. The results indicated that ∼10% of initial RNA-synthesizing activity (Fig. 3A, top), but only ∼1% of core particles (i.e., proteins) (Fig. 3A, bottom), were recovered in the bead-associated fraction. In other words, affinity-isolated, transcriptionally active cores in this experiment showed ∼10 times more transcriptional activity per particle than did the original sample. Since ∼1% of cores account for ∼10% of RNA-synthesizing activity, we extrapolate that with improved recovery from this protocol, ∼10% of cores would account for 100% of RNA-synthesizing activity. We thus conclude that only ∼10% of cores in a standard preparation are constitutively active and able to produce elongated transcripts in the absence of DMSO. This finding is consistent with previous reports suggesting that only 8 to 14% of cores are active at producing transcripts (18, 26) and thus also with the observation that DMSO can activate another fraction.
FIG. 3.
Initial evidence that DMSO activates a separate fraction of MRV cores. All experiments were performed with cores of MRV strain T1L unless otherwise indicated. When present, the activator concentrations were 6% DMSO and 1 M TMG. Representative results are shown from a total of two or three experiments in each case. (A) What portion of a standard core preparation is constitutively active for transcription? For more details of the affinity-isolation procedure following transcription in the presence of BrUTP (BrU), see Results and Materials and Methods. At top, the transcription efficiencies of different supernatant (sup) and immunoprecipitated (IP) fractions from the affinity-isolation procedure are compared. Yields were analyzed by liquid scintillation counting of [α-32P]CTP incorporated into acid-insoluble material and are expressed as a percentage relative to that obtained from the original preparation of MRV type 2 Jones cores (left, no BrU sup). Relative transcription efficiencies (in percentages) are indicated by numbers above the bars. At the bottom, the number of core particles in the BrUTP-labeled, affinity-isolated sample (right) was estimated by denaturing gel electrophoresis and immunoblot analysis relative to that in the original core preparation (left). The λ core proteins were detected using polyclonal anti-core serum. Volumes of loaded samples are indicated (in μl) for each lane. (B) Can C-fraction cores be D fraction? Either the original preparation of cores (left) or affinity-isolated, C-fraction cores (right) were allowed to transcribe for 1 h in the presence or absence of DMSO or TMG as indicated. Yields were analyzed by liquid scintillation counting of [α-32P]CTP incorporated into acid-insoluble material and expressed as a percentage relative to that obtained from the original core preparation in the absence of DMSO or TMG. (C) Can C and D fractions interconvert? Cores were allowed to transcribe for 1 h in the absence or presence of DMSO or TMG as indicated. In addition, a control sample (no pretranscription) was incubated on ice in the absence of NTPs. The reaction media in all samples were then exchanged for fresh transcription mix containing [α-32P]CTP, and transcription was continued for 1 h in the absence or presence of DMSO as indicated. Yields were analyzed by liquid scintillation counting of [α-32P]CTP incorporated into acid-insoluble material and expressed as a percentage relative to that obtained in the absence of DMSO for each set of pretranscription conditions. (D) Heat inactivation of C- and D-fraction cores. Cores in transcription buffer were heated at 65°C for the indicated times and then subjected to 1-h transcription in the absence or presence of DMSO as indicated. Yields were analyzed by liquid scintillation counting of [α-32P]CTP incorporated into acid-insoluble material and are expressed as a percentage relative to that obtained in the absence of DMSO or heating. (E) Abortive synthesis. Same as in panel D, but the results were analyzed by using a 10% sequencing polyacrylamide gel to detect abortive transcripts as described for Fig. 1D.
We next investigated whether or not the preceding fraction of affinity-isolated, constitutively active cores are activated with DMSO or TMG. In fact, little or no such activation was observed with these cores (Fig. 3B, right). These results indicate that the activation by DMSO or TMG observed with the original core preparation (Fig. 3B, left; see also Fig. 1B) is largely attributable to a separate, previously inactive fraction (i.e., the D fraction). Moreover, this fraction must represent separate core particles and not just other transcriptase complexes within the same, previously active core particles because such other transcriptase complexes should have been activated. Lastly, these results indicate that the constitutively active fraction (i.e., the C fraction) in the original core preparation does not readily or rapidly transform, either spontaneously or due to transcription, into D-fraction cores.
We also investigated whether D-fraction cores may transform into C-fraction ones. Occurrence of that transition would be expected to lead to depletion of D-fraction cores from a standard preparation, either spontaneously or during transcription, reducing the extent of DMSO activation over time. In fact, however, if pretranscribed for 1 h, cores showed a similar extent of DMSO activation as ones without pretranscription (Fig. 3C). The data therefore indicate that both fractions are stable and show either no or very slow interconversion. Other implications of these results are that the effects of DMSO are reversible and that the requirement for DMSO remains in effect through each successive round of transcription by these cores.
We also obtained evidence for separate C and D fractions by analyzing the relative stability of MRV transcription at an elevated temperature. Cores were incubated at 65°C for each indicated time, followed by transcription in the absence or presence of DMSO. C-fraction cores (active in the absence of DMSO) resisted heat inactivation for 8 to 10 min at 65°C, after which they were slowly inactivated (Fig. 3D). D-fraction cores (active only in the presence of DMSO), in contrast, began undergoing inactivation almost immediately and were almost fully inactivated by 10 min at 65°C (Fig. 3D). Notably, the inactivations observed for both C and D fractions over the 20-min time course were not due to inactivation of phosphodiester bond formation by the core-associated RdRps, in that all of the heat-treated samples remained active at synthesizing abortive transcripts (Fig. 3E). These results furthermore indicate that transcript initiation, per se, was not affected in either fraction by incubation at 65°C.
DMSO activates promoter escape in another fraction of MRV cores.
Having found that physically separate core particles are activated by DMSO, we sought to pinpoint the step in transcription at which this activation occurs. In this experiment (Fig. 4A), cores were allowed to transcribe for only short periods: 20 s (lanes 1 and 3) or 40 s (lanes 2 and 4) in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of DMSO. The results revealed that transcription is stimulated by DMSO over each of these short periods (compare the relative intensities in lanes 1 and 3 and in lanes 2 and 4). Since no complete transcription cycle is possible within these times and since previous evidence indicated that DMSO is not activating initiation (see Fig. 1D and 3E), it thus appears that DMSO is targeting promoter escape and/or elongation. However, in each pair of lanes representing the same transcription period (20 or 40 s), it is also apparent that the relative lengths of the partial transcripts are approximately the same in the absence or presence of DMSO, shorter for 20 s (lanes 1 and 3) than for 40 s (lanes 2 and 4). Since these transcript lengths are an indication of elongation rate in each sample, the results thus suggest that elongation rates are not substantially increased by DMSO and that DMSO is instead primarily targeting promoter escape. By pinpointing the effects of DMSO in this way, these results provide further evidence that DMSO is activating a distinct fraction of cores in the preparation.
Another variation of this experiment provided further evidence that DMSO is activating promoter escape in a distinct fraction of cores. For Fig. 4A, lane 5, cores were transcribed for 20 s, DMSO was then added, and transcription was continued for another 20 s. In this case, by adding DMSO at the 20-s time point, we would expect—since DMSO is not stimulating elongation (Fig. 4A, lanes 1 to 4; see also Fig. 4B and C below)—to see the partial transcripts elongate over the next 20 s to the same, “normal” length for 40-s transcripts as seen after 40 s in the absence of DMSO (lane 2). At the same time, if DMSO is activating promoter escape in another fraction of cores, we would expect to see those newly initiated transcripts elongate over the next 20 s to the same, “normal” length for 20-s transcripts as seen after 20 s in the presence of DMSO (lane 3). Indeed, these are the observations in Fig. 4A, lane 5.
Since unwinding of long duplex regions must occur during elongation and since a known effect of DMSO is to destabilize RNA duplexes, elongation is a logical step at which DMSO may activate MRV transcription. A technique similar to that used to study spermidine effects above was therefore used to test more carefully whether DMSO may also activate elongation in this system. Samples were transcribed with [α-32P]CTP in the presence of DMSO for 10 to 50 s, at which times the reactions were stopped and media were removed. Each sample was then divided into thirds, one of which was immediately prepared for electrophoresis (no elongation), while the others were resuspended in nonlabeling transcription mix without or with DMSO. Samples were incubated for 10 min to allow completion of synthesis of the 5′-labeled transcripts. If DMSO is also required to activate elongation by D-fraction cores, cores in no-DMSO samples would not be able to elongate all of the previously initiated, 5′-labeled transcripts, which would be reflected as reduced amounts of full-length transcripts on the gel compared to those observed in plus-DMSO samples. In contrast, if DMSO is not required to activate elongation in D-fraction cores, the amounts of full-length transcripts in no- and plus-DMSO samples would be the same. No differences were observed between these corresponding samples (Fig. 4B), indicating that all cores can complete synthesis of previously initiated transcripts in the absence of DMSO and thus that DMSO is not required to activate elongation in D-fraction cores.
An effect of DMSO to increase the rate of elongation would likely not have been detected in the preceding experiment because of the long time allowed to complete elongation of the 5′-labeled transcripts. To confirm the lack of effect of DMSO on elongation rate, as was suggested earlier in Fig. 4A, we analyzed a set of short transcription reactions, from 1 to 10 min, in the absence or presence of DMSO. Little or no difference in the timing of first appearance of the S-, M-, or L-class transcripts, in the absence or presence of DMSO, was detected in this experiment (Fig. 4C), a finding consistent with the conclusion that DMSO does not increase elongation rate. Most of the experiments in Fig. 4 were repeated with TMG, and the results were essentially the same as those obtained with DMSO.
Other unaffected properties of D-fraction cores.
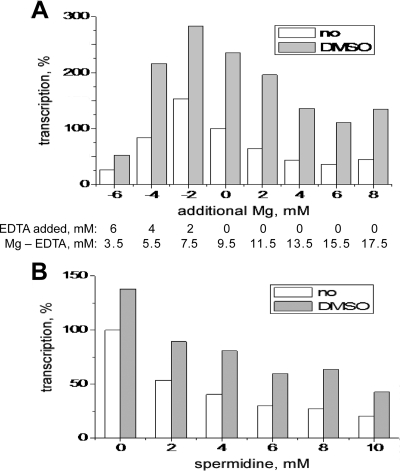
Over the course of the present study, we compared additional properties of C- and D-fraction cores. We first tested whether DMSO affects the Mg2+ dependence of MRV transcription. Mg2+ dependence is a complex result of different structural and catalytic effects. Mg2+ binding stabilizes nucleic acid duplexes (12). In addition, two Mg2+ ions bind in the polymerase active center and are the direct, catalytic moieties (27). As a result of these effects, Mg2+ dependence is characteristic of a particular enzyme, and changes in optimal Mg2+ concentration may reflect changes in polymerase structure (24). Thus, by measuring the Mg2+ dependence of a transcription reaction, one indirectly measures many underlying effects and processes. Both C- and D-fraction cores showed very similar profiles of Mg2+ dependence (Fig. 5A), suggesting that these fractions are largely similar and differ in only some specific, and perhaps even minor, aspect. This conclusion is further supported by the observation that the two fractions showed essentially the same inhibitory response to different concentrations of spermidine (Fig. 5B).
FIG. 5.
Other unaffected properties of D-fraction cores. Cores of MRV strain T1L were subjected to 1-h transcriptions with either no DMSO or 6% DMSO. Representative results are shown from a total of two experiments in each case. (A) Mg2+ dependence. Variations relative to standard Mg2+ concentration (9.5 mM, shown as 0 on the x axis) were achieved by adding either EDTA or additional MgCl2 to the standard transcription mix. Transcription yields are expressed as a percentage relative to that with no DMSO and standard Mg2+ concentration. (B) Spermidine sensitivity. Transcription yields are expressed as a percentage relative to that with no DMSO and no spermidine.
DISCUSSION
In this report we show that spermidine, spermine, DMSO, and TMG, agents known to affect RNA duplex stability, also affect MRV transcription, suggesting the importance of conformational changes in the dsRNA templates at one or more steps in the transcription cycle. The known effects of these small molecules on nucleic acids has led us to conclude that their effects on MRV transcription occur mostly at the RNA level. However, because the effects of these agents on proteins are also known (1, 36, 50), we cannot exclude the possibility that protein-based mechanisms may contribute to their effects in this report. Still, major protein-based effects seem less likely in this case because all four of these molecules are mostly reported to stabilize proteins, whereas their effects on MRV transcription are opposite: inhibitory for spermidine and spermine but stimulatory for DMSO and TMG. In addition, their effects are not specific to MRV, in that rotavirus transcription is similarly affected by each respective agent. This supports our interpretation that the effects are largely at the RNA level and leads us to speculate that they may also be seen with other, more divergent dsRNA viruses. In initial experiments on transcription by partitivirus virions (a gift from S. A. Ghabrial, University of Kentucky), for example, we have seen only limited inhibition by spermidine or activation by DMSO but substantial activation by TMG (unpublished data). Because partitivirus transcription is semiconservative (38), not conservative like reovirus transcription, observed differences in the effects of small molecules might be useful for distinguishing such fundamental differences in the transcription mechanisms of different dsRNA viruses.
Mechanism of action by polyamines.
Spermidine and spermine showed no inhibition of either initiation leading to abortive synthesis or elongation, but inhibited one or more of the events between the end of elongation in one transcription round and initiation in the next round. These intervening events include (i) termination, (ii) repositioning of the 3′ end of minus strand to reenter the template-entry channel of the RdRp, and (iii) migration of the 3′ end of minus strand through the template entry channel and into the catalytic active site (45). Steps ii and iii appear to be especially complex in involving specific translocations of the minus-strand 3′ region and also in likely depending on simultaneous unwinding of 3′ sequences of minus strand from the 5′ sequences of plus strand. Spermidine inhibition of any of steps i, ii, or iii would yield the results observed in our experiments. However, based on the knowledge that polyamines stabilize and compact RNA duplexes, it seems more likely that steps ii and iii, both of which involve RNA movements and changes in topology (probably including the initial stages of duplex unwinding that then continue through the elongation stage) are the main targets of spermidine action.
Mechanism of action by DMSO or TMG and physical basis of C/D difference.
Unlike the polyamines, DMSO had no effect on constitutive transcription, but turned on additional transcriptase complexes that were otherwise silent. Morever, our results show that these DMSO-activated complexes are located in a separate fraction of core particles (the “D” fraction). Thus, in our standard core preparations, a much larger number of particles is actively transcribing in the presence of DMSO than in its absence. Our results furthermore indicate that there is little or no transformation between the constitutively active fraction (the “C” fraction) and the D fraction over time. The reason why C- and D-like transcriptase complexes appear not to be mixed within the same core particle is unknown but is discussed further below.
Our findings indicate that the C and D fractions have similar capacities to continue elongation of nascent transcripts but differ in their capacities to begin elongation, i.e., to mediate promoter escape (see especially Fig. 4B). D-fraction cores depend on DMSO for promoter escape through each successive round of transcription, and their associated machinery for this step is much less resistant to heat inactivation. Unfortunately, the molecular basis of promoter escape in MRV cores is largely unknown, but it must involve further unwinding of the dsRNA template, which according to the current model occurs near the template entry channel on the outer surface of the RdRp (45). In better-studied systems, promoter escape has been shown to involve conformational changes such as “scrunching” in the duplex template (41), conformational changes in the RNA polymerase (51), and/or NTP hydrolysis by auxiliary proteins (9). Thus, effects of DMSO and TMG on either RNA- or protein-based targets during promoter escape in MRV transcription remain possible. The NTP phosphohydrolase(s) known to be present in MRV cores (6, 28, 33, 34), for example, might play a specific role in promoter escape (15). However, since we found that 6% DMSO or 1 M TMG has little or no effect on in vitro NTP hydrolysis by MRV cores (data not shown), that particular target appears unlikely.
The fact that each core particle appears to have all of its transcriptase complexes in the same, C or D, state, which can furthermore not be readily interconverted, suggests to us that some global effect is responsible for the C/D difference. By “global” we mean an effect concertedly conveyed to all 10 to 12 transcriptase complexes within a particular core. One possibility is that the main defect of D-fraction cores relates to some difference in their genomic RNA conformation or arrangement. The nature of such a difference is unknown but would need to impact the local structure around the 3′ end of template minus strand. For example, it might relate to higher-order structures that may need to form between different RNA segments to ensure proper assortment and packaging during core assembly (35). Another possibility is that the main defect of D-fraction cores relates to some difference in their protein content or conformation. It seems hard to explain why all copies of a protein would be missing or conformationally altered in the D fraction. Nonetheless, it may be that a particular protein (or proteins) is packaged into the core in a highly cooperative mode, such that if the first copy is occasionally missing or altered, other copies will be missing or altered as well. Additional experiments to discern the underlying physical difference(s) between C- and D-fraction cores are clearly warranted and could yield fundamental insights.
Elongation or promoter escape as the rate-limiting step in MRV transcription.
That the transition to elongation (i.e., promoter escape) is the rate-limiting step in MRV transcription has long been suggested, based largely on the observation that initiator oligonucleotides (i.e., abortive transcripts) are produced in vast excess to full-length transcripts (15, 49, 52). To explain this observation, it has been hypothesized that the viral RdRps spend a disproportionate amount of time in initiation complexes. Abortive initiations are then the more frequent events in MRV transcription, and initiation-to-elongation transitions, required for giving rise to full-size transcripts, happen at only low frequency.
Our data suggest that the preceding hypothesis is incorrect for C-fraction cores. For standard core preparations, the measured rate of full-length transcript accumulation in our reactions at 45°C is ∼1/core/h (data not shown). Theoretically, the rate should be much higher: assuming that synthesis of an average MRV mRNA takes ∼4 min (based on the reported elongation rate of 7 to 12 nucleotides/s [2, 3, 42] and our own estimate of 10 to 12 nucleotides/s [data not shown]), the predicted rate of full-length transcript accumulation is ∼15/core/h. In the present study, we were able to isolate a fraction of cores (the C fraction) with ∼10-times-higher specific transcription activity than the original preparation, and for this fraction the measured rate of full-length transcript accumulation (∼10/core/h) is therefore much closer to the theoretical prediction. This makes elongation the rate-limiting step in transcription by C-fraction cores and leaves a smaller amount of time for abortive initiation and other steps.
As for D-fraction cores, the results suggest that when these particles are activated by DMSO or TMG, elongation is probably again the rate-limiting step in their transcription cycles. In the absence of activation, however, these particles are incapable of promoter escape, which is thereby the rate-liming step for their transcription. For the remaining, non-C/non-D fraction that makes up 70 to 80% of cores in standard preparations, the results suggest that those particles are likewise incapable of promoter escape, even in the presence of DMSO or TMG, and thus promoter escape is likely rate limiting for their transcription as well.
Other implications of the small C fraction in core preparations.
The great prevalence of abortive over full-length transcripts can be largely explained by postulating that ∼90% of cores in standard preparations, in the absence of DMSO or TMG, are able to initiate transcription but are unable to mediate promoter escape. According to this interpretation, full-length transcripts are then synthesized only by C-fraction cores in the preparation, which constitute only ∼10% of the total, while abortive transcripts are synthesized by all particles, including the remaining ∼90%. In fact, the small decrease in abortive transcripts synthesized in the presence of DMSO (see Fig. 1D and 3E) may reflect that with DMSO activation of promoter escape in another 10 to 20% of cores in the preparation (i.e., D-fraction cores), the transcriptases in those cores spend substantially more time in elongation complexes and less time in initiation complexes.
In hindsight, one should also ask why the synthesis of abortive transcripts is not decreased over a 1-h reaction in the presence of spermidine as shown in Fig. 1D. If formation of new initiation complexes after termination of the first transcription round is blocked by spermidine, then the synthesis of abortive transcripts should also be decreased. The apparent explanation is that the transcriptases in ∼90% of cores in standard preparations (in all but C-fraction cores) never exit the initiation complexes (i.e., never undergo promoter escape) and thus continue synthesizing abortive transcripts throughout the reaction irrespective of the presence of spermidine.
Acknowledgments
We thank Ann Hochschild for helpful suggestions about the manuscript, other members of our lab for discussions, and Elaine Freimont for technical assistance. We also thank Dick Bellamy, Shane Trask, Scott Aoki, and Steve Harrison for the gift of rotavirus double-layered particles and Eugene Makeyev for the gift of TMG.
This study was supported in part by the Bill and Melinda Gates Foundation through a Collaboration for AIDS Vaccine Discovery grant to a Vaccine Discovery Consortium (including M.L.N.) headed by Tim Zamb at the International AIDS Vaccine Initiative (New York, NY).
Footnotes
Published ahead of print on 18 March 2009.
REFERENCES
- 1.Arakawa, T., and S. N. Timasheff. 1985. The stabilization of proteins by osmolytes. Biophys. J. 47411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, A. K., and A. J. Shatkin. 1970. Transcription in vitro by reovirus-associated ribonucleic acid-dependent polymerase. J. Virol. 61-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, N., S. Gillies, S. Bullivant, and A. R. Bellamy. 1974. Electron microscopy study of reovirus reaction cores. J. Virol. 14315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baskaran, N., R. Kandpal, A. Bhargava, M. Glynn, A. Bale, and S. Weissman. 1996. Uniform amplification of a mixture of deoxyribonucleic acids with a varying GC content. Genome Res. 6633-638. [DOI] [PubMed] [Google Scholar]
- 5.Bellamy, A. R., J. L. Nichols, and W. K. Joklik. 1972. Nucleotide sequences of reovirus oligonucleotides: evidence for abortive RNA synthesis during virus maturation. Nat. New Biol. 23849-51. [DOI] [PubMed] [Google Scholar]
- 6.Bisaillon, M., J. Bergeron, and G. Lemay. 1997. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus λ1 protein. J. Biol. Chem. 27218298-18303. [DOI] [PubMed] [Google Scholar]
- 7.Bloomfield, V. 1997. DNA Condensation by multivalent cations. Biopolymers 44269-282. [DOI] [PubMed] [Google Scholar]
- 8.Chandran, K., S. B. Walker, Y. Chen, C. M. Contreras, L. A. Schiff, T. S. Baker, and M. L. Nibert. 1999. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 733941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25375-380. [DOI] [PubMed] [Google Scholar]
- 10.Dryden, K. A., D. L. Farsetta, G. Wang, J. M. Keegan, B. N. Fields, T. S. Baker, and M. L. Nibert. 1998. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 24533-46. [DOI] [PubMed] [Google Scholar]
- 11.Dryden, K. A., G. Wang, M. Yeager, M. L. Nibert, K. M. Coombs, D. B. Furlong, B. N. Fields, and T. S. Baker. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1221023-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichhorn, G. L., and Y. A. Shin. 1968. Interaction of metal ions with polynucleotides and related compounds. XII. The relative effect of various metal ions on DNA helicity. J. Am. Chem. Soc. 907323-7328. [DOI] [PubMed] [Google Scholar]
- 13.England, T., and O. Uhlenbeck. 1978. 3′-terminal labeling of RNA with T4 RNA ligase. Nature 275560-561. [DOI] [PubMed] [Google Scholar]
- 14.Escara, J. F., and J. R. Hutton. 1980. Thermal stability and renaturation of DNA in dimethyl sulfoxide solutions: acceleration of the renaturation rate. Biopolymers 191315-1327. [DOI] [PubMed] [Google Scholar]
- 15.Farsetta, D. L., K. Chandran, and M. L. Nibert. 2000. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J. Biol. Chem. 27539693-39701. [DOI] [PubMed] [Google Scholar]
- 16.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuichi, Y., S. Muthukrishnan, J. Tomasz, and A. J. Shatkin. 1976. Mechanism of formation of reovirus mRNA 5′-terminal blocked and methylated sequence, m7GpppGmpC. J. Biol. Chem. 2515043-5053. [PubMed] [Google Scholar]
- 18.Gillies, S., S. Bullivant, and A. R. Bellamy. 1971. Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science 174694-696. [DOI] [PubMed] [Google Scholar]
- 19.Harvey, J. D., A. R. Bellamy, W. C. Earnshaw, and C. Schutt. 1981. Biophysical studies of reovirus type 3. IV. Low-angle x-ray diffraction studies. Virology 112240-249. [DOI] [PubMed] [Google Scholar]
- 20.He, X., J. Zhou, M. Bartlam, R. Zhang, J. Ma, Z. Lou, X. Li, J. Li, A. Joachimiak, Z. Zeng, R. Ge, Z. Rao, and Y. Liu. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 4541123-1126. [DOI] [PubMed] [Google Scholar]
- 21.Henke, W., K. Herdel, K. Jung, D. Schnorr, and S. Loening. 1997. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 253957-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou, M. H., S. B. Lin, J. M. Yuann, W. C. Lin, A. H. Wang, and L. S. Kan. 2001. Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res. 295121-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutton, J. R., and J. G. Wetmur. 1975. Activity of endonuclease S1 in denaturing solvents: dimethysulfoxide, dimethylformamide, formamide and formaldehyde. Biochem. Biophys. Res. Commun. 66942-948. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, R. A., and C. C. Richardson. 1987. Enzymatic properties of a proteolytically nicked RNA polymerase of bacteriophage T7. J. Biol. Chem. 2623790-3799. [PubMed] [Google Scholar]
- 25.Ivanovic, T., M. A. Agosto, K. Chandran, and M. L. Nibert. 2007. A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J. Biol. Chem. 28212210-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joklik, W. K. 1970. The molecular biology of reovirus. J. Cell Physiol. 76289-301. [DOI] [PubMed] [Google Scholar]
- 27.Joyce, C. M., and T. A. Steitz. 1995. Polymerase structures and function: variations on a theme? J. Bacteriol. 1776321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J., J. S. L. Parker, K. E. Murray, and M. L. Nibert. 2004. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 2794394-4403. [DOI] [PubMed] [Google Scholar]
- 29.Lehrach, H., D. Diamond, J. M. Wozney, and H. Boedtker. 1977. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry 164743-4751. [DOI] [PubMed] [Google Scholar]
- 30.Levin, D. H., N. Mendelsohn, M. Schonberg, H. Klett, S. Silverstein, A. M. Kapuler, and G. Acs. 1970. Properties of RNA transcriptase in reovirus subviral particles. Proc. Natl. Acad. Sci. USA 66890-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, X., S. M. McDonald, M. A. Tortorici, Y. J. Tao, R. Vasquez-DelCaprio, M. L. Nibert, J. T. Patton, and S. C. Harrison. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 161678-1688. [DOI] [PMC free article] [PubMed]
- 32.Luongo, C. L., X. Zhang, S. B. Walker, Y. Chen, T. J. Broering, D. L. Farsetta, V. D. Bowman, T. S. Baker, and M. L. Nibert. 2002. Loss of activities for mRNA synthesis accompanies loss of λ2 spikes from reovirus cores: an effect of λ2 on λ1 shell structure. Virology 29624-38. [DOI] [PubMed] [Google Scholar]
- 33.Noble, S., and M. L. Nibert. 1997. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J. Virol. 712182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 717728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277217-225. [DOI] [PubMed] [Google Scholar]
- 36.Powroznik, B., M. Gharbi, G. Dandrifosse, and O. Peulen. 2004. Enhancement of lysozyme stability and activity by polyamines. Biochimie 86651-656. [DOI] [PubMed] [Google Scholar]
- 37.Ralser, M., R. Querfurth, H. Warnatz, H. Lehrach, M. L. Yaspo, and S. Krobitsch. 2006. An efficient and economic enhancer mix for PCR. Biochem. Biophys. Res. Commun. 347747-751. [DOI] [PubMed] [Google Scholar]
- 38.Ratti, G., and K. W. Buck. 1978. Semi-conservative transcription in particles of a double-stranded RNA mycovirus. Nucleic Acids Res. 53843-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rees, W., T. Yager, J. Korte, and P. von Hippel. 1993. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry 32137-144. [DOI] [PubMed] [Google Scholar]
- 40.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404960-967. [DOI] [PubMed] [Google Scholar]
- 41.Revyakin, A., C. Liu, R. H. Ebright, and T. R. Strick. 2006. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 3141139-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skehel, J. J., and W. K. Joklik. 1969. Studies on the in vitro transcription of reovirus RNA catalyzed by reovirus cores. Virology 39822-831. [DOI] [PubMed] [Google Scholar]
- 43.Steitz, T. A. 2004. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 144-9. [DOI] [PubMed] [Google Scholar]
- 44.Tabor, C. W., and H. Tabor. 1976. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu. Rev. Biochem. 45285-306. [DOI] [PubMed] [Google Scholar]
- 45.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage: structural studies of reovirus polymerase λ3. Cell 27733-745. [DOI] [PubMed] [Google Scholar]
- 46.Taraporewala, Z. F., and J. T. Patton. 2004. Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae. Virus Res. 10157-66. [DOI] [PubMed] [Google Scholar]
- 47.Vijayanathan, V., T. Thomas, A. Shirahata, and T. J. Thomas. 2001. DNA condensation by polyamines: a laser light scattering study of structural effects. Biochemistry 4013644-13651. [DOI] [PubMed] [Google Scholar]
- 48.Wiener, J. R., and W. K. Joklik. 1989. The sequences of the reovirus serotype 1, 2, and 3 L1 genome segments and analysis of the mode of divergence of the reovirus serotypes. Virology 169194-203. [DOI] [PubMed] [Google Scholar]
- 49.Yamakawa, M., Y. Furuichi, K. Nakashima, A. J. LaFiandra, and A. J. Shatkin. 1981. Excess synthesis of viral mRNA 5′-terminal oligonucleotides by reovirus transcriptase. J. Biol. Chem. 2566507-6514. [PubMed] [Google Scholar]
- 50.Yang, Z. W., S. W. Tendian, W. M. Carson, W. J. Brouillette, L. J. Delucas, and C. G. Brouillette. 2004. Dimethyl sulfoxide at 2.5% (v/v) alters the structural cooperativity and unfolding mechanism of dimeric bacterial NAD+ synthetase. Protein Sci. 13830-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin, Y. W., and T. A. Steitz. 2002. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 2981387-1395. [DOI] [PubMed] [Google Scholar]
- 52.Zarbl, H., K. E. Hastings, and S. Millward. 1980. Reovirus core particles synthesize capped oligonucleotides as a result of abortive transcription. Arch. Biochem. Biophys. 202348-360. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, X., S. B. Walker, P. R. Chipman, M. L. Nibert, and T. S. Baker. 2003. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 101011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]